Abstract
Over the past decade there has been a growing acknowledgement that a large proportion of proteins within most proteomes contain disordered regions. Disordered regions are segments of the protein chain which do not adopt a stable structure. Recognition of disordered regions in a protein is of great importance for protein structure prediction, protein structure determination and function annotation as these regions have a close relationship with protein expression and functionality. As a result, a great many protein disorder prediction methods have been developed so far. Here, we present an overview of current protein disorder prediction methods including an analysis of their advantages and shortcomings. In order to help users to select alternative tools under different circumstances, we also evaluate 23 disorder predictors on the benchmark data of the most recent round of the Critical Assessment of protein Structure Prediction (CASP) and assess their accuracy using several complementary measures.
Introduction
For many years, a crucial underpinning of structural biology has been that a protein’s function is determined by its structure. While this important structure-to-function paradigm remains largely intact, it has evolved and been reassessed.1 These changes were spurred in part due to the post-genomic age and the accompanying influx of data. Large scale analysis of sequence data revealed that many proteins are comprised completely or in part of low complexity segments which are frequently associated with non-globular regions.2 Additional work, notably that of Tompa, summarized and outlined the potential usefulness of flexibility in the three dimensional structure of a protein with respect to function.3 The end result has been that while the link between function and structure remains, a well defined stable structure is not necessary for a protein to perform particular functions.
Proteins or segments of the protein chain which do not adopt a stable structure are known by many names. Earlier work often referred to these proteins as intrinsically unstructured proteins (IUPs).1,3 More recently, other terms such as intrinsically disordered proteins (IDP) or regions have also been used.4 In this work we will use the term disordered regions.
There has been much interest in characterizing disordered regions in proteins and this is for many reasons. From a practical standpoint, protein disordered regions can hinder protein analysis. Disordered regions in a protein have a biased amino acid composition5 that may give rise to inaccurate sequence alignments to unrelated proteins.6 By recognizing disordered regions, one can avoid aligning disordered regions with ordered regions and thus increase the accuracy of sequence similarity analysis. Furthermore, disordered regions often make the purification and crystallization of a protein difficult.7 The identification of a protein as highly disordered could save valuable time as researchers would not spend time attempting to determine a structure which does not exist. Proteins with disordered regions also play important functional roles. The literature documents disordered proteins participating in functions such as protein–DNA binding, phosphorylation, signalling and regulation.5,8,9
Given the prevalence of disordered proteins and the growing acknowledgement of the functional relevance of these proteins, considerable effort has been made by the bioinformatics community to provide tools to predict protein disorder. To date, more than 50 disorder predictors have been implemented.10 Aiming at enhancing the development of disorder prediction, the Critical Assessment of protein Structure Prediction (CASP),47 a biannual, community-wide blind competition launched in 1994 (http://predictioncenter.org/), has a section devoted to the assessment of such predictors since 2004. In this study, we outline typical approaches employed by protein disorder prediction methods and discuss some advantages and shortcomings of several implementations. In order to provide users some information to choose alternative tools according to different criteria, we also benchmark the 23 predictors that participated in the CASP9 experiments using several complementary measures. However, for the official assessment (e.g., ranking) of these methods, readers should refer to the disorder prediction assessment paper to be published in the CASP9 supplemental issue of the journal Proteins and the CASP9 web site (http://predictioncenter.org/casp9/).
Disorder prediction methods
A number of approaches have been developed to predict protein disorder regions. These methods can be broadly classified into four different categories: (1) ab initio, or sequence only, (2) clustering, (3) template based and (4) meta or consensus. There are also a few implementations which cannot be easily placed in one of these four categories and these we will call hybrid methods. Sixteen currently existing disorder prediction methods are listed in Table 1, including their availability and method categories. We now discuss the basis for each category and briefly discuss some implementations.
Ab initio methods
The distinguishing feature of ab initio approaches is that they depend almost exclusively on sequence information. That is to say that to make a prediction, nothing other than the primary sequence is needed. Disordered regions in proteins are predicted using features extracted from the primary sequence in conjunction with statistical models. Oftentimes these models include machine learning techniques such as support vector machines and neural networks. There are many protein disorder predictors that make use of ab initio methods and in CASP8 and CASP9, a large quantity of predictors adopted such an approach.
DISOPRED11 is a web service for ab initio disordered region prediction. It was trained on a large non-redundant set of sequences with high resolution X-ray structures. A sequence profile for each protein target was generated using a PSI-BLAST12 search against a filtered sequence database. The procedure is based on the premise that the disordered residues may appear in the records that are consistently missing. The data were used to train linear support vector machines, and the input vector for each residue was constructed from the profiles of a symmetric window of fifteen residue positions.13 The prediction accuracy may decrease if there are few homologues for the target protein.
PreDisorder14 is an ab initio protein disorder predictor designed by our group and based on a recursive neural network.
First, an input profile is generated for the target protein sequence after the sequence has been aligned against several template profiles using PSI-BLAST.12 Later, this profile along with the predicted secondary structure and solvent accessibility are fed into a 1D recursive neural network to make the disorder predictions.
POODLE (Prediction Of Order and Disorder by machine Learning) is an integrated system that predicts disordered regions and consists of three predictors, POODLE-S,15 POODLE-L16 and POODLE-W.17 Each predictor specializes in making short disordered region predictions, long disordered region predictions or unfolded proteins. POODLE-L predicts long disordered regions via support vector machines using ten physicochemical properties of amino acids. It combines the results of 10 two-level SVM predictors and generates the final prediction result. POODLE-S predicts the short disordered regions from amino acid sequences based on physicochemical properties and a position specific scoring matrix. In contrast, POODLE-W is a semi-supervised learning approach for classifying proteins as either mostly disordered or mostly ordered. As a semi-supervised learning approach, POODLE-W can use protein sequences with unknown structure to increase the accuracy.
SPINE-D18 is another ab initio method based on a neural network with two hidden layers. Five independent predictors were trained, and a final prediction is made on their consensus. The input nodes contain residue-level information, window-level information, and one terminal tag. The residue-level information includes seven physical parameters, a position specific scoring matrix vector generated from PSI-BLAST profiles, predicted secondary structure and solvent accessibility from SPINE-X,19 and predicted torsion-angle fluctuations. The window-level information includes amino acid composition, local compositional complexity, and predicted secondary structure. The terminal tag marks residues on both N- and C-termini.
IUPred20,21 makes a prediction based on the assumption that globular proteins depend on the stabilizing energy of a large quantity of inter-residue interactions, whereas IUPs (intrinsically unfolded proteins) fail to have the capacity to form sufficient inter-residue interactions. The estimated energies of IUPs correspond to less favourable energies in comparison with globular proteins. Consequently, prediction is carried out according to the estimated capacity of polypeptides to form stabilizing contacts. IUPred has a limitation in that it can only be used on proteins without disulfide bonds or metal-binding regions.
Several feed-forward neural network predictors called PONDRs22–24 were trained by using the back-propagation learning algorithm. They take ten selected attributes including the fractional composition of particular amino acids, two different hydropath scales, and so on as inputs from windows of generally 21 amino acids. Then the neural network with a fully connected hidden layer of ten neurons has been trained on a specific set of ordered and disordered sequences and outputs a value for the central amino acid in the window.
GlobProt25 is a simple approach based on the propensity for a given amino acid to be in a random coil or in a regular secondary structure. It is used to identify regions of globularity and disorder within protein sequences.
FoldIndex©26 is a convenient web server to predict if a target protein sequence is intrinsically unfolded based on the theory that folding of a protein is governed by a balance between attractive and repulsive forces. The original version of this method can just distinguish globular proteins from the unstructured ones when the protein does not consist of both ordered and disordered regions. Later, it solves this problem by computing the ratio of the net charge versus the hydropath along the protein. Sliding windows are able to identify large regions with folding propensities within a protein, instead of verifying the folding propensity of the whole protein. However, this method cannot perform on the N- and C-termini, and so it may not be applicable to the small proteins.
RONN27,28 is a novel ab initio approach of detecting natively disordered regions in proteins based on a bio-basis function neural network trained on disordered proteins. The decision about the probability of disorder is based on alignments to an ensemble of sequences (ordered, disordered, or a mixture of both). The training of the neural network is performed on the calculation of “distances”, as determined by sequence alignment, from a subset of an ensemble of well-characterized prototype sequences. However, it may give rise to inaccuracies when RONN is applied in the detection of short disordered regions, or in the first and last residues of disordered regions.
NORSp fulfils the prediction of disordered regions based on the belief that long regions predicted to be No Ordered Regular Secondary Structure (NORS) are more likely unstructured.29 This method predicts long regions with NORS by combining the secondary structure, membrane helices and coiled-coil information into the calculation of the structural content for each sequence window. Those regions with low structural content (i.e., below the given threshold) are predicted to be NORS regions. An issue of this method is that it cannot handle some special cases when some highly mobile regions have a more stable predicted secondary structure.
PreLink30 identifies unstructured regions in a protein based on biased amino acid composition and low hydrophobic clusters. The probability for a given sequence fragment to be part of a structured or an unstructured region and the distance for each amino acid to the nearest hydrophobic cluster are used in the prediction process. This approach only requires the primary sequence, instead of a multiple sequence alignment.
FoldUnfold is yet another ab initio method and it detects protein disordered regions based on a parameter termed the mean packing density of residues.31,32 Firstly, mean packing density (i.e., the average number of spatially nearby residues whose heavy atoms are within 8 Å, excluding the neighbouring residues in the sequence) for each amino acid residue is calculated from a database of 5829 three-dimensional structures. Then, the average packing density observed for a certain type of residue in a globular state is assigned as the expected packing density for each residue. The average of these gained numbers inside the window is assigned to the central residue. Consequently, the profile of the expected packing density for the target protein sequence is constructed. Regions with weak expected packing density are more probable to be disordered regions.
The final ab initio method we will mention here is Spritz.33 It predicts disorder residues by two probabilistic soft margin support vector machines SVM-LD and SVM-SD. Both of these SVMs are specialized to predict either short (i.e., less than 30 residues in length) or long disordered regions.
Clustering methods
In clustering methods, tertiary structure models are predicted for the target protein, and then these models are superimposed by carrying out structural alignments. This is done to calculate the approximate posterior probability of a residue being disordered for the target protein. One clustering approach which performs well according to our previous assessment is DISOclust.35 It is a protein disorder prediction approach based on the analysis of three dimensional models using ModFOLDclust13,34,35,45,46 or the latest version IntFOLD-DR.49 It operates on the belief that the positions of ordered residues within a protein target should be conserved in three dimensional conformations of multiple structure models, whereas the disordered residues may appear where there is local variation in multiple models. The DISOclust server combines both the DISOclust method and an in-house version of the DISOPRED method in order to improve predictions.
Meta methods
Meta predictors make their predictions by combining the output of other disorder predictors. This averaging effect usually results in a slight to moderate increase in accuracy and thus it is also a popular method among predictors.
metaPrDOS36 is a meta disorder prediction approach combining disorder prediction results from PrDOS,37 DISOPRED,11,13 DisEMBL,38 DisProt,39 DISpro,7 IUpred,20,21 POODLE-S15 and DISOclust.34
GSmetaDisorder is a meta method combining the results from 12 major disorder predictors including DisEMBL,38 DISOPRED2,13 DISpro,7 GlobPlot,25 iPDA,40 IUPred,20,21 Pdisorder, POODLE-S,15 PrDOS,37 Spritz,33 DisPSSMP,41 and RONN27 and produces a final consensus disorder prediction result.
MFDp42 is a meta approach that combines disorder predictions from three complementary predictors: DISOPRED2,13 DISOclust,34 and IUPred.20,21 Different from other meta methods, MFDp also takes a variety of information such as PSSM, predicted secondary structure, solvent accessibility, residue flexibility, back-bone dihedral torsion angles, and globular domains as input. Three subsets of selected input features are fed into three support vector machines specialized for short disorder, long disorder and complete disorder predictions. The maximal value among the three SVM outputs is taken as the predicted probability of disorder.
Template based methods
Template based methods predict disordered regions for proteins using homologous template structures. These methods attempt to find homologous proteins which have a known structure (i.e., templates) using fold recognition methods or by searching a template database. Once templates have been identified, they can be aligned or further manipulated and disordered regions identified. In this review, we do not discuss the details of any particular implementation as this method is no longer commonly used as the sole basis of a disorder predictor. We do however mention this category because it is represented by some hybrid methods (e.g., PrDOS37).
Hybrid methods
Besides the above methods, there are also some predictors using a hybrid approach. These are methods which combine two or more of the previously described categories. PrDOS37 for example is an approach combining both ab initio and template based methods. Firstly, a position-specific scoring matrix (PSSM) or profile is generated for the target amino acid sequence after two-rounds of PSI-BLAST searches against a non-redundant sequence database. Then, two predictions are carried out simultaneously. One is based on local amino acid sequence information using a support vector machine and the other one is based on template based prediction using the alignments of homologues with structures.
Benchmark of disorder predictors on a CASP9 dataset
Evaluation metrics
Comparing the performance of disorder predictors is a difficult task. First, the predictors often use different approaches and have methods developed with different flavours. Some, as described, focus more on structural features while others rely more on sequence derived data. Undoubtedly, these design choices can bias predictors towards one dataset over another. Difficulties also arise from the fact that while the notion of a disordered protein or region is well understood, a precise definition for whether or not a particular residue is ordered or disordered has not been solidified across the community.
Here, we chose to base our benchmark of disorder predictors on those that participated in the ninth instalment of the Critical Assessment of protein Structure Prediction (CASP9). CASP is a community wide blind assessment of various protein structure prediction methods. Over the course of several months, prediction targets are released on a daily basis and predictors have a period of a couple of days to submit their predictions to the prediction centre where they are stored. For each of the 23 disorder predictors which participated in CASP9, we downloaded their disorder predictions from the prediction centre (http://predictioncenter.org/casp9/). These predictors made predictions for 26 335 residues on 117 protein targets. When evaluating the disorder predictions against the protein targets, residues which were missing coordinates in the target file (i.e., a file containing three-dimensional coordinates for residues) or with variation of spatial positions of corresponding residues in chains (X-ray) or models (NMR) by more than 3.5 Å were considered to be disordered. We recognize that not all protein disorder predictors participate in CASP and so the benchmark should not be considered exhaustive.
The goal of our evaluation is to provide some information for users to select complementary tools according to different needs instead of ranking methods. Therefore, in our evaluation, we used a number of widely used measurements in the bioinformatics field and CASP experiments to assess the performances of our method and other disorder predictors. One of them is the AUC score, which represents the area under the Receiver Operating Characteristic (ROC) curve. The standard errors of AUC scores were calculated as in ref. 48. This score measures the performance of a classifier system and its dependence upon its discrimination threshold. We also calculated the positive sensitivity (TP/(TP + FN)), positive specificity (TP/(TP + FP)), negative sensitivity (TN/(TN + FP)), negative specificity (TN/(TN + FN)), and the false positive rate (FP/(TN + FP)). Based on these values, we also evaluated the overall accuracy (ACC) score43 measured as the average of the positive sensitivity and negative sensitivity. Considering that different methods may use different criteria to set a probability threshold to make order/disorder decisions, we calculated the break-even score of each method and its corresponding decision threshold. A break-even score is the value when the positive sensitivity is equal to positive specificity at a particular threshold. Moreover, aiming at integrating all the above measurements, we adopted the product of positive sensitivity and negative sensitivity and the harmonic mean, or F-measure, of the positive sensitivity and positive specificity as additional measures. The final assessment metric is a weighted score first introduced in CASP644 and is defined as (TP × Wdisorder − FP × Worder + TN × Worder − FN × Wdisorder)/(number of residues) where Wdisorder is set to 92.63 and Worder to 7.37. This measure places more emphasis on the correct classification of disordered residues.
Results
Table 2 reports the ACC scores, AUC scores and their standard errors, weighted scores, break-even scores and their decision thresholds, false positive rate, positive sensitivity, negative specificity, negative sensitivity, positive specificity, product of positive sensitivity and negative sensitivity, and F-measure for the disorder predictors. Moreover, Table 2 also shows the total number of residues predicted by each predictor respectively. The results of our evaluation of the predictors according to ACC scores are consistent with the official CASP9 evaluation results posted on the prediction centre’s website (http://predictioncenter.org/casp9/).
Table 2.
Results for protein disorder predictors that participated in CASP9 on 117 targets
| Disorder predictor | ACC score | AUC score(SE) | Weighted score | Break-even(ths) | False pos. | Pos. sens. | Pos. spec. | Neg. sens. | Neg. spec. | Sens. prod. | F-meas. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Prdos2a,d | 0.752 | 0.852(0.005) | 7.153 | 0.48(0.57) | 0.103 | 0.608 | 0.375 | 0.897 | 0.957 | 0.545 | 0.464 |
| MULTICOM-REF INE/PreDisordera | 0.748 | 0.819(0.005) | 7.187 | 0.45(0.55) | 0.154 | 0.650 | 0.300 | 0.846 | 0.960 | 0.550 | 0.410 |
| biomine_DR_pdbc | 0.739 | 0.818(0.005) | 6.763 | 0.48(0.54) | 0.119 | 0.597 | 0.338 | 0.881 | 0.956 | 0.526 | 0.432 |
| GSmetaDisorderMDc | 0.736 | 0.813(0.005) | 6.906 | 0.40(0.89) | 0.184 | 0.657 | 0.266 | 0.816 | 0.959 | 0.536 | 0.378 |
| masona | 0.730 | 0.740(0.006) | 6.297 | — | 0.077 | 0.537 | 0.416 | 0.923 | 0.952 | 0.496 | 0.469 |
| ZHOU-SPINE-Da | 0.729 | 0.829(0.005) | 6.411 | 0.46(0.54) | 0.122 | 0.579 | 0.326 | 0.878 | 0.954 | 0.509 | 0.417 |
| GSmetaserverc | 0.713 | 0.811(0.005) | 5.982 | 0.40(0.85) | 0.151 | 0.577 | 0.279 | 0.849 | 0.952 | 0.490 | 0.376 |
| ZHOU-SPINE-DMc | 0.705 | 0.789(0.006) | 5.621 | 0.42(0.54) | 0.125 | 0.535 | 0.303 | 0.875 | 0.949 | 0.468 | 0.387 |
| Distill-Punch1a | 0.701 | 0.797(0.006) | 5.392 | — | 0.103 | 0.505 | 0.338 | 0.897 | 0.946 | 0.453 | 0.405 |
| GSmetaDisorderc | 0.694 | 0.793(0.005) | 5.268 | 0.40(0.63) | 0.131 | 0.519 | 0.287 | 0.869 | 0.947 | 0.451 | 0.370 |
| OnD-CRFa,d | 0.694 | 0.733(0.005) | 5.513 | — | 0.198 | 0.586 | 0.231 | 0.802 | 0.950 | 0.470 | 0.332 |
| CBRC_POODLEa | 0.693 | 0.828(0.005) | 4.958 | 0.43(0.51) | 0.061 | 0.447 | 0.425 | 0.939 | 0.944 | 0.419 | 0.435 |
| MULTICOMc | 0.687 | 0.852(0.005) | 4.723 | 0.45(0.48) | 0.045 | 0.419 | 0.481 | 0.955 | 0.942 | 0.400 | 0.448 |
| IntFOLD-DRb | 0.683 | 0.794(0.005) | 4.831 | 0.38(0.57) | 0.115 | 0.481 | 0.299 | 0.885 | 0.944 | 0.426 | 0.369 |
| Biomine_DR_mixedc | 0.683 | 0.769(0.006) | 4.901 | 0.35(0.62) | 0.135 | 0.501 | 0.274 | 0.865 | 0.945 | 0.433 | 0.354 |
| Spritz3c | 0.683 | 0.751(0.006) | 4.732 | 0.39(0.63) | 0.091 | 0.457 | 0.336 | 0.909 | 0.943 | 0.415 | 0.387 |
| DISOPRED3Ca | 0.669 | 0.851(0.005) | 3.975 | 0.55(0.17) | 0.010 | 0.349 | 0.775 | 0.990 | 0.937 | 0.345 | 0.481 |
| GSmetaDisorder3Dc | 0.669 | 0.781(0.006) | 4.142 | 0.40(0.5) | 0.061 | 0.398 | 0.399 | 0.939 | 0.939 | 0.374 | 0.399 |
| biomine_DR_pdb_cc | 0.659 | 0.815(0.005) | 3.647 | 0.48(0.18) | 0.015 | 0.333 | 0.696 | 0.985 | 0.936 | 0.328 | 0.451 |
| OnD-CRF-pruneda,d | 0.659 | 0.707(0.006) | 4.358 | 0.32(0.75) | 0.208 | 0.526 | 0.205 | 0.792 | 0.943 | 0.417 | 0.295 |
| Distilla,b,d | 0.654 | 0.693(0.006) | 4.152 | 0.29(0.79) | 0.202 | 0.510 | 0.204 | 0.798 | 0.941 | 0.407 | 0.291 |
| ULg-GIGA | 0.589 | 0.718(0.006) | 1.302 | 0.35(0.11) | 0.012 | 0.191 | 0.608 | 0.988 | 0.924 | 0.188 | 0.290 |
| Biomine_DR_mixed_cc | 0.572 | 0.769(0.006) | 0.644 | 0.35(0.30) | 0.008 | 0.152 | 0.647 | 0.992 | 0.920 | 0.151 | 0.247 |
Results of our evaluation of all the protein disorder predictors ordered by the ACC score. For the AUC and the break-even point, the standard error and decision threshold at with the break-even point occurred are shown, respectively, in parentheses. False pos. is the false positive rate. Pos. spec. and Neg. spec. are the positive and negative specificities. Pos. sens. and Neg. sens. are the positive and negative sensitivities. F-meas. is the F-measure of the positive sensitivity and positive specificity and Sens. prod. is the product of the positive sensitivity and negative sensitivity. MULTICOM_REFINE is the predictor name of our method PreDisorder in CASP9.
Denotes ab initio methods.
Denotes clustering methods.
Denotes meta methods.
Denotes template-based methods.
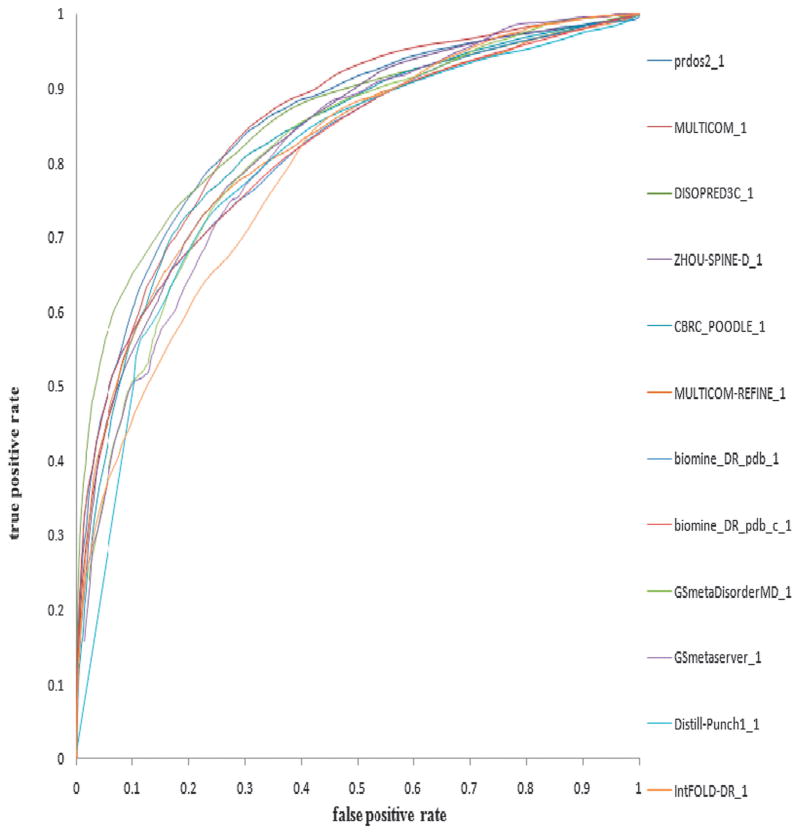
Fig. 1 shows the ROC curves for the top 12 predictors in terms of the AUC score. The predictors are ordered by AUC scores since the AUC measure is probably the most balanced measurement.
Fig. 1.
ROC curves of top 12 CASP9 predictors (ordered by AUC score) on the CASP9 dataset which consisted of 117 protein targets.
Most of the disorder prediction methods which participated in CASP9 have already been discussed and are classified into four main categories. Analyzing the results, no single category or type of method decisively outperforms the others. The ab initio methods MULTICOM_REFINE (i.e., PreDisorder), mason, SPINE-D and Distill-Punch, the meta-method biomine_DR_pdb, GSmetaDisorderMD, and the hybrid method PrDOS (i.e., combining both ab initio and template-based methods) are all among the top with respect to ACC scores on the one hand. On the other hand, PrDOS, MULTICOM (i.e., our meta disorder prediction method), DISOPRED3C, ZHOU-SPINE-D, CBRC_POODLE, MULTICOM_REFINE (i.e., PreDisorder), and biomine_DR_pdb performed better according to the AUC score. Moreover, all the predictors did a good job in terms of negative specificity and negative sensitivity. This is not hard to deduce though, since most of the residues in a protein are ordered resulting in ratios between true negatives (TN) and true negatives plus false positives (TN + FP) and true negatives (TN) and true negatives plus false negatives (TN + FN) which are very close to 1.
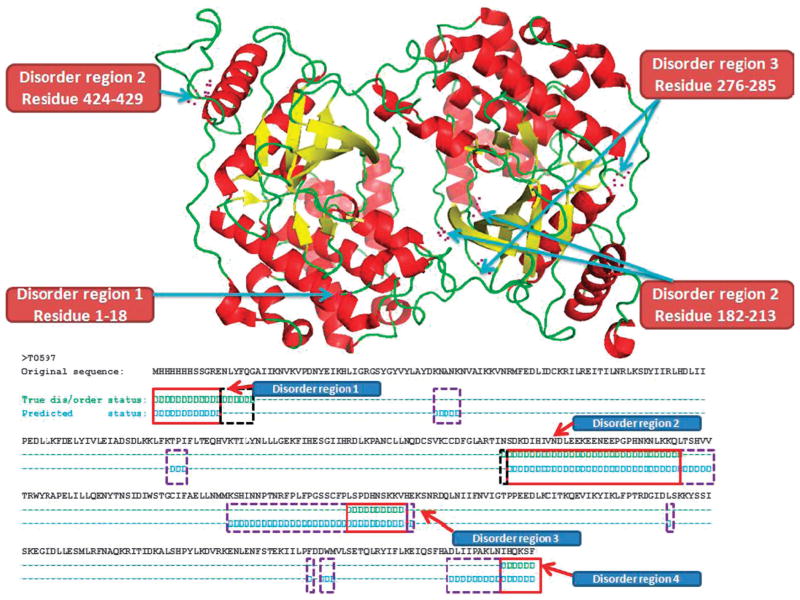
Fig. 2 is an example showing the disorder regions for target T0597 as predicted by our ab initio predictor PreDisoder (named MULTICOM_REFINE in CASP9). The three dimensional structure on the top of the figure is the experimentally determined structure of the target. The example shows that four disordered regions within and at the ends of the protein were rather accurately predicted.
Fig. 2.
Disorder regions predicted by our ab initio predictor PreDisorder (named MULTICOM_REFINE in CASP9) for one CASP9 target T0597. The length of the target protein is 429 AA. The real 3D structure of T0597 is shown on the top of this figure. The helix is shown in red colour, sheet in yellow, and loop in green. Disorder regions of the target protein are 1–18, 182–213, 276–285, and 424–429, and are also identified in the figure. Below the 3D structure, correct predictions are identified by red thick rectangles, the regions predicted as ordered but experimentally disordered are identified by virtual black rectangles, and the regions predicted as disordered but experimentally ordered are identified by virtual purple rectangles. TP is 59, TN is 314, FP is 49, FN is 7, positive sensitivity is 0.894, positive specificity is 0.546, negative sensitivity is 0.865, and negative specificity is 0.978.
Conclusions and future work
In response to the need of quickly and easily identifying intrinsically disordered protein regions for biomedical research, the bioinformatics community has developed an array of complementary computational methods to rather reliably predict protein disordered regions from a protein sequence. These tools are playing and will continue to play an important role in protein structure analysis and prediction, protein structure determination, protein interaction study, and protein function annotation.
Further development of protein disorder prediction methods is important and possible. According to our benchmarking, no single category of methods performs decisively better than others. Combining complementary disorder prediction methods and/or multiple sources of information such as homologous structure templates, multiple sequence alignment, secondary structure, solvent accessibility and other new features may improve the accuracy of disorder prediction. Furthermore, current best performing ab initio disorder prediction methods largely adopt a black-box approach based on machine learning methods such as neural networks and support vector machines,10 which do not reveal the biophysical relationship between a disorder region and its amino acid sequence. Integrating molecular dynamic simulations with disorder prediction methods may help elucidate why and how an intrinsically disordered protein sequence folds into an unstructured ensemble rather than a more deterministic three-dimensional structure.
Table 1.
List of some currently popular disorder prediction methods
Acknowledgments
We thank Zheng Wang for his help with data preparation used in the evaluation of disorder predictions. The work was partially supported by a NIH grant (1R01GM093123) to JC.
Footnotes
Published as part of a Molecular BioSystems themed issue on Intrinsically Disordered Proteins. Guest Editor: M. Madan Babu.
Notes and references
- 1.Wright PE, Dyson HJ. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 2.Wootton JC. Comput Chem. 1994;18:269–285. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- 3.Tompa P. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 4.Melamud E, Moult J. Proteins: Struct, Funct, Genet. 2003;53(suppl 6):561–565. doi: 10.1002/prot.10533. [DOI] [PubMed] [Google Scholar]
- 5.Dyson HJ, Wright PE. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 6.Ferron F, Longhi S, Canard B, Karlin D. Proteins: Struct, Funct, Bioinf. 2006;65:1–14. doi: 10.1002/prot.21075. [DOI] [PubMed] [Google Scholar]
- 7.Cheng J, Sweredoski MJ, Baldi P. Data Min Knowl Discovery. 2005;11:213–222. [Google Scholar]
- 8.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 9.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 10.He B, Wang K, Liu Y, Xue B, Uversky VN, Dunker AK. Cell Res. 2009;19:929–949. doi: 10.1038/cr.2009.87. [DOI] [PubMed] [Google Scholar]
- 11.Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. Bioinformatics. 2004;20:2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 12.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Deng X, Eickholt J, Cheng J. BMC Bioinf. 2009;10:436. doi: 10.1186/1471-2105-10-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimizu K, Hirose S, Noguchi T. Bioinformatics. 2007;23:2337–2338. doi: 10.1093/bioinformatics/btm330. [DOI] [PubMed] [Google Scholar]
- 16.Hirose S, Shimizu K, Kanai S, Kuroda Y, Noguchi T. Bioinformatics. 2007;23:2046–2053. doi: 10.1093/bioinformatics/btm302. [DOI] [PubMed] [Google Scholar]
- 17.Shimizu K, Muraoka Y, Hirose S, Tomii K, Noguchi T. BMC Bioinf. 2007;8:78. doi: 10.1186/1471-2105-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Faraggi E. In: Introduction to Protein Structure Prediction: Methods and Algorithms. Rangwala H, Karypis G, editors. Wiley; 2010. [Google Scholar]
- 19.Faraggi E, Yang Y, Zhang S, Zhou Y. Structure (London) 2009;17:1515–1527. doi: 10.1016/j.str.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dosztanyi Z, Csizmok V, Tompa P, Simon I. J Mol Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 21.Dosztanyi Z, Csizmok V, Tompa P, Simon I. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 22.Romero P, Obradovic Z, Dunker K. Genome Inf Ser. 1997;8:110–124. [PubMed] [Google Scholar]
- 23.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Genome Inf Ser. 1999;10:30–40. [PubMed] [Google Scholar]
- 24.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Proteins: Struct, Funct, Genet. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 25.Linding R, Russell RB, Neduva V, Gibson TJ. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 27.Yang ZR, Thomson R, McNeil P, Esnouf RM. Bioinformatics. 2005;21:3369–3376. doi: 10.1093/bioinformatics/bti534. [DOI] [PubMed] [Google Scholar]
- 28.Thomson R, Hodgman TC, Yang ZR, Doyle AK. Bioinformatics. 2003;19:1741–1747. doi: 10.1093/bioinformatics/btg237. [DOI] [PubMed] [Google Scholar]
- 29.Liu J, Rost B. Nucleic Acids Res. 2003;31:3833–3835. doi: 10.1093/nar/gkg515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coeytaux K, Poupon A. Bioinformatics. 2005;21:1891–1900. doi: 10.1093/bioinformatics/bti266. [DOI] [PubMed] [Google Scholar]
- 31.Garbuzynskiy SO, Lobanov MY, Galzitskaya OV. Protein Sci. 2004;13:2871–2877. doi: 10.1110/ps.04881304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galzitskaia OV, Garbuzinskii SA, Lobanov M. Mol Biol (Moscow) 2006;40:341–348. [PubMed] [Google Scholar]
- 33.Vullo A, Bortolami O, Pollastri G, Tosatto SC. Nucleic Acids Res. 2006;34:W164–W168. doi: 10.1093/nar/gkl166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGuffin LJ. Bioinformatics. 2008;24:1798–1804. doi: 10.1093/bioinformatics/btn326. [DOI] [PubMed] [Google Scholar]
- 35.McGuffin LJ. Bioinformatics. 2008;24:586–587. doi: 10.1093/bioinformatics/btn014. [DOI] [PubMed] [Google Scholar]
- 36.Ishida T, Kinoshita K. Bioinformatics. 2008;24:1344–1348. doi: 10.1093/bioinformatics/btn195. [DOI] [PubMed] [Google Scholar]
- 37.Ishida T, Kinoshita K. Nucleic Acids Res. 2007;35:W460–W464. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Structure (London) 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, Obradovic Z, Dunker AK. Nucleic Acids Res. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su CT, Chen CY, Hsu CM. Nucleic Acids Res. 2007;35:W465–W472. doi: 10.1093/nar/gkm353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su CT, Chen CY, Ou YY. BMC Bioinf. 2006;7:319. doi: 10.1186/1471-2105-7-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizianty MJ, Stach W, Chen K, Kedarisetti KD, Disfani FM, Kurgan L. Bioinformatics. 2010;26:i489–i496. doi: 10.1093/bioinformatics/btq373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. BMC Bioinf. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin Y, Dunbrack RL., Jr Proteins: Struct, Funct, Bioinf. 2005;61(suppl 7):167–175. doi: 10.1002/prot.20734. [DOI] [PubMed] [Google Scholar]
- 45.McGuffin LJ. Proteins: Struct, Funct, Bioinf. 2009;77:185–190. doi: 10.1002/prot.22491. [DOI] [PubMed] [Google Scholar]
- 46.McGuffin LJ, Roche DB. Bioinformatics. 2010;26:182–188. doi: 10.1093/bioinformatics/btp629. [DOI] [PubMed] [Google Scholar]
- 47.Mount J, Fidelis K, Kryshtafovych A, Rose B, Tramontano A. Proteins. 2009;77(suppl 9):1–4. doi: 10.1002/prot.22589. [DOI] [PubMed] [Google Scholar]
- 48.Hanley JA, McNeil BJ. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 49.Roche DB, Buenavista MT, Tetchner SJ, McGuffin LJ. Nucleic Acids Res. 2011;39(suppl 2):W171–W176. doi: 10.1093/nar/gkr184. [DOI] [PMC free article] [PubMed] [Google Scholar]