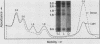Abstract
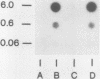
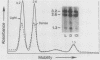
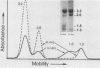
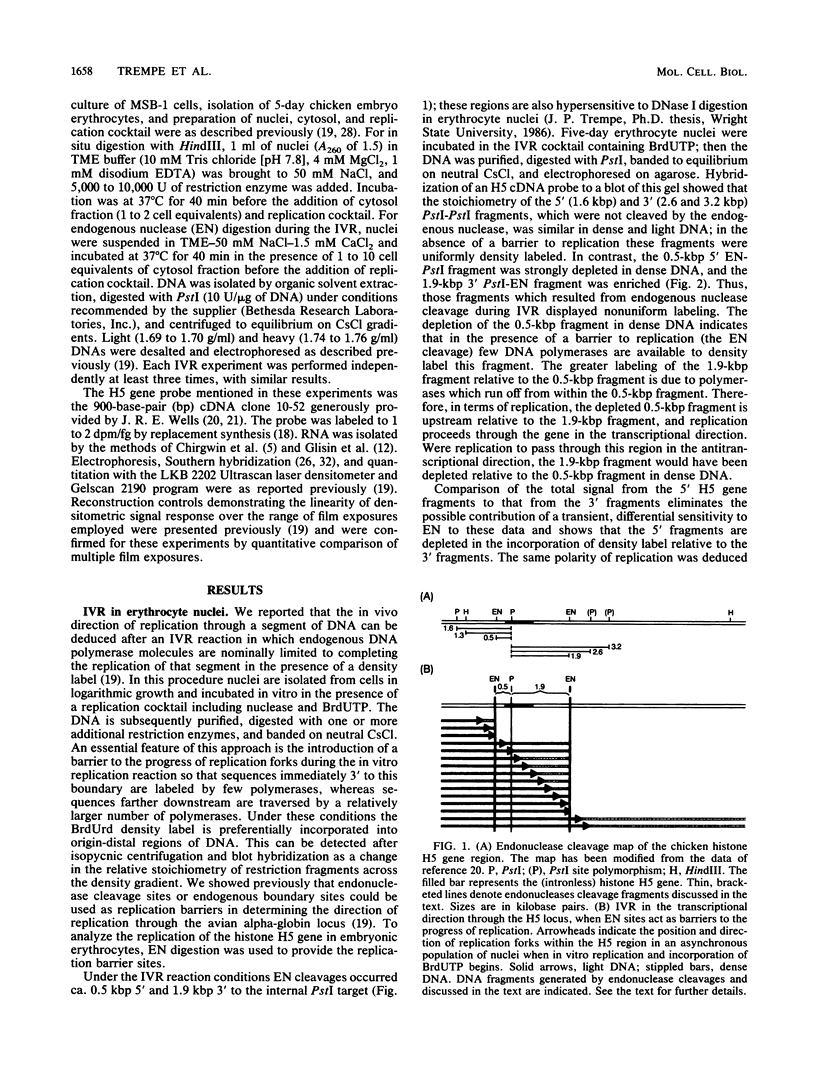
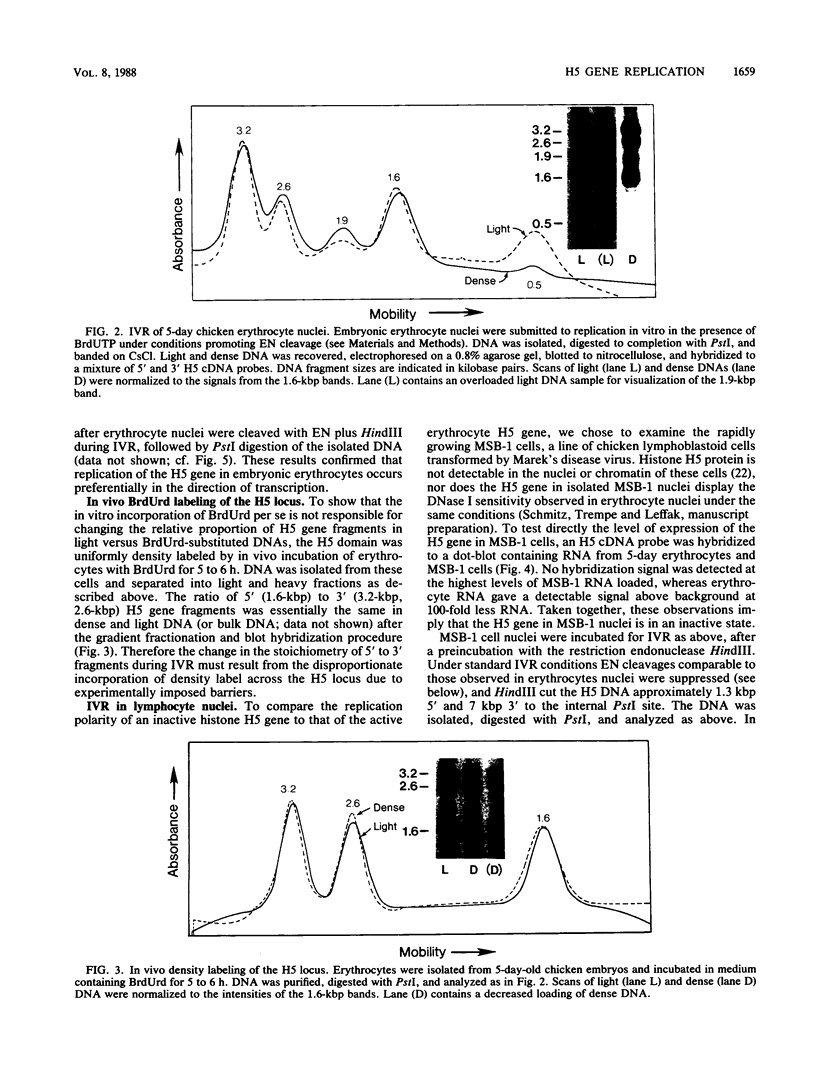
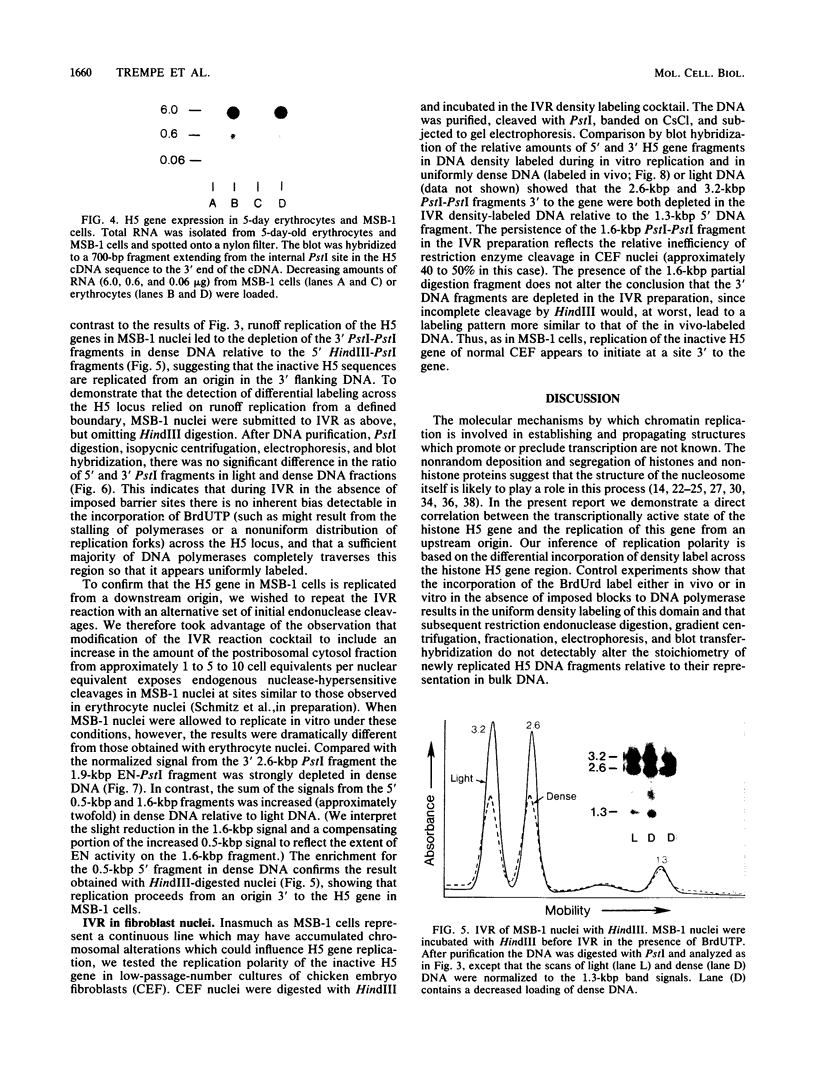
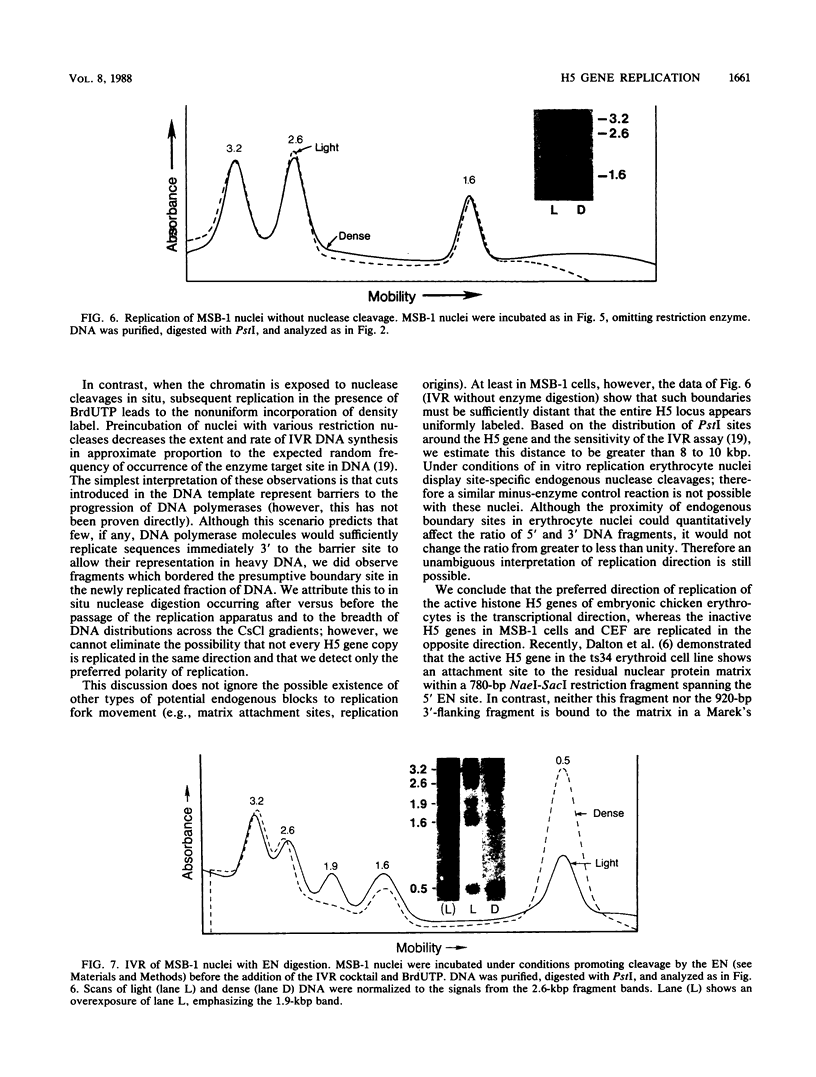
We used an in vitro nuclear runoff replication assay to analyze the direction of replication of the active and inactive histone H5 genes in avian cells. In embryonic erythrocytes the transcribed histone H5 gene displayed sensitivity to endogenous nuclease cleavage. In contrast, this gene was insensitive to endogenous nuclease digestion under the same conditions in nuclei of the lymphoblastoid cell line MSB-1, and histone H5 gene transcripts were not detectable by dot-blot analysis of MSB-1 cell RNA. When nuclei were isolated from embryonic erythrocytes and incubated with bromodeoxyuridine triphosphate, runoff replication from endogenous nuclease cleavage sites led to a relative enrichment for fragments near the 3' end of the histone H5 gene in the density-labeled DNA. In nuclei of MSB-1 cells or chicken embryo fibroblasts, however, runoff replication from restriction enzyme-cut sites (or induced endogenous nuclease-cut sites in MSB-1 nuclei) led to a relative enrichment for fragments near the 5' end of the H5 gene in dense DNA. Based on the enhanced incorporation of bromodeoxyuridine into origin-distal regions of DNA during the in vitro runoff replication assay, we conclude that the active histone H5 gene in embryonic erythrocytes is preferentially replicated in the transcriptional direction from an origin in the 5'-flanking DNA, whereas its inactive counterparts in MSB-1 cells and chicken embryo fibroblasts are preferentially replicated in the opposite direction.
Full text
PDF
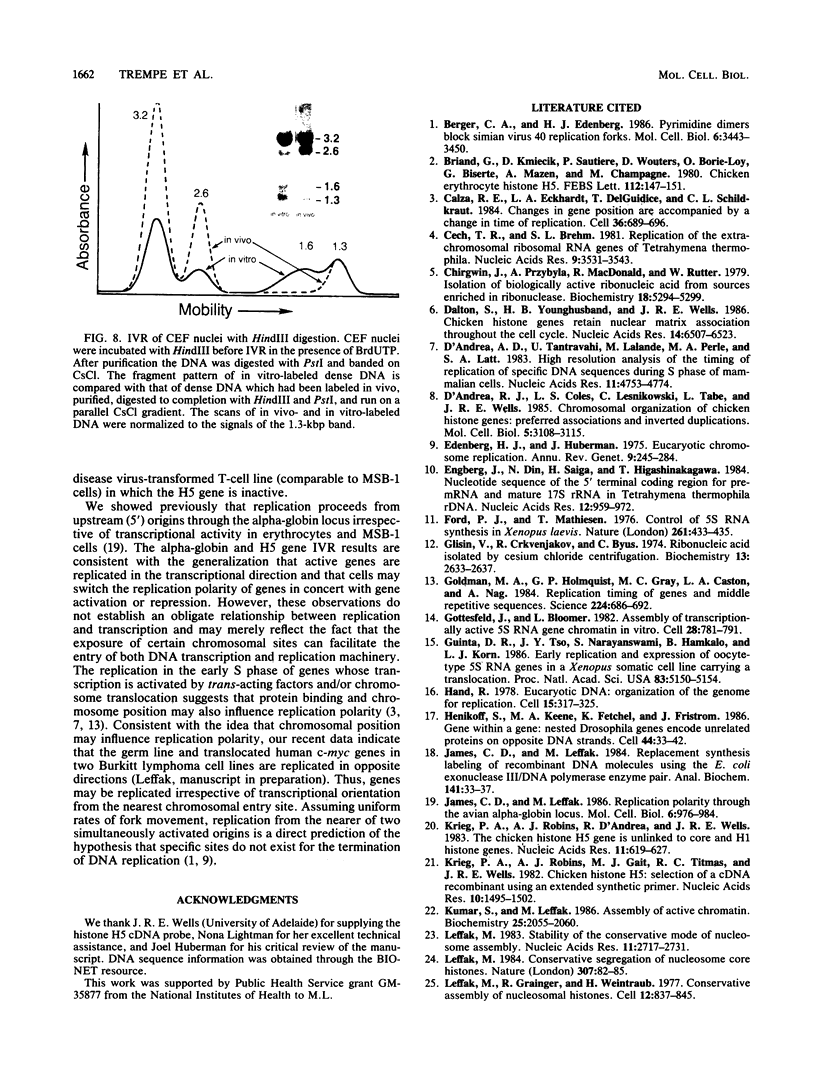

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berger C. A., Edenberg H. J. Pyrimidine dimers block simian virus 40 replication forks. Mol Cell Biol. 1986 Oct;6(10):3443–3450. doi: 10.1128/mcb.6.10.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briand G., Kmiecik D., Sautiere P., Wouters D., Borie-Loy O., Biserte G., Mazen A., Champagne M. Chicken erythrocyte histone H5. IV. Sequence of the carboxy-termined half of the molecule (96 residues) and complete sequence. FEBS Lett. 1980 Apr 7;112(2):147–151. doi: 10.1016/0014-5793(80)80167-0. [DOI] [PubMed] [Google Scholar]
- Calza R. E., Eckhardt L. A., DelGiudice T., Schildkraut C. L. Changes in gene position are accompanied by a change in time of replication. Cell. 1984 Mar;36(3):689–696. doi: 10.1016/0092-8674(84)90349-0. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Brehm S. L. Replication of the extrachromosomal ribosomal RNA genes of Tetrahymena thermophilia. Nucleic Acids Res. 1981 Jul 24;9(14):3531–3543. doi: 10.1093/nar/9.14.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- D'Andrea A. D., Tantravahi U., Lalande M., Perle M. A., Latt S. A. High resolution analysis of the timing of replication of specific DNA sequences during S phase of mammalian cells. Nucleic Acids Res. 1983 Jul 25;11(14):4753–4774. doi: 10.1093/nar/11.14.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea R. J., Coles L. S., Lesnikowski C., Tabe L., Wells J. R. Chromosomal organization of chicken histone genes: preferred associations and inverted duplications. Mol Cell Biol. 1985 Nov;5(11):3108–3115. doi: 10.1128/mcb.5.11.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton S., Younghusband H. B., Wells J. R. Chicken histone genes retain nuclear matrix association throughout the cell cycle. Nucleic Acids Res. 1986 Aug 26;14(16):6507–6523. doi: 10.1093/nar/14.16.6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg H. J., Huberman J. A. Eukaryotic chromosome replication. Annu Rev Genet. 1975;9:245–284. doi: 10.1146/annurev.ge.09.120175.001333. [DOI] [PubMed] [Google Scholar]
- Engberg J., Din N., Saiga H., Higashinakagawa T. Nucleotide sequence of the 5'-terminal coding region for pre-rRNA and mature 17S rRNA in Tetrahymena thermophila rDNA. Nucleic Acids Res. 1984 Jan 25;12(2):959–972. doi: 10.1093/nar/12.2.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford P. J., Mathieson T. Control of 5S RNA synthesis in Xenopus laevis. Nature. 1976 Jun 3;261(5559):433–435. doi: 10.1038/261433a0. [DOI] [PubMed] [Google Scholar]
- Glisin V., Crkvenjakov R., Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974 Jun 4;13(12):2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Goldman M. A., Holmquist G. P., Gray M. C., Caston L. A., Nag A. Replication timing of genes and middle repetitive sequences. Science. 1984 May 18;224(4650):686–692. doi: 10.1126/science.6719109. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J., Bloomer L. S. Assembly of transcriptionally active 5S RNA gene chromatin in vitro. Cell. 1982 Apr;28(4):781–791. doi: 10.1016/0092-8674(82)90057-5. [DOI] [PubMed] [Google Scholar]
- Guinta D. R., Tso J. Y., Narayanswami S., Hamkalo B. A., Korn L. J. Early replication and expression of oocyte-type 5S RNA genes in a Xenopus somatic cell line carrying a translocation. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5150–5154. doi: 10.1073/pnas.83.14.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand R. Eucaryotic DNA: organization of the genome for replication. Cell. 1978 Oct;15(2):317–325. doi: 10.1016/0092-8674(78)90001-6. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Keene M. A., Fechtel K., Fristrom J. W. Gene within a gene: nested Drosophila genes encode unrelated proteins on opposite DNA strands. Cell. 1986 Jan 17;44(1):33–42. doi: 10.1016/0092-8674(86)90482-4. [DOI] [PubMed] [Google Scholar]
- James C. D., Leffak I. M. Replacement synthesis labeling of DNA molecules in vitro using the Escherichia coli exonuclease III/DNA polymerase I enzyme pair. Anal Biochem. 1984 Aug 15;141(1):33–37. doi: 10.1016/0003-2697(84)90421-4. [DOI] [PubMed] [Google Scholar]
- James C. D., Leffak M. Polarity of DNA replication through the avian alpha-globin locus. Mol Cell Biol. 1986 Apr;6(4):976–984. doi: 10.1128/mcb.6.4.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A., Robins A. J., D'Andrea R., Wells J. R. The chicken H5 gene is unlinked to core and H1 histone genes. Nucleic Acids Res. 1983 Feb 11;11(3):619–627. doi: 10.1093/nar/11.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A., Robins A. J., Gait M. J., Titmas R. C., Wells J. R. Chicken histone H5: selection of a cDNA recombinant using an extended synthetic primer. Nucleic Acids Res. 1982 Mar 11;10(5):1495–1502. doi: 10.1093/nar/10.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Leffak M. Assembly of active chromatin. Biochemistry. 1986 Apr 22;25(8):2055–2060. doi: 10.1021/bi00356a033. [DOI] [PubMed] [Google Scholar]
- Leffak I. M. Conservative segregation of nucleosome core histones. Nature. 1984 Jan 5;307(5946):82–85. doi: 10.1038/307082a0. [DOI] [PubMed] [Google Scholar]
- Leffak I. M., Grainger R., Weintraub H. Conservative assembly and segregation of nucleosomal histones. Cell. 1977 Nov;12(3):837–845. doi: 10.1016/0092-8674(77)90282-3. [DOI] [PubMed] [Google Scholar]
- Leffak I. M. Stability of the conservative mode of nucleosome assembly. Nucleic Acids Res. 1983 May 11;11(9):2717–2732. doi: 10.1093/nar/11.9.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffak M., Trempe J. P. Histone H1 and HMG 14/17 are deposited nonrandomly in the nucleus. Nucleic Acids Res. 1985 Jul 11;13(13):4853–4869. doi: 10.1093/nar/13.13.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser T., Thacher T., Rechsteiner M. Arginine-rich histones do not exchange between human and mouse chromosomes in hybrid cells. Cell. 1980 Apr;19(4):993–1003. doi: 10.1016/0092-8674(80)90090-2. [DOI] [PubMed] [Google Scholar]
- Muller M. T., Kajiwara K., Mueller G. C. Role of cytosol proteins in DNA chain growth and chromatin replication in Friend erythroleukemia cell nuclei. Biochim Biophys Acta. 1981 May 29;653(3):391–407. doi: 10.1016/0005-2787(81)90195-7. [DOI] [PubMed] [Google Scholar]
- Niles E. G., Sutiphong J., Haque S. Structure of the Tetrahymena pyriformis rRNA gene. Nucleotide sequence of the transcription initiation region. J Biol Chem. 1981 Dec 25;256(24):12849–12856. [PubMed] [Google Scholar]
- Prior C. P., Cantor C. R., Johnson E. M., Allfrey V. G. Incorporation of exogenous pyrene-labeled histone into Physarum chromatin: a system for studying changes in nucleosomes assembled in vivo. Cell. 1980 Jul;20(3):597–608. doi: 10.1016/0092-8674(80)90306-2. [DOI] [PubMed] [Google Scholar]
- Smithies O. The control of globin and other eukaryotic genes. J Cell Physiol Suppl. 1982;1:137–143. doi: 10.1002/jcp.1041130421. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Truett M. A., Gall J. G. The replication of ribosomal DNA in the macronucleus of Tetrahymena. Chromosoma. 1977 Dec 6;64(4):295–303. doi: 10.1007/BF00294937. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Flint S. J., Leffak I. M., Groudine M., Grainger R. M. The generation and propagation of variegated chromosome structures. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):401–407. doi: 10.1101/sqb.1978.042.01.042. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Wormington W. M., Schlissel M., Brown D. D. Developmental regulation of Xenopus 5S RNA genes. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):879–884. doi: 10.1101/sqb.1983.047.01.101. [DOI] [PubMed] [Google Scholar]