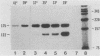
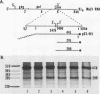
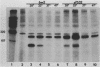
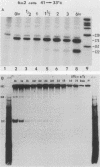
Abstract
The spliced form of MuSVts110 viral RNA is approximately 20-fold more abundant at growth temperatures of 33 degrees C or lower than at 37 to 41 degrees C. This difference is due to changes in the efficiency of MuSVts110 RNA splicing rather than selective thermolability of the spliced species at 37 to 41 degrees C or general thermosensitivity of RNA splicing in MuSVts110-infected cells. Moreover, RNA transcribed from MuSVts110 DNA introduced into a variety of cell lines is spliced in a temperature-sensitive fashion, suggesting that the structure of the viral RNA controls the efficiency of the event. We exploited this novel splicing event to study the cleavage and ligation events during splicing in vivo. No spliced viral mRNA or splicing intermediates were observed in MuSVts110-infected cells (6m2 cells) at 39 degrees C. However, after a short (about 30-min) lag following a shift to 33 degrees C, viral pre-mRNA cleaved at the 5' splice site began to accumulate. Ligated exons were not detected until about 60 min following the initial detection of cleavage at the 5' splice site, suggesting that these two splicing reactions did not occur concurrently. Splicing of viral RNA in the MuSVts110 revertant 54-5A4, which lacks the sequence -AG/TGT- at the usual 3' splice site, was studied. Cleavage at the 5' splice site in the revertant viral RNA proceeded in a temperature-sensitive fashion. No novel cryptic 3' splice sites were activated; however, splicing at an alternate upstream 3' splice site used at low efficiency in normal MuSVts110 RNA was increased to a level close to that of 5'-splice-site cleavage in the revertant viral RNA. Increased splicing at this site in 54-5A4 viral RNA is probably driven by the unavailability of the usual 3' splice site for exon ligation. The thermosensitivity of this alternate splice event suggests that the sequences governing the thermodependence of MuSVts110 RNA splicing do not involve any particular 3' splice site or branch point sequence, but rather lie near the 5' end of the intron.
Full text
PDF


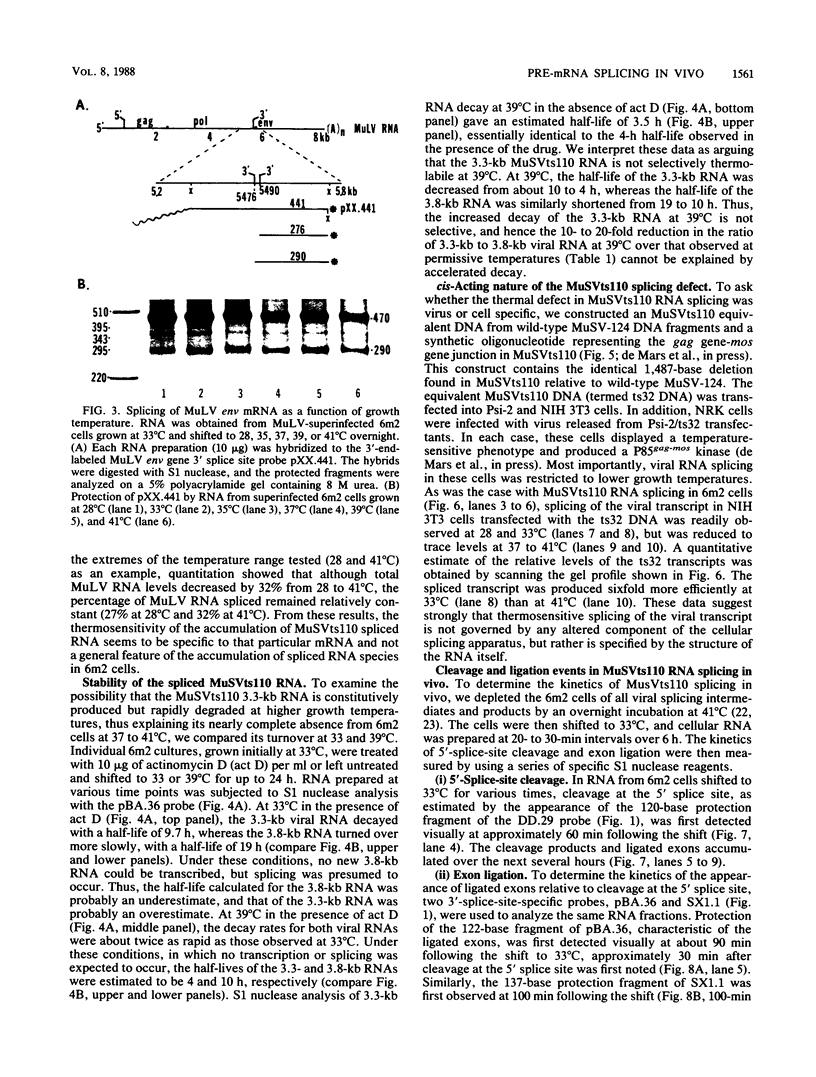
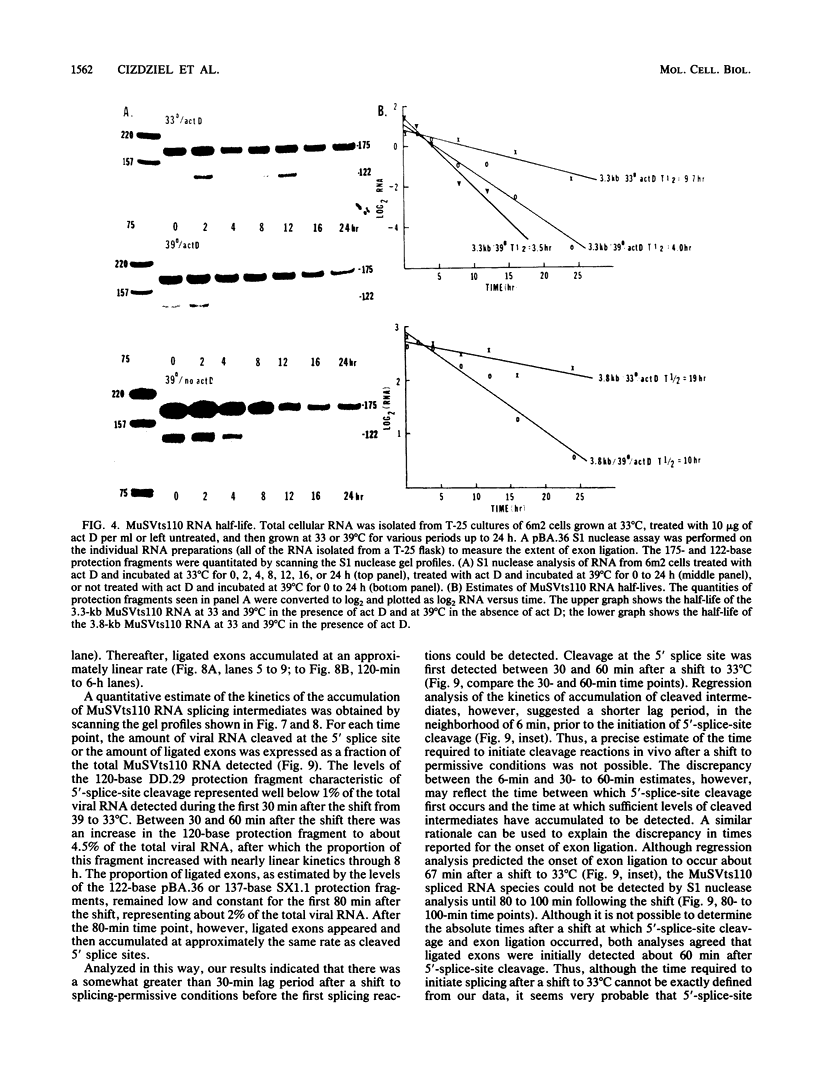




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aebi M., Hornig H., Padgett R. A., Reiser J., Weissmann C. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell. 1986 Nov 21;47(4):555–565. doi: 10.1016/0092-8674(86)90620-3. [DOI] [PubMed] [Google Scholar]
- Biggart N. W., Gallick G. E., Murphy E. C., Jr Nickel-induced heritable alterations in retroviral transforming gene expression. J Virol. 1987 Aug;61(8):2378–2388. doi: 10.1128/jvi.61.8.2378-2388.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair D. G., Hull M. A., Finch E. A. The isolation and preliminary characterization of temperature-sensitive transformation mutants of Moloney sarcoma virus. Virology. 1979 Jun;95(2):303–316. doi: 10.1016/0042-6822(79)90486-0. [DOI] [PubMed] [Google Scholar]
- Brown R. L., Horn J. P., Wible L., Arlinghaus R. B., Brinkley B. R. Sequence of events in the transformation process in cells infected with a temperature-sensitive transformation mutant of Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5593–5597. doi: 10.1073/pnas.78.9.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizdziel P. E., Nash M. A., Blair D. G., Murphy E. C., Jr Molecular basis underlying phenotypic revertants of Moloney murine sarcoma virus MuSVts110. J Virol. 1986 Jan;57(1):310–317. doi: 10.1128/jvi.57.1.310-317.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski P. J., Padgett R. A., Sharp P. A. Messenger RNA splicing in vitro: an excised intervening sequence and a potential intermediate. Cell. 1984 Jun;37(2):415–427. doi: 10.1016/0092-8674(84)90372-6. [DOI] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Hamelin R., Brizzard B. L., Nash M. A., Murphy E. C., Jr, Arlinghaus R. B. Temperature-sensitive viral RNA expression in Moloney murine sarcoma virus ts110-infected cells. J Virol. 1985 Feb;53(2):616–623. doi: 10.1128/jvi.53.2.616-623.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelin R., Chan E. K., Tan E. M., Arlinghaus R. B. Antibodies against small nuclear ribonucleoproteins immunoprecipitate complexes containing ts110 Moloney murine sarcoma virus genomic and messenger RNAs. Virology. 1986 Jul 15;152(1):87–99. doi: 10.1016/0042-6822(86)90374-0. [DOI] [PubMed] [Google Scholar]
- Hamelin R., Kabat K., Blair D., Arlinghaus R. B. Temperature-sensitive splicing defect of ts110 Moloney murine sarcoma virus is virus encoded. J Virol. 1986 Jan;57(1):301–309. doi: 10.1128/jvi.57.1.301-309.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn J. P., Wood T. G., Murphy E. C., Jr, Blair D. G., Arlinghaus R. B. A selective temperature-sensitive defect in viral RNA expression in cells infected with a ts transformation mutant of murine sarcoma virus. Cell. 1981 Jul;25(1):37–46. doi: 10.1016/0092-8674(81)90229-4. [DOI] [PubMed] [Google Scholar]
- Hwang L. S., Park J., Gilboa E. Role of intron-contained sequences in formation of moloney murine leukemia virus env mRNA. Mol Cell Biol. 1984 Nov;4(11):2289–2297. doi: 10.1128/mcb.4.11.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junghans R. P., Murphy E. C., Jr, Arlinghaus R. B. Electron microscopic analysis of ts1 10 Moloney mouse sarcoma virus, a variant of wild-type virus with two RNAs containing large deletions. J Mol Biol. 1982 Oct 25;161(2):229–250. doi: 10.1016/0022-2836(82)90150-4. [DOI] [PubMed] [Google Scholar]
- Keller W. The RNA lariat: a new ring to the splicing of mRNA precursors. Cell. 1984 Dec;39(3 Pt 2):423–425. doi: 10.1016/0092-8674(84)90449-5. [DOI] [PubMed] [Google Scholar]
- Kloetzer W. S., Maxwell S. A., Arlinghaus R. B. Further characterization of the P85gag-mos -associated protein kinase activity. Virology. 1984 Oct 15;138(1):143–155. doi: 10.1016/0042-6822(84)90154-5. [DOI] [PubMed] [Google Scholar]
- Kloetzer W. S., Maxwell S. A., Arlinghaus R. B. P85gag-mos encoded by ts110 Moloney murine sarcoma virus has an associated protein kinase activity. Proc Natl Acad Sci U S A. 1983 Jan;80(2):412–416. doi: 10.1073/pnas.80.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M. M., Grabowski P. J., Padgett R. A., Sharp P. A. Characterization of the branch site in lariat RNAs produced by splicing of mRNA precursors. Nature. 1985 Feb 14;313(6003):552–557. doi: 10.1038/313552a0. [DOI] [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T., Ruskin B., Green M. R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984 Apr;36(4):993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Lazo P. A., Prasad V., Tsichlis P. N. Splice acceptor site for the env message of Moloney murine leukemia virus. J Virol. 1987 Jun;61(6):2038–2041. doi: 10.1128/jvi.61.6.2038-2041.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987 Feb 19;325(6106):673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Mann R., Baltimore D. Varying the position of a retrovirus packaging sequence results in the encapsidation of both unspliced and spliced RNAs. J Virol. 1985 May;54(2):401–407. doi: 10.1128/jvi.54.2.401-407.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash M. A., Brizzard B. L., Wong J. L., Murphy E. C., Jr Murine sarcoma virus ts110 RNA transcripts: origin from a single proviral DNA and sequence of the gag-mos junctions in both the precursor and spliced viral RNAs. J Virol. 1985 Feb;53(2):624–633. doi: 10.1128/jvi.53.2.624-633.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash M., Brown N. V., Wong J. L., Arlinghaus R. B., Murphy E. C., Jr S1 nuclease mapping of viral RNAs from a temperature-sensitive transformation mutant of murine sarcoma virus. J Virol. 1984 May;50(2):478–488. doi: 10.1128/jvi.50.2.478-488.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. J., Lin R. J., Cheng S. C., Abelson J. Molecular consequences of specific intron mutations on yeast mRNA splicing in vivo and in vitro. Cell. 1985 Aug;42(1):335–344. doi: 10.1016/s0092-8674(85)80129-x. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Grabowski P. J., Hardy S. F., Sharp P. A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984 Aug 31;225(4665):898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985 May;41(1):95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Role of the 3' splice site consensus sequence in mammalian pre-mRNA splicing. Nature. 1985 Oct 24;317(6039):732–734. doi: 10.1038/317732a0. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Rymond B. C., Rosbash M. Cleavage of 5' splice site and lariat formation are independent of 3' splice site in yeast mRNA splicing. Nature. 1985 Oct 24;317(6039):735–737. doi: 10.1038/317735a0. [DOI] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Stanker L. H., Horn J. P., Gallick G. E., Kloetzer W. S., Murphy E. C., Jr, Blair D. G., Arlinghaus R. B. Gag-mos Polyproteins encoded by variants of the Moloney strain of mouse sarcoma virus. Virology. 1983 Apr 15;126(1):336–347. doi: 10.1016/0042-6822(83)90483-x. [DOI] [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983 Apr 14;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Van Beveren C., van Straaten F., Galleshaw J. A., Verma I. M. Nucleotide sequence of the genome of a murine sarcoma virus. Cell. 1981 Nov;27(1 Pt 2):97–108. doi: 10.1016/0092-8674(81)90364-0. [DOI] [PubMed] [Google Scholar]
- Zeitlin S., Efstratiadis A. In vivo splicing products of the rabbit beta-globin pre-mRNA. Cell. 1984 Dec;39(3 Pt 2):589–602. doi: 10.1016/0092-8674(84)90466-5. [DOI] [PubMed] [Google Scholar]
- Zeitlin S., Parent A., Silverstein S., Efstratiadis A. Pre-mRNA splicing and the nuclear matrix. Mol Cell Biol. 1987 Jan;7(1):111–120. doi: 10.1128/mcb.7.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]