Abstract
Background aims
Immunotherapy targeting MAGE-A3 in multiple myeloma (MM) could eradicate highly aggressive and proliferative clonal cell populations responsible for relapse. However, expression of many cancer-testis antigens, including MAGE-A3, can be heterogeneous, leading to the potential for tumor escape despite MAGE-A3-induced immunity. We hypothesized that a combination of the hypomethylating agent 5-azacitidine (5AC) and the histone deacetylase inhibitor (HDACi) MGCD0103 (MGC) could induce MAGE-A3 expression in MAGE-A3-negative MM, resulting in recognition and killing of MM cells by MAGE-A3-specific cytotoxic T lymphocytes (CTL).
Methods
Gene expression analyses of MAGE-A3 expression in primary MM patient samples at diagnosis and relapse were completed to identify populations that would benefit from MAGE-A3 immunotherapy. MM cell lines were treated with 5AC and MGC. Real-time polymerase chain reaction (PCR) and Western blotting were performed to assess MAGE-A3 RNA and protein levels, respectively. Chromium-release assays and interferon (IFN) secretion assays were employed to ascertain MAGE-A3 CTL specificity against treated targets.
Results
Gene expression analysis revealed that MAGE-A3 is expressed in MM patients at diagnosis (25%) and at relapse (49%). We observed de novo expression of MAGE-A3 RNA and protein in MAGE-A3-negative cell lines treated with 5AC. MGC treatment alone did not induce expression but sequential 5AC/MGC treatment led to enhanced expression and augmented recognition by MAGE-A3-specific CTL, as assessed by 51Cr-release assays (P = 0.047) and enzyme-linked immunosorbent assay (ELISA) for IFN-γ secretion (P = 0.004).
Conclusions
MAGE-A3 is an attractive target for immunotherapy of MM and epigenetic modulation by 5AC, and MGC can induce MAGE-A3 expression and facilitate killing by MAGE-A3-specific CTL.
Keywords: 5-azacitidine, cancer-testis antigen, demethylation, epigenetics, histone deactylase inhibitor, hypomethylation, MAGE-A3, MGCD0103, multiple myeloma
Introduction
Currently available therapies offer poor long-term outcomes for multiple myeloma (MM) patients with a high-risk genetic signature (1). This group accounts for 15% of newly diagnosed patients and 75% of those with relapsed disease. Approaches to intensify therapies in this group of patients have led to cumulative toxicity and host exhaustion and have not improved overall survival (OS). Therefore, new therapeutic approaches that are both non-toxic and non-cross-resistant with chemotherapy are desperately needed for these patients. One potential answer lies in immune therapies targeting tumor-specific antigens, which may eradicate chemoresistant tumor-cell clones without inducing significant toxicities. For example, vaccination with tumor-specific antigens and transfer of tumor-specific T cells is safe and has induced clinical responses in lung cancer and melanoma (2,3).
Targets of particular interest are the cancer-testis antigens (CT-Ag), whose expression in normal tissues is restricted to immunoprivileged sites such as the testes, ensuring that immune responses generated toward these antigens will be non-toxic to normal tissues. CT-Ag expression is common in MM (4–8) and has been linked to poor prognosis (9,10). CT-Ag expression in cancer is probably the result of global hypomethylation, specifically of CpG islands at promoter sites (11–13).This phenomenon may also explain reports of coordinate expression of multiple CT-Ag in malignancies, including MM (8,14–16).
One potential concern in targeting CT-Ag for immunotherapy is that CT-Ag-negative clones could lead to tumor escape (17–20). Interestingly, de novo induction of CT-Ag expression has been achieved with hypomethylating agents such as 5-aza 2-deoxy-cytidine (DAC) and its nucleoside analog 5-azacitidine (5AC) (20–27), which incorporates into RNA and, to a lesser extent, DNA (12,28). 5AC has been approved by the food and drug administration (FDA) for use in myelodysplastic syndrome (MDS). Phase I and II clinical trials investigating the use of 5AC in MM have been initiated (protocol ID NCT00761722 and NCT00412919). Further increases in hypomethylating agent-induced gene expression have been achieved with histone deactylase inhibitors (HDACi) such as trichostatin A, valproic acid and MGCD0103 (MGC) via hyperacetylation of the histone core (12,29,30).
We have reported previously that potent immune responses to the CT-Ag MAGE-A3 can be induced by vaccination of a MM patient with MAGE-A3-positive disease with MAGE-A3 recombinant protein (31). We therefore wished to study whether the combination of demethylating agents and HDACi could optimize such therapy. We first analyzed expression of the CT-Ag MAGE-A3 in MM patients and correlated expression with validated disease subgroups identified by gene-expression profiling (GEP) (32) and survival. We then studied whether 5AC induced expression of MAGE-A3 in MM cells and whether any up-regulation could be enhanced with MGC. Finally, we assessed whether MAGE-A3/HLA-A*6801 -specific cytotoxic T lymphocytes (CTL) could kill 5AC/MGC-treated targets.
Methods
Subject samples, GEP, subgroup and survival curve analyses
Normal tissue RNA (plasma cell, lung, uterus, kidney, stomach, brain, breast, spleen, prostate, skeletal muscle, testis, thymus, liver, ovary, heart and small intestine) were obtained from Clontech (Mountain View, CA, USA). Bone marrow was collected from healthy donors and patients with MM, after informed consent, in accordance with the Declaration of Helsinki. Approval was obtained from the University of Arkansas for Medical Sciences (UAMS, Little Rock, AR, USA) Institutional Review Board for sample procurement. CD 138-positive plasma cells were purified using CD 138 antibody (Ab)-coated magnetic beads (Miltenyi Biotec Inc., Auburn, CA, USA), as described previously (33). GEP (33), molecular subgroup (32), high-risk group classifications (34) and survival curve (32) analyses were performed as reported previously using samples obtained from MM patients uniformly treated with our clinical protocols UARK 98-026 [total therapy (TT) 2] and UARK 2003-033 (TT3) (35,36).
Cell lines and drug treatment
MM cell lines ANBL-6, OCI-MY1 and OCI-MY5 were kindly provided by Michael Kuehl, MD (Genetics Department, Medicine Branch, National Cancer Institute, Bethesda, MD, USA). RPMI-8226 was obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). LP1 was kindly provided by Facet Biotech (Redwood City, CA, USA) and ARK was developed in our laboratories at UAMS. The colorectal cell line COLO205, included in some experiments as a positive control for 5AC-induced MAGE-A3 expression (15), was obtained from ATCC. Cell lines were treated with vehicle (phosphate-buffered saline, PBS), 5AC (Cel-gene Corporation, Summit, NJ, USA) and/or MGC (Methylgene, Montreal, Quebec, Canada) for the treatment times and doses indicated.
Flow cytometry
Viability after treatment was assessed by staining with annexin V and propidium iodide as per the manufacturer’s instructions (Vybrant apoptosis assay kit number 3; Invitrogen, Carlsbad, CA, USA). HLA-A*6801 cell-surface expression on the transfected LP1 cells was verified with an HLA-A*2/28 Ab directly conjugated to fluorescein isothiocyanate (FITQ One Lambda, Canoga Park, CA, USA), which recognizes HLA-A*6801. An appropriate iso-type control was included. Intracellular staining for MAGE-A3 was performed after fixation and per-meabilization of 106 LP1 A68 cells treated with and without 5AC, MGC or a combination of both, as per the manufacturer’s instructions (BD Cytofix/Cytoperm fixation/permeabilization kit; BD Biosciences, San Jose, CA, USA). The MAGE-A3 protein was detected using an indirect labeling method with 5 µg purified primary murine IgGl monoclonal Ab 57B, specific for MAGE-A3 protein (kindly gifted from Dr G. Spagnoli, University Hospital, Basel, Switzerland) and 0.5 µg secondary Ab rat anti-mouse IgGl-phycoerythrin (PE) (clone A85-1; BD Biosciences). Controls included a non-specific isotype murine IgGl primary Ab with a secondary rat anti-mouse IgGl-PE Ab to confirm the absence of non-specific staining. The cell line ARK was included as a positive control for MAGE-A3 expression, while untreated LP1 served as a negative control. Stained cells were run immediately on a FACSCalibur flow cytometer (BD Biosciences) and analyzed using FCS Express software (DeNovo, Los Angeles, CA, USA).
Real-time polymerase chain reaction analysis
Total RNA (1 µg) was isolated using an RNeasy minikit (Qiagen, Valencia, CA, USA) and used to synthesize cDNA with a Superscript III kit (Invitrogen). Real-time polymerase chain reactions (PCR) were performed on an ABI Prism 7000 sequence detection system, using a standard 40-cycle protocol and predesigned Taqman primer/probes (Applied Biosystems, Foster City, CA, USA) for MAGE-A3 (Hs00366532_ml) and the endogenous control Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) (Hs99999905_ml).The fold change in MAGE-A3 expression relative to the mock-treated control was assessed using a comparative cycle threshold (Ct) method, in which the target gene (MAGE-A3) was normalized to the reference gene (GAPDH) (37). The following equation was used to determine the value: MAGE-A3-fold change = 2−ddCt, where ddCt = [(treated sample CtMAGE-A3 − CtGAPDH)] − (mock-treated sample CtMAGE-A3 − CtGAPDH)].
Western blot analysis
Cells (105) for Western blot analysis were lysed with 1X Laemmli-SDS buffer (Sigma, St Louis, MO, USA), subjected to sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE) using 4–20% Tris-Glycine gels (Invitrogen), transferred onto nitrocellulose membranes and then probed with a l:105 dilution of GAPDH (clone 2D487; Abcam, Cambridge, MA, USA), 1 µg/mL MAGE-A3 57B or 1 µg/mL acetylated histone H3 (acH3) (clone AH3-120; Abcam)-specific Ab. The detection system utilized a sheep-anti-mouse horseradish peroxidase (HRP) Ab and an advanced enhanced chemiluminescent (ECL) substrate (VWR, Suwanee, GA, USA).
Bisulfite conversion and methylation-specific PCR
Genomic DNA (1 µg) was extracted using a DNeasy kit (Qiagen) and subjected to sodium bisulfite conversion using an Epitect bisulfite kit (Qiagen) as per the kit instructions. Five microliters of each reaction were subjected to methylation-specific (MS)-PCR using the forward and reverse primers specific for the methylated (MG3M) and unmethylated (MG3U) sequences reported by Honda et al. (24). The 228-bp MS control amplicon was generated using Jurkat genomic DNA (New England Biolabs, Beverly, MA, USA) treated with the CpG methyl-transferase SSSI. Testis genomic DNA (BioChain, Hayward, CA, USA) served as an unmethylated PCR-positive control, also generating a 228-bp amplicon.
Transfection of LP1 with HLA-A*6801 (LP1 A68)
Transfection of 5 × 106 LP1 cells was performed with 2 µg plasmid encoding HLA-A*6801 and neoR (Genscript, Piscataway, NJ, USA) using the Amaxa cell line nucleofection kit R and nucleofector device II as per the manufacturer’s instructions (Lonza, Walkersville, MD, USA). Acceptable transfection efficiency (>10% HLA*A6801+) was confirmed 24 h post-transfection via flow cytometry prior to 2 weeks of selection with 1 µg/mL G418 (Invitrogen) to obtain >95% HLA-A*6801 + stable transfectants.
Chromium-release assays
Standard 4-h 51CrO4 release assays were employed to determine the percentage specific lysis of treated targets using previously established MAGE-A3/HLA-A*6801 -specific CTL (10). Percentage specific lysis was calculated as: (test CPM release − spontaneous CPM release)/(maximal CPM release − spontaneous CPM release) × 100%. Untreated target cells pre-incubated for 90 min with irrelevant or MAGE-A3/HLA-A*6801 peptide served as negative and positive controls, respectively. As an additional positive control, the MAGE-A3-and HLA-A*6801-positive MM cell line ARK was included. The natural killer (NK) cell-sensitive leukemia cell line K562 was included to confirm the absence of NK cell activity.
Interferon-γ enzyme-linked immunosorbent assay
Measurement of secreted interferon (IFN)-γ in CTL/MM target co-cultures was performed using a Quantikine IFN-γ immunoassay kit (R&D Systems, Minneapolis, MN, USA) according to the manufacturer’s instructions. The minimal detectable concentration of IFN-γ with this assay is 8 pg/mL. Mean IFN-γ levels for MAGE-A3 CTL stimulated by LP1 A68 treated with 5AC/MGC were compared with unstimulated MAGE-A3 CTL or MAGE-A3 CTL co-cultured with untreated LP1 A68 (negative controls). MAGE-A3 CTL stimulated by ARK (MAGE-A3- and HLA-A*6801-positive) served as a positive control. Cells were co-cultured at a CTL:target ratio of 5:1 and allowed to incubate for 4 h before harvesting supernatants.
Results
AIAGE-A3 is frequently expressed in high-risk MM
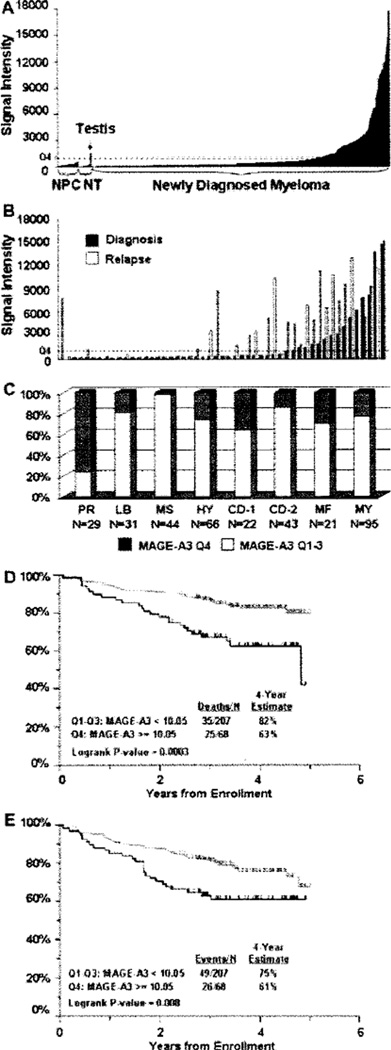
We observed high (quartile 4, Q4) MAGE-A3 expression in 25% of 565 newly diagnosed MM patients (Figure 1A). We have previously reported complete concordance between MAGE-A3 gene expression by GEP and protein expression by immunohistochemistry (38). MAGE-A3 expression was not detected in normal plasma cells (n = 26) nor in normal donor-derived tissues (n = 16), with the exception of testis (Figure 1A). A paired analysis of 51 patients showed that MAGE-A3 (Q4) is more frequently expressed at relapse (49% versus 31%, P < 0.001; Figure 1B). We have recently identified seven genetically distinct subgroups with differing clinical features and prognosis (32). High MAGE-A3 expression was most prevalent in the group characterized by a proliferation signature (76%, P < 10−10), which is associated with poor clinical outcome (Figure 1C). In the context of a second prognostic model (70-gene model) (1,34) analyzing 351 untreated patients, baseline MAGE-A3 expression (Q4) was present in 52% of patients with high-risk MM versus 21% with low-risk MM (P < 0.001). OS for patients expressing MAGE-A3 at baseline in our TT3 trial was significantly lower compared with patients that did not express MAGE-A3 at baseline (P < 0.0003, n = 275; Figure 1D). Analysis of event-free survival (EFS) for patients expressing MAGE-A3 at baseline in our TT3 trial also revealed a significant difference from patients that did not express MAGE-A3 at baseline (P < 0.008, n = 275; Figure 1E).
Figure 1.
MAGE-A3 expression in MM is associated with relapsed and proliferative disease as well as poorer outcome. (A) MAGE-A3 is not expressed in normal plasma cells (NPC) or in normal tissues (NT) except for testis, but is expressed (≥quartile 4, Q4, dashed line) in 25% of MM patients at diagnosis. (B) A paired analysis of MAGE-A3 expression in 51 patients at diagnosis versus relapse showed that MAGE-A3 is more frequently detected at relapse (49% versus 31 %, P < 0.001).A paired sample t-test was performed to assess significance. (C) The expression of MAGH-A3 is associated with a proliferation signature (PR) compared with the other groups, P < 10−10: LB, low bone disease; MS, multiple myeloma set domain (MMSET); HY, hyperdiploid, CD-1, cyclin d1 (CCND1), CD-2, CCND2, MF, c-maf/MAFB genes. A Pearson chi-square was performed for the significance test. (D) A Kaplan-Meier plot showing the overall survival of MM patients enrolled in TT3 (n = 275). MAGE-A3 expression in Q4 is associated with decreased overall survival. (E) EFS is significantly decreased in TT3 patients expressing MAGE-A3 in Q4 (n = 275).
MAGE-A3 gene expression was induced by 5AC and enhanced by MGC
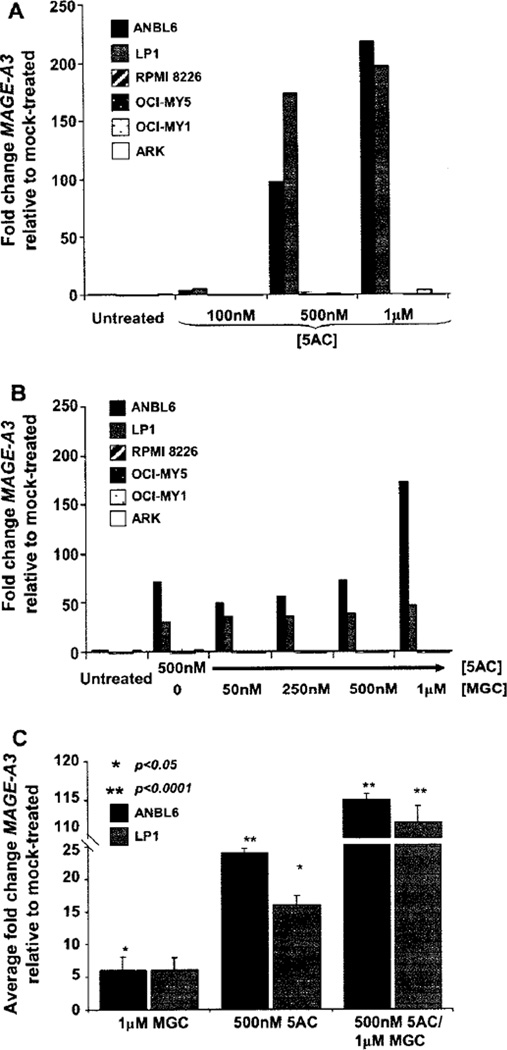
We determined by reverse transcriptase-PCR (RT-PCR) and GEP that MAGE-A3 is detectable in cell lines RPMI-8226, OCI-MY5, OCI-MY1 and ARK, whereas ANBL6 and LP1 are negative (data not shown). A dose-dependent negative effect on cell viability was observed when the lines were treated with 5AC concentrations ranging from 100 nm to 10 µm (Supplementary Figure 1A can be found at: http://www.informahealthcare/cyt/10.3109/14653249.2010.529893). MAGE-A3 expression was induced in the ANBL6 and LP1 lines following incubation with 5AC for 3 days at concentrations of 500 nm and 1 µm (Figure 2A). 5AC concentrations greater than 1 µm significantly decreased viability (<60%) and are therefore not shown. We also tested the effect of 24-h MGC treatment alone on viability and MAGE-A3 expression and observed that five of six MM cell lines were sensitive to apoptosis at MGC concentrations > 1 µm (Supplementary Figure 1B can be found at: http://www.informahealthcare/cyt/10.3109/14653249.2010.529893). Only low levels of MAGE-A3 up-regulation (<8 fold) were observed and only at MGC concentrations > 1 µm, where viability was poor, and therefore these data are not shown.
Figure 2.
(A) MAGE-A3 induction after 5AC treatment occurred in a dose-dependent fashion in MM cell lines ANBL6 and LP1. Cells were treated for 3 days with 5AC at the indicated concentrations followed by normal media for 1 additional day. MAGE-A3 expression was determined by relative real-time PCR. (B) Sequential 5AC/MGC increased MAGE-A3 further in ANBL6 and LP1. Cells were treated for 3 days with vehicle or 500 nm 5AC followed by 1 day with vehicle or MGC at the concentrations indicated. Viability was poor (<60%) at >1 µm 5AC or MGC for five of the six cell lines tested, and hence is not shown. (C) De novo induction of MAGE-A3 was reproducible in MAGE-A3-negative MM cell lines ANBL6 and LP1. Bars indicate average values for fold-change from mock-treated for at least three replicate experiments, and the standard error of the mean (SEM) was calculated using the formula SEM = (standard deviation of the delta CT(MAGE-A3 − GAPDH))/square root (n). An unpaired t-test was performed on the delta CT(MAGE-A3 − GAPDH) values for all independent experiments to ascertain the significance compared with mock-treated.
Additional experiments were performed to investigate the impact of sequential 5AC/MGC treatment on viability and MAGE-A3 expression using a 500 nm concentration of 5AC and varying concentrations of MGC. High concentrations of MGC after 500 nm 5AC treatment had a significant negative impact on cell viability (Supplementary Figure 1C can be found at: http://www.informahealthcare/cyt/10.3109/14653249.2010.529893) and resulted in levels of MAGE-A3 expression that were higher than those observed with 5AC treatment alone for lines ANBL6 and LP1 (Figure 2B). MAGE-A3 induction by 5AC and enhanced expression with sequential 5AC/MGC were reproducible and significant for ANBL6 and LP1 (Figure 2C, Supplementary Figure 3 can be found at: http://www.informahealthcare/cyt/10.3109/14653249.2010.529893). In other experiments, treatment times and/ or concentrations of 5AC and MGC were adjusted to accommodate experimental goals. Cells could not be treated for more than 2 days with MGC because of toxicity and thus longer time–course experiments were not implemented with MGC.
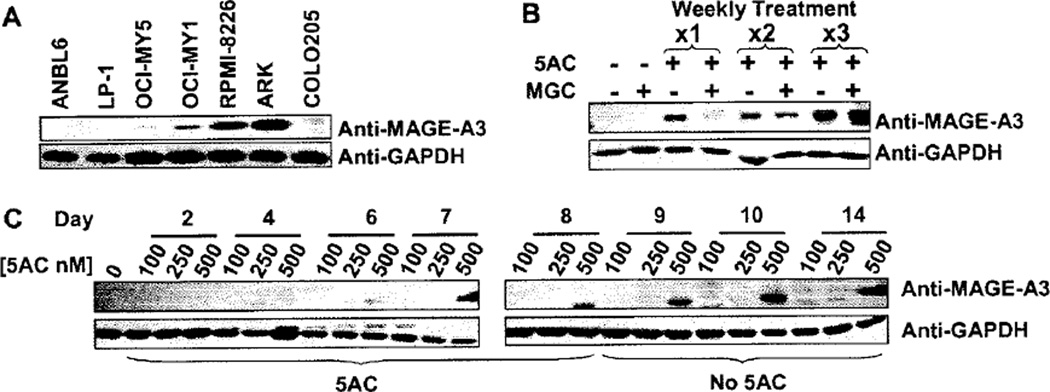
MAGE-A3 protein expression was induced by 5AC and enhanced by MGC
We next confirmed that induction of MAGE-A3 gene expression resulted in the production of MAGE-A3 protein. We were able to confirm the presence of MAGE-A3 protein in MM cell lines RPMI, OCI-MY5, OCI-MY1 and ARK (Figure 3A). As expected, MAGE-A3 protein was not detected in untreated ANBL6, LP1 or COLO205. We then compared protein expression in ANBL6, LP1 and COLO205 in untreated, 5AC alone, MGC alone and sequential 5AC/MGC treatment. MAGE-A3 protein was barely detectable in 5AC-treated ANBL6 and COLO205 cells and not detected in the 5AC-treated LP1 cell line (data not shown). We investigated whether protein expression in ANBL6 and LP1 could be induced to detectable levels by increasing the 5AC exposure time to 8 days, followed by 6 days of culture in the absence of drug. A range of 5AC concentrations (0, 100, 250 and 500 nm) was used in case longer treatment times significantly decreased viability. MAGE-A3 expression was induced in ANBL6 cells after 6 days of 500 nm 5AC treatment and the level of expression increased further after 3 additional days of culture in the absence of 5AC (Figure 3B). In another time-course experiment, ANBL6 was treated for 3 weeks; each weekly treatment consisted of the daily addition of 500 nm 5AC for 5 days followed by 2 days with vehicle or MGC. MAGE-A3 protein levels increased with each weekly treatment with 5AC, and were enhanced after the addition of MGC (Figure 3C).
Figure 3.
De novo induction of MAGE-A3 protein in ANBL6. (A) MAGE-A3 protein was detected in untreated MM lines RPMI-8226, OCI-MY5, OCI-MY1 and ARK by Western blot analysis but not in ANBL6 or LP1. (B) ANBL6 was treated with 100 nm, 250 nm or 500 nm 5AC daily for 8 days then allowed to culture for an additional 6 days without drugs. Only 500 nm 5AC induced MAGE-A3 protein expression. (C) MAGE-A3 protein expression in ANBL6 was enhanced by repeated weekly treatment with 500 nm 5AC. Treatment with 500 nm MGC for an additional 2 days after 1, 2 or 3 weekly 5AC treatments also enhanced MAGE-A3 protein expression. Prolonged treatment with MGC was avoided because of toxicity issues.
Recognition of MAGE-A3-induced MM by MAGE-A3-specific T cells
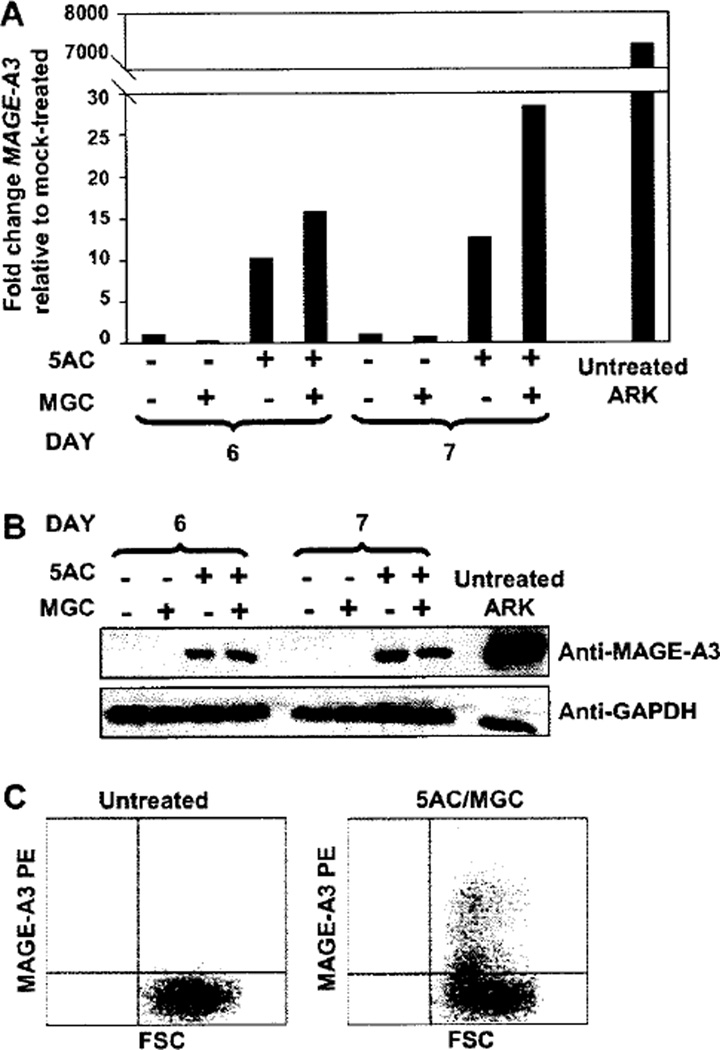
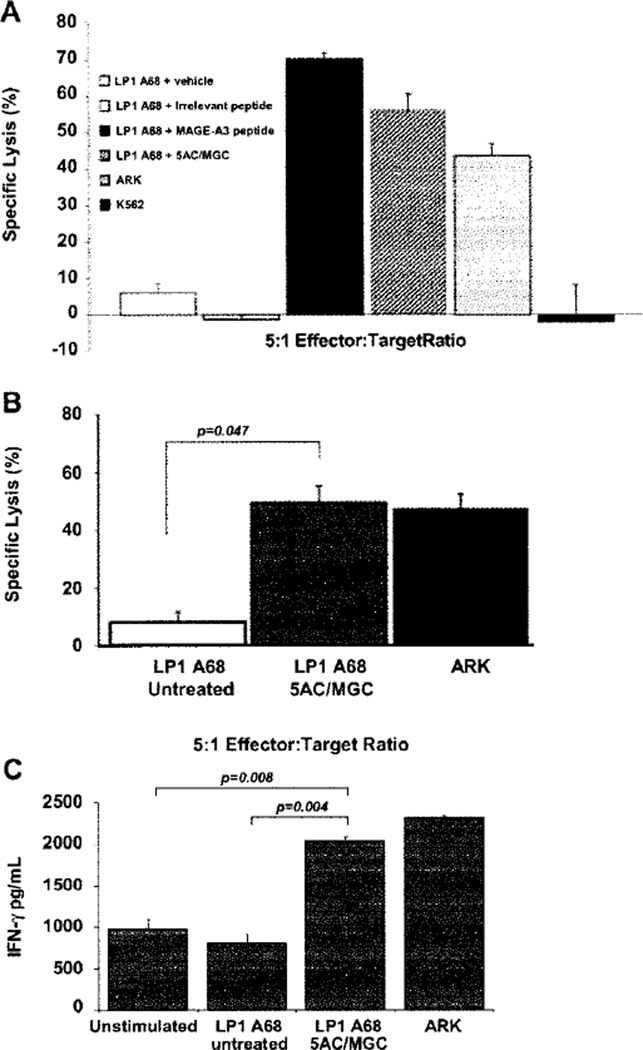
In order to determine whether the up-regulation of MAGE-A3 protein by 5AC/MGC was sufficient to allow recognition and killing by MAGE-A3/HLA-A*6801-specific T cells, we engineered a MM cell target that expressed HLA-A*6801 .The cell line LP1 was successfully transfected with HLA-A*6801 (>95% HLA-A*6801+, after obtaining stable transfectants) and the transfection did not affect MAGE-A3 expression compared with non-transfected LP1 (Supplementary Figure 2 can be found at: http://www.informahealth-care/cyt/10.3109/14653249.2010.529893). LP1 A68 cells were treated with 500 nm 5AC for 5 days followed by 1 or 2 days of vehicle or 500 nm MGC. Relative real-time PCR confirmed induction of MAGE-A3 expression in LP1 A68 after treatment with 5AC or sequential 5AC/MGC (Figure 4A). A strong MAGE-A3 protein band was observed in the 5AC-treated LP1 A68 cells and with sequential 5AC/MGC (Figure 4B). Intracellular flow cytometry detected a population of MAGE-A3-positive cells for 5AC/MGC-treated LP1 A68, but not untreated or MGC-treated LP1 A68 cells (Figure 4C). MGC- and 5AC-treated targets are not shown as the degree of MAGE-A3 expression in each was similar to untreated and 5AC/MGC-treated cells, respectively. In three independent experiments, MAGE-A3-specific CTL exhibited an average of 49% specific lysis (22%, 56% and 70%; P = 0.047) against 5AC/MGC-treated targets (Figure 5B), whereas targets treated with 5AC had minimal specific lysis over untreated controls (data not shown). Killing was also not observed when targets were treated with 5AC/MGC for less than 7 days (data not shown). Figure 5A demonstrates the CTL specificity for MAGE-A3/HLA-A*6801. The CTL killed the MAGE-A3- and HLA-A*6801-positive cell line ARK, representing recognition of the naturally processed and presented MAGE-A3115–123 epitope on MM cells by these CTL. K562 targets were not killed, thus ruling out NK cell activity. Furthermore, killing of untreated LP1 A68 was not observed unless this cell line was pulsed with MAGE-A3115–123 peptide (Figure 5A). In addition, IFN-γ was detected at a significantly higher level in the supernatants of the effector/target co-culture when the target was treated with 5AC/MGC, as opposed to the untreated target (P = 0.004) or when the MAGE-A3-positive cell line ARK was used as a target (Figure 5C).
Figure 4.
5AC and MGC induce MAGE-A3 RNA and protein expression in HLA-A68-transfected LP1 cells (LP1 A68). LP1 A68 cells were treated for 5 days with 500 nm 5AC, washed, then cultured for an additional 1 (A and B) or 2 (A, B and C) days in the presence of vehicle or 500 nm MGC. MAGE-A3 expression of RNA was detected by relative real-time PCR (A). MAGE-A3 protein expression was detected by Western blot (B) or intracellular flow cytometry (C). In (C), a population of MAGIS-A3-expressing cells were detected in 5AC/MGC-treated LP1 A68 (right plot) but not in untreated LP1 A68 (left plot). MAGE-A3-PE is plotted against forward scatter (FSC).
Figure 5.
Recognition of 5AC/MGC-treated MM by MAGE-A3-specific CTL. (A) LP1 A68 cells were treated with vehicle or 500 nm 5AC for 5 days followed by an additional 2 days with vehicle or 500 nm MGC prior to use as targets for MAGE-A3/HIA-A*6801 CTL effectors in a standard 4-h 51CrO4 release assay. Untreated target cells were pre-incubated with irrelevant or MAGE-A3/HLA-A*6801 peptide (negative and positive controls, respectively) for 90 min before the assay. As an additional positive control, the MAGE-A3- and HLA-A*6801-positive MM cell line ARK was included. The NK cell-sensitive leukemia cell line K562 was included to confirm the absence of alloreactivity. (B) Bars indicate average values calculated from three individual experiments for untreated and 5AC/MGC-treated targets. (C) Supernatants collected from MAGE-A3 CTL:target co-cultures were assayed for IFN-γ levels using an enzyme-linked immunosorbent assay (ELISA). INF-γ levels were elevated in the supernatants of MAGE-A3 CTL stimulated with 5AC/MGC-treated LP1 A68 and ARK compared with the supernatants from unstimulated MAGE-A3 CTL or CTL stimulated with untreated LP1 A68. Unpaired t-tests were performed to ascertain the significance for (B) and (C).
Reduced methylation of the MAGE-A3 promoter was observed after 5AC treatment and correlates with de novo expression of MAGE-A3
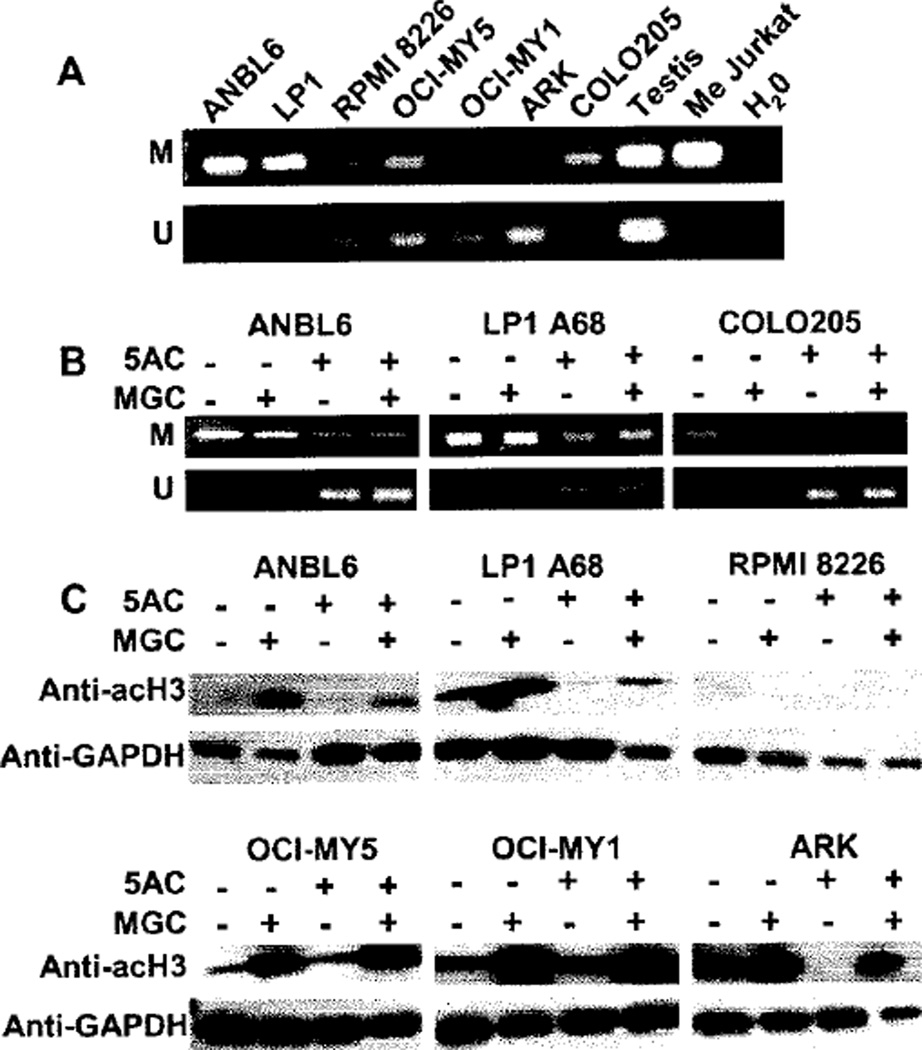
To support the hypothesis that MAGE-A3 up-regulation after 5AC treatment was induced by promoter hypomethylation, we performed MS-PCR for the MAGE-A3 promoter. ANBL6, LP1 and COLO205 cells produced high-intensity bands with the primers specific for the methylated sequence (Figure 6A), which was expected as we did not detect MAGE-A3 expression by RT-PCR for those lines. In particular, ARK cells produced an intense band with the primers for the unmethylated sequence, whereas no band was observed with the methylated primers, suggesting that in ARK the MAGE-A3 promoter is demethylated to a high degree. We then performed MS-PCR for ANBL6, LP1, COLO205 and LP1 A68 cells treated with 500 nm 5AC or vehicle for 4 days followed by 500 nm MGC or vehicle for 2 days (Figure 6B). We observed unmethylated bands for the cells treated with 5AC and 5AC/MGC, supporting the hypothesis that 5AC-induced MAGE-A3 gene expression was caused by demethylation of the MAGE-A3 promoter.
Figure 6.
Detection of MAGE-A3 promoter demethylation and histone H3 hyperacetylation after treatment with 5AC and MGC, respectively. (A) Baseline mcthylation status of the MAGE-A3 promoter in six MM cell lines and the colorectal line COLO205 using PCR primers specific for methylated (M) or unmethylated (U) sequences. Testis was included as an unmethylated positive control and Jurkat methylated enzymatically by SSS1 (Me Jurkat) was included as a methylated positive control. H2O was added in lieu of DNA template as a negative control for PCR contamination. (B) MAGE-A3 promoter demethylation was observed in MAGE-A3-negative cell lines after treatment with 5AC/MGC. ANBL6, COLO-205 and LP1 A68 cells were treated for 4 days with 500 nm 5AC or vehicle followed by 2 days of vehicle or 500 nm MGC and then subjected to methylation-specific PCR. (C) Western blot analysis of hyperacetylated histone H3 subunits. Anti-acH3 (acetyl K9) showed a greater intensity 17-kDa band in MM cell lines treated with MGC, with and without prior 5AC treatment, relative to untreated cells. GAPDH served as a loading control.
Hyperacetylation of H3 in MGC-treated MM cell lines
Western blot analysis to determine the acetylation status of the histone subunit H3 (acH3) demonstrated an increase in acH3 subunits in MGC-treated cell lines with and without prior 5AC treatment (Figure 6C).
Discussion
In our analysis of 565 newly diagnosed patients, MAGE-A3 expression was observed in 25% of patients. However, MAGE-A3 expression was much higher in relapsed patients (50%) and higher still in patients with highly proliferative disease (80%). Further, there was a correlation of adverse survival (OS and EFS) with MAGE-A3 expression. Our data are supported by other studies in which MAGE-A3 expression has been correlated with advanced MM, MM progression, MM burden, a high plasma cell proliferation index and cytogenetic abnormalities (6,8,38–41). The increased frequency of MAGE family gene expression in aggressive disease may be because of a functional role in cell-cycle progression and drug resistance (42–48). These features make MAGE-A3 an attractive target for MM immuno-therapy. MAGE-A3 is also immunogenic and not expressed by normal tissues surveyed by the immune system. We and others have previously reported that MAGE-A3 protein vaccination induces broad, potent and long-lasting immune responses (31,49). A concern is that several studies have reported heterogeneous expression of CT-Ag in tumors, which raises the specter of tumor escape by MAGE-A3-negative MM clones (17,19,50–54). We therefore hypothesized that epigenetic modulation could induce MAGE-A3 expression, thus permitting recognition and killing of MM by MAGE-A3-specific CTL.
We were indeed able to induce MAGE-A3 expression by exposing the MAGE-A3-negative MM cell lines LP1 and ANBL6 and the colorectal cell line COLO205 to the methyltransferase inhibitor 5AC at an in vitro concentration of 500 nm, which is comparable to a clinically achievable dose of 12.5 mg/m2 (28). Baseline MAGE-A3 expression was correlated with MAGE-A3 promoter methylation status. Untreated cell lines that lacked expression of MAGE-A3 exhibited methylated promoter sequences as opposed to MAGE-A3-positive cell lines that had unmethylated sequences detected. Furthermore, MS-PCR of ANBL6 and LP1 indicated that the MAGE-A3 promoters were hypermethylated prior to treatment and hypomethylated after treatment with 5AC. These results implicate MAGE-A3 promoter demethylation as a mechanism of MAGE-A3 induction that can be modulated by 5AC (15,24,26,55).
We also investigated the effect of the HDACi MGC on MAGE-A3 gene expression. MGC alone did not appreciably increase MAGE-A3 expression; however, sequential treatment with 5AC followed by MGC led to enhanced MAGE-A3 gene expression, suggesting that histone deacetylase (HDAC) activity may play a modest role in the transcriptional regulation of MAGE-A3.
De novo MAGE-A3 protein expression was confirmed in ANBL6 cells and LP1 cells transfected with HLA-A*6801 after 5AC treatment by Western blotting and flow cytometry. When ANBL6 was subjected to longer treatment with 5AC, we observed enhanced MAGE-A3 protein expression compared with shorter incubations, suggesting that the maximal effect on MAGE-A3 promoter demethylation had not been reached. Killing of LP1 A68 targets after treatment with 5AC/MGC by MAGE-A3-specific CTL also occurred only after prolonged exposures. Failed target lysis could be the result of insufficient density of MAGE-A3 peptide/HLA complexes on the target cell surface, or differences in MAGE-A3 intracellular processing, that can affect the context in which the processed peptides are displayed on the cell surface (56). The addition of MGC may have enhanced MAGE-A3 protein expression while longer treatment periods may allow additional time for optimal protein turnover, thus increasing peptide availability enough to cross the threshold needed for recognition by MAGE-A3 CTL. Synergism between MGC and 5AC was observed at the RNA level. Combinatorial or synergistic activity in terms of cytolysis was not formally tested because of limited MAGE-A3-specific CTL availability.
In summary, MAGE-A3 and other CT-Ag have been investigated extensively as a target for immunotherapy. Heterogeneous tumor antigen expression must be addressed when designing immunotherapeutic treatments to prevent escape by selection of tumor antigen-negative cancer cells. Combining vaccination or adoptive transfer of MAGE-specific T cells with an agent that epigenetically ‘turns on’ MAGE-A3 production could lead to immune recognition and eradication of both highly proliferative antigen-positive clones as well as residual, previously antigen-negative, MM cells. A potential disadvantage of the use of hypomethylating agents is the enhancement of genomic instability. Furthermore, up-regulation of MAGE-A3 could potentially have other negative consequences, including anti-apoptotic effects, promotion of the survival of clonogenic precursors and the induction of drug resistance (48).
Clinical studies are therefore warranted to determine whether effective up-regulation of MAGE-A3 protein is sufficiently sustained for T-cell expansion and cytolysis of MM cells to occur, which can then negate the possible adverse effects of the induction of MAGE-A3 expression. This study provides proof of the principle that MAGE-A3-expressing MM can be recognized by MAGE-A3-specific CTL generated from CTL precursors isolated from a MAGE-A3-vaccinated MM patient. A combination regimen of hypomethylation, HDAC inhibition and vaccine therapy in patients with MAGE-A3-positive MM, either in remission or with stable disease, after therapy, could be a first clinical trial to test the hypothesis that MAGE-A3-specific immunosurveillance may have therapeutic benefit.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the faculty and staff of the Myeloma Institute for Research and Treatment. Celgene Corporation provided 5AC. Methyl Gene provided MGC.
This work was supported by funding from Celgene Corporation and NIH/NCI grant CA 055819.
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Supplementary material available online
The supplementary figures can be found at: http://www.informahealthcare/cyt/10.3109/14653249.2010.529893
Supplementary Figure 1.
Supplementary Figure 2.
Supplementary Figure 3.
References
- 1.Shaughnessy JD, Jr, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109:2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 2.Vansteenkiste J, Zielinski M, Dahabre J, Linder A, Malinowski W, Jassem J, et al. Multi-center, double blind, randomized, placebo-controlled phase II study to assess the efficacy of recombinant MAGE-A3 vaccine as adjuvant therapy in stage IB/II MAGE-A3 positive, completely resected, non-small cell lung cancer (NSCLC) J Clin Oncol. 2006;24:7019. [Google Scholar]
- 3.Dudley ME, Wunderlich JR, Yang JC, Sherry RM, Topalian SL, Restifo NP, et al. Adoptive cell transfer therapy following non-myeloablative but lymphodepleting chemotherapy for the treatment of patients with refractory metastatic melanoma. J Clin Oncol. 2005;23:2346–2357. doi: 10.1200/JCO.2005.00.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pellat-Deceunynck C, Mellerin MP, Labarriere N, Jego G, Moreau-Aubry A, Harousseau JL, et al. The cancer germ-line genes MAGE-1, MAGE-3 and PRAME are commonly expressed by human myeloma cells. Eur J Immunol. 2000;30:803–809. doi: 10.1002/1521-4141(200003)30:3<803::AID-IMMU803>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 5.van Baren N, Brasseur F, Godelaine D, Hames G, Ferrant A, Lehmann F, et al. Genes encoding tumor-specific antigens are expressed in human myeloma cells. Blood. 1999;94:1156–1164. [PubMed] [Google Scholar]
- 6.Jungbluth AA, Ely S, DiLiberto M, Niesvizky R, Williamson B, Frosina D, et al. The cancer-testis antigens CT7 (MAGE- C1) and MAGE-A3/6 are commonly expressed in multiple myeloma and correlate with plasma-cell proliferation. Blood. 2005;106:167–174. doi: 10.1182/blood-2004-12-4931. [DOI] [PubMed] [Google Scholar]
- 7.van Rhee F, Szmania SM, Zhan F, Gupta SK, Pomtree M, Lin P, et al. NY-ESO-1 is highly expressed in poor-prognosis multiple myeloma and induces spontaneous humoral and cellular immune responses. Blood. 2005;105:3939–3914. doi: 10.1182/blood-2004-09-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Condomines M, Hose D, Raynaud P, Hundemer M, De Vos J, Baudard M, et al. Cancer/testis genes in multiple myeloma: expression patterns and prognosis value determined by micro-array analysis. J Immunol. 2007;178:3307–3315. doi: 10.4049/jimmunol.178.5.3307. [DOI] [PubMed] [Google Scholar]
- 9.Andrade VC, Vettore AL, Felix RS, Almeida MS, Carvalho F, Oliveira JS, et al. Prognostic impact of cancer/testis antigen expression in advanced stage multiple myeloma patients. Cancer Immun. 2008;8:2. [PMC free article] [PubMed] [Google Scholar]
- 10.Atanackovic D, Luetkens T, Hildebrandt Y, Arfsten J, Bartels K, Horn C, et al. Longitudinal analysis and prognostic effect of cancer-testis antigen expression in multiple myeloma. Clin Cancer Res. 2009;15:1343–1352. doi: 10.1158/1078-0432.CCR-08-0989. [DOI] [PubMed] [Google Scholar]
- 11.De Smet C, Lurquin C, Lethe B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mole Cell Biol. 1999;19:7327–7335. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 13.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 14.Gure AO, Chua R, Williamson B, Gonen M, Ferrera CA, Gnjatic S, et al. Cancer-testis genes are coordinately expressed and are markers of poor outcome in non-small cell lung cancer. Clin Cancer Res. 2005;11:8055–8062. doi: 10.1158/1078-0432.CCR-05-1203. [DOI] [PubMed] [Google Scholar]
- 15.Wischnewski F, Pantel K, Schwarzenbach H. Promoter demethylation and histone acetylation mediate gene expression of MAGE-A1, -A2, -A3, and -A12 in human cancer cells. Mol Cancer Res MCR. 2006;4:339–349. doi: 10.1158/1541-7786.MCR-05-0229. [DOI] [PubMed] [Google Scholar]
- 16.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Castillejo JA, Navarro G, San Jose-Eneriz E, et al. Epigenetic regulation of human cancer/testis antigen gene, HAGE, in chronic myeloid leukemia. Haematologica. 2007;92:153–162. doi: 10.3324/haematol.10782. [DOI] [PubMed] [Google Scholar]
- 17.Jungbluth AA, Chen YT, Stockert E, Busam KJ, Kolb D, Iversen K, et al. Immunohistochemical analysis of NY-ESO-1 antigen expression in normal and malignant human tissues. Int J Cancer. 2001;92:856–860. doi: 10.1002/ijc.1282. [DOI] [PubMed] [Google Scholar]
- 18.Jungbluth AA, Busam KJ, Kolb D, Iversen K, Coplan K, Chen YT, et al. Expression of MAGE-antigens in normal tissues and cancer. Int J Cancer. 2000;85:460–465. [PubMed] [Google Scholar]
- 19.dos Santos NR, Torensma R, de Vries TJ, Schreurs MW, de Bruijn DR, Kater-Baats E, et al. Heterogeneous expression of the SSX cancer/testis antigens in human melanoma lesions and cell lines. Cancer Res. 2000;60:1654–1662. [PubMed] [Google Scholar]
- 20.Sigalorti L, Fratta E, Coral S, Tanzarella S, Danielli R, Colizzi F, et al. Intratumor heterogeneity of cancer/testis antigens expression in human cutaneous melanoma is methylation-regulated and functionally reverted by 5-aza-2′-deoxycytidine. Cancer Res. 2004;64:9167–9171. doi: 10.1158/0008-5472.CAN-04-1442. [DOI] [PubMed] [Google Scholar]
- 21.Coral S, Sigalotti L, Colizzi F, Spessotto A, Nardi G, Cortini E, et al. Phenotypic and functional changes of human melanoma xenografts induced by DNA hypomethylation: immunotherapeutic implications. J Cell Physiol. 2006;207:58–66. doi: 10.1002/jcp.20540. [DOI] [PubMed] [Google Scholar]
- 22.Dubovsky JA, McNeel DG. Inducible expression of a prostate cancer-testis antigen, SSX-2, following treatment with a DNA methylation inhibitor. Prostate. 2007;67:1781–1790. doi: 10.1002/pros.20665. [DOI] [PubMed] [Google Scholar]
- 23.Guo ZS, Hong JA, Irvine KR, Chen GA, Spiess PJ, Liu Y, et al. De novo induction of a cancer/testis antigen by 5-aza-2′-deoxycytidine augments adoptive immunotherapy in a murine tumor model [see Comment] Cancer Res. 2006;66:1105–1113. doi: 10.1158/0008-5472.CAN-05-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honda T, Tamura G, Waki T, Kawata S, Terashima M, Nishizuka S, et al. Demethylation of MAGE promoters during gastric cancer progression. Br J Cancer. 2004;90:838–843. doi: 10.1038/sj.bjc.6601600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Natsume A, Wakabayashi T, Tsujimura K, Shimato S, Ito M, Kuzushima K, et al. The DNA demethylating agent 5-aza-2′-deoxycytidine activates NY-ESO-1 antigenicity in orthotopic human glioma. Int J Cancer. 2008;122:2542–2553. doi: 10.1002/ijc.23407. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Zhang J, Zhang Y, Lim SH. SPAN-Xb expression in myeloma cells is dependent on promoter hypomethylation and can be upregulated pharmacologically. Int J Cancer. 2006;118:1436–1444. doi: 10.1002/ijc.21499. [DOI] [PubMed] [Google Scholar]
- 27.Goodyear OC, Agathanggelou A, Ryan G, Novitsky-Basso I, Stankovic T, Moss P, et al. The epigenetic therapies azacitidine and sodium valproatc augment immune responses to the MAGE cancer testis antigen in acute myeloid leukemia and myeloma. ASH Annual Meeting Abstracts. 2009;114:823–824. [Google Scholar]
- 28.Issa J-PJ, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res. 2009;15:3938–3946. doi: 10.1158/1078-0432.CCR-08-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villar-Garea A, Esteller M. Histone deacetylase inhibitors: understanding a new wave of anticancer agents. Int J Cancer. 2004;112:171–178. doi: 10.1002/ijc.20372. [DOI] [PubMed] [Google Scholar]
- 30.Acharya MR, Sparreboom A, Venitz J, Figg WD. Rational development of histone deacetylase inhibitors as anticancer agents: a review. Mol Pharmacol. 2005;68:917–932. doi: 10.1124/mol.105.014167. [DOI] [PubMed] [Google Scholar]
- 31.Szmania S, Gnjatic S, Tricot G, Stone K, Zhan F, Moreno A, et al. Immunization with a recombinant MAGE-A3 protein after high-dose therapy for myeloma. J Immunother. 2007;30:847–854. doi: 10.1097/CJI.0b013e318158fcff. [DOI] [PubMed] [Google Scholar]
- 32.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006;108:2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhan F, Hardin J, Kordsmeier B, Bumm K, Zheng M, Tian E, et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99:1745–1757. doi: 10.1182/blood.v99.5.1745. [DOI] [PubMed] [Google Scholar]
- 34.Shaughnessy J, Haessler J, Zeldis J, Huang Y, Zhan F, Sawyer J, et al. An update on the role of thalidomide (THAL) in total therapy 2 (TT2) for newly diagnosed patients with multiple myeloma (MM): analysis of subgroups defined by standard prognostic factors (SPF) and gene expression profiling (GEP) derived subgroups. Blood. 2006;108:968a. [Google Scholar]
- 35.Barlogie B, Tricot G, Anaissie E, Shaughnessy J, Rasmussen E, van Rhee F, et al. Thalidomide and hematopoietic-cell transplantation for multiple myeloma. N Engl J Med. 2006;354:1021–1030. doi: 10.1056/NEJMoa053583. [DOI] [PubMed] [Google Scholar]
- 36.Barlogie B, Anaissie E, van Rhee F, Haessler J, Hollmig K, Pineda-Roman M, et al. Incorporating bortezomib into upfront treatment for multiple myeloma: early results of total therapy 3. Br J Haematol. 2007;138:176–185. doi: 10.1111/j.1365-2141.2007.06639.x. [DOI] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Gupta S, Zhan F, Lin P, Droogenbroeck J, Batchu R, Nollet F, et al. NY-ESO-1 and MAGE-A3 are highly expressed in myeloma patients with abnormal cytogenetics and/or relapse. Hematol J. 2003;4:S267–S268. [Google Scholar]
- 39.Goodyear O, Piper K, Khan N, Starczynski J, Mahendra P, Pratt G, et al. CD8+ T cells specific for cancer germline gene antigens are found in many patients with multiple myeloma, and their frequency correlates with disease burden. Blood. 2005;106:4217–4224. doi: 10.1182/blood-2005-02-0563. [DOI] [PubMed] [Google Scholar]
- 40.Atanackovic D, Arfsen J, Cao Y, Gnjatic S, Schneiders F, Bartels K, et al. Cancer-testis antigens are commonly expressed in multiple myeloma and induce systemic immunity following allogeneic stem cell transplantation. Blood. 2007;109:1103–1112. doi: 10.1182/blood-2006-04-014480. [DOI] [PubMed] [Google Scholar]
- 41.Cho H, Ely S, Austin W, Niesvizky R, Pearse R, Coleman M, et al. Differential expression of type I MAGE in new and relapsed multiple myeloma: evidence for association with proliferation and progression of disease. Blood. 2006;108:970a. [Google Scholar]
- 42.Kasuga C, Nakahara Y, Ueda S, Hawkins C, Taylor MD, Smith CA, et al. Expression of MAGE and GAGE genes in medulloblastoma and modulation of resistance to chemo therapy. Laboratory investigation. J Neurosurg Pediatrics. 2008;1:305–313. doi: 10.3171/PED/2008/1/4/305. [DOI] [PubMed] [Google Scholar]
- 43.Barker PA, Salehi A. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J Neurosci Res. 2002;67:705–712. doi: 10.1002/jnr.10160. [DOI] [PubMed] [Google Scholar]
- 44.Monte M, Simonatto M, Peche LY, Bublik DR, Gobessi S, Pierotti MA, et al. MAGE-A tumor antigens target p53 trans-activation function through histone deacetylase recruitment and confer resistance to chemotherapeutic agents. Proc Natl Acad Sci USA. 2006;103:11160–11165. doi: 10.1073/pnas.0510834103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duan Z, Duan Y, Lamendola DE, Yusuf RZ, Naeem R, Penson RT, et al. Overexpression of MAGE/GAGE genes in paclitaxel/doxorubicin-resistant human cancer cell lines. Clin Cancer Res. 2003;9:2778–2785. [PubMed] [Google Scholar]
- 46.Yang B, O’Herrin S, Wu J, Reagan-Shaw S, Ma Y, Nihal M, et al. Select cancer testes antigens of the MAGE-A, -B, and -C families are expressed in mast cell lines and promote cell viability in vitro and in vivo. J Invest Dermatol. 2007;127:267–275. doi: 10.1038/sj.jid.5700548. [DOI] [PubMed] [Google Scholar]
- 47.Yang B, O’Herrin SM, Wu J, Reagan-Shaw S, Ma Y, Bhat KM, et al. MAGE-A, mMage-b, and MAGE-C proteins form complexes with KAP1 and suppress p53-dependent apopto sis in MAGE-positive cell lines. Cancer Res. 2007;67:9954–9962. doi: 10.1158/0008-5472.CAN-07-1478. [DOI] [PubMed] [Google Scholar]
- 48.Atanackovic D, Hildebrandt Y, Jadczak A, Cao Y, Luetkens T, Meyer S, et al. Cancer-testis antigens MAGE-C 1/CT7 and MAGE-A3 promote the survival of multiple myeloma cells. Haematologica. 2010;95:785–793. doi: 10.3324/haematol.2009.014464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atanackovic D, Altorki NK, Cao Y, Ritter E, Ferrara CA, Ritter G, et al. Booster vaccination of cancer patients with MAGE-A3 protein reveals long-term immunological memory or tolerance depending on priming. Proc Natl Acad Sci USA. 2008;105:1650–1655. doi: 10.1073/pnas.0707140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta S, Pei L, Droogenbroeck J, Szmania S, Yaccoby S, Batchu RB, et al. Intra-and intertumoral variation in the expression of cancer testis antigens, MAGE-3 and NY-ESO-1 in multiple myeloma. Blood. 2002;100:603a. [Google Scholar]
- 51.Jungbluth AA, Stockert E, Chen YT, Kolb D, Iversen K, Coplan K, et al. Monoclonal antibody MA454 reveals a heterogeneous expression pattern of MAGE-1 antigen in forma lin-fixed paraffin embedded lung tumours. Br J Cancer. 2000;83:493–497. doi: 10.1054/bjoc.2000.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akcakanat A, Kanda T, Tanabe T, Komukai S, Yajima K, Nakagawa S, et al. Heterogeneous expression of GAGE, NY-ESO-1, MAGH-A and SSX proteins in esophageal cancer: implications for immunotherapy. Int J Cancer. 2006;118:123–128. doi: 10.1002/ijc.21219. [DOI] [PubMed] [Google Scholar]
- 53.Dhodapkar MV, Osman K, Teruya-Feldstein J, Filippa D, Hedvat CV, Iversen K, et al. Expression of cancer/testis (CT) antigens MAGE-A 1, MAGE-A3, MAGE-A4, CT-7, and NY-ESO-1 in malignant gammopathies is heterogeneous and correlates with site, stage and risk status of disease. Cancer Immun. 2003;3:9. [PubMed] [Google Scholar]
- 54.Kufer P, Zippelius A, Lutterbuse R, Mecklenburg I, Enzmann T, Montag A, et al. Heterogeneous expression of MAGE-A genes in occult disseminated tumor cells: a novel multimarker reverse transcription-polymerase chain reaction for diagnosis of micrometastatic disease. Cancer Res. 2002;62:251–261. [PubMed] [Google Scholar]
- 55.Serrano A, Tanzarella S, Lionello I, Mendez R, Traversari C, Ruiz-Cabello F, et al. Expression of HLA class I antigens and restoration of antigen-specific CTL response in melanoma cells following 5-aza-2′-deoxycytidine treatment. Int J Cancer. 2001;94:243–251. doi: 10.1002/ijc.1452. [DOI] [PubMed] [Google Scholar]
- 56.Hirano N, Butler MO, Xia Z, Berezovskaya A, Murray AP, Ansen S, et al. Efficient presentation of naturally processed HLA class I peptides by artificial antigen-presenting cells for the generation of effective antitumor responses. Clin Cancer Res. 2006;12:2967–2975. doi: 10.1158/1078-0432.CCR-05-2791. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.