Abstract
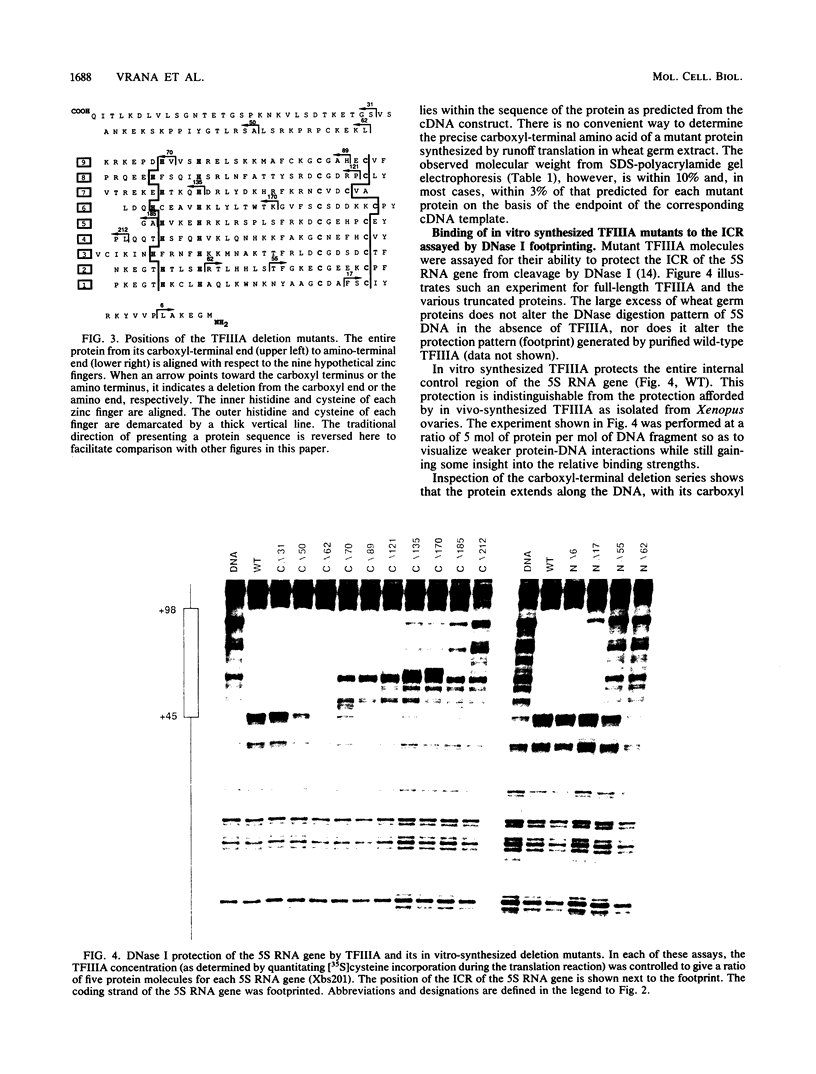
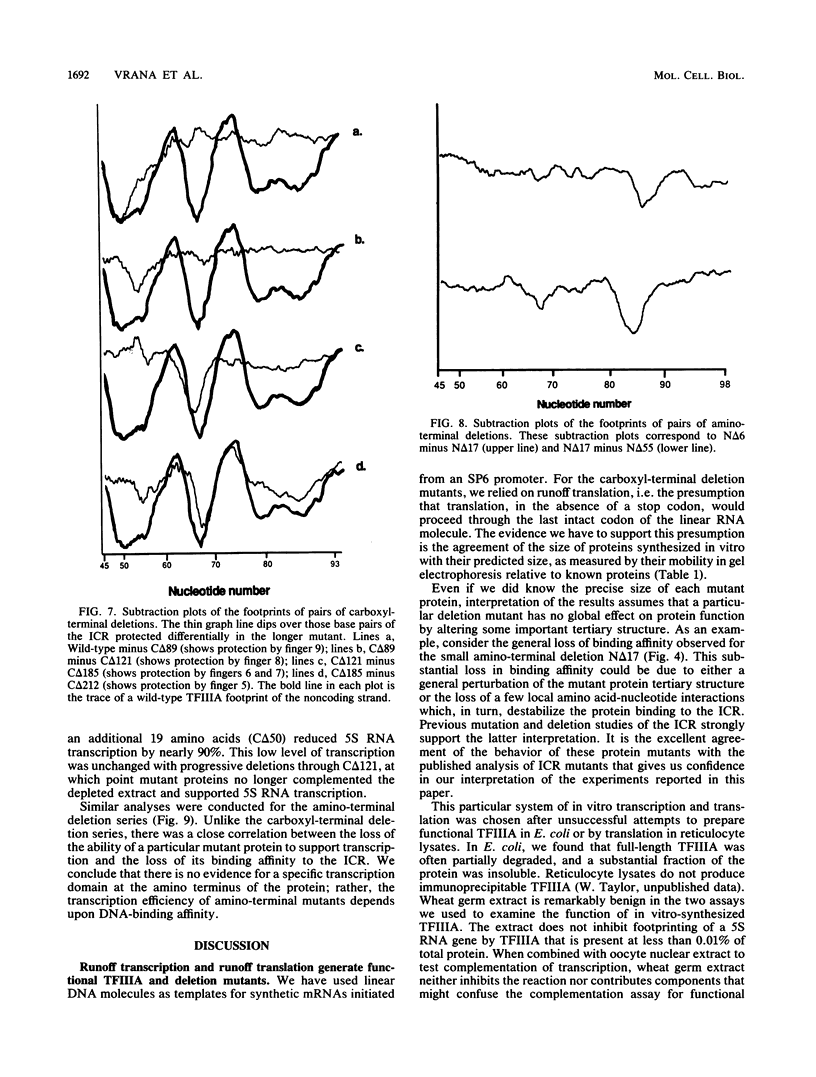
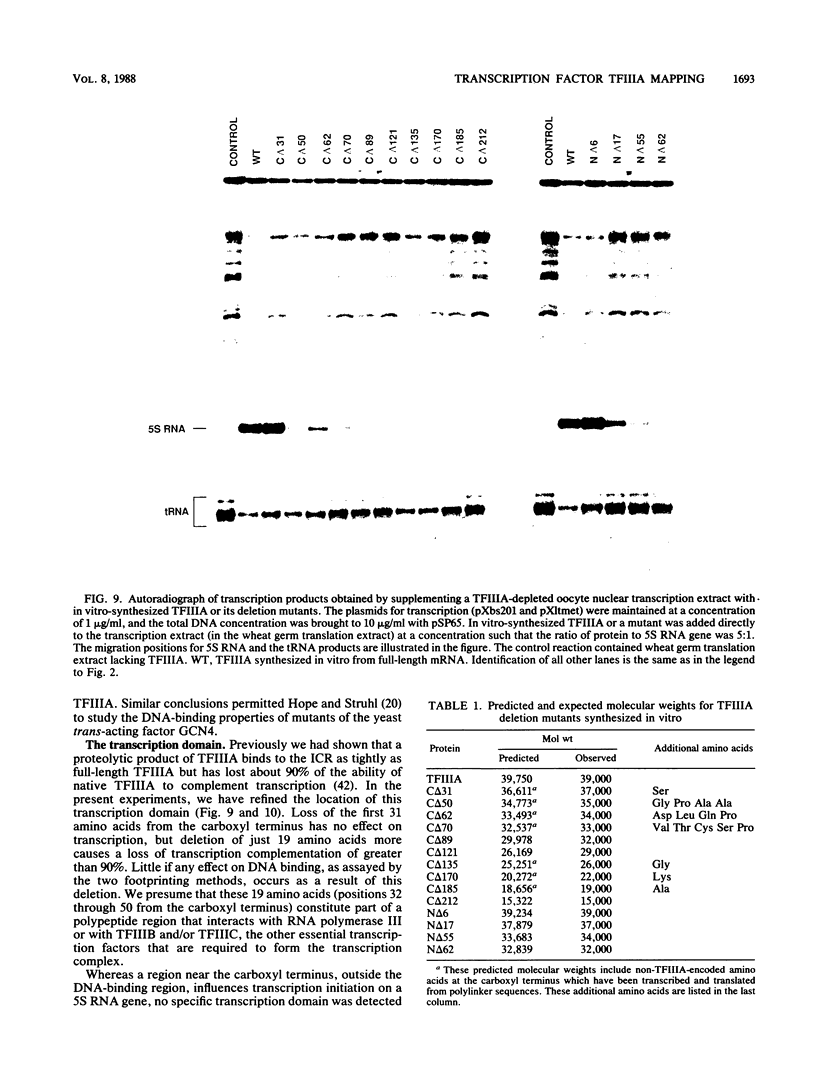
Functional deletion mutants of the trans-acting factor TFIIIA, truncated at both ends of the molecule, have been expressed by in vitro transcription of a cDNA clone and subsequent cell-free translation of the synthetic mRNAs. A region of TFIIIA 19 amino acids or less, near the carboxyl terminus, is critical for maximal transcription and lies outside the DNA-binding domain. The elongated protein can be aligned over the internal control region (ICR) of the Xenopus 5S RNA gene with its carboxyl terminus oriented toward the 5' end of the gene and its amino terminus oriented toward the 3' end of the gene. The nine "zinc fingers" and the linkers that separate them comprise 80% of the protein mass and correspond to the DNA-binding domain of TFIIIA. The zinc fingers near the amino terminus of the protein contribute more to the overall binding energy of the protein to the ICR than do the zinc fingers near the carboxyl end. The most striking feature of TFIIIA is its modular structure. This is demonstrated by the fact that each zinc finger binds to just one of three short nucleotide sequences within the ICR.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. E., Ptashne M., Harrison S. C. Structure of the repressor-operator complex of bacteriophage 434. 1987 Apr 30-May 6Nature. 326(6116):846–852. doi: 10.1038/326846a0. [DOI] [PubMed] [Google Scholar]
- Bieker J. J., Roeder R. G. Physical properties and DNA-binding stoichiometry of a 5 S gene-specific transcription factor. J Biol Chem. 1984 May 25;259(10):6158–6164. [PubMed] [Google Scholar]
- Birkenmeier E. H., Brown D. D., Jordan E. A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell. 1978 Nov;15(3):1077–1086. doi: 10.1016/0092-8674(78)90291-x. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Wormington W. M., Brown D. D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982 Feb;28(2):413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- Brenowitz M., Senear D. F., Shea M. A., Ackers G. K. "Footprint" titrations yield valid thermodynamic isotherms. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8462–8466. doi: 10.1073/pnas.83.22.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. D., Schlissel M. S. A positive transcription factor controls the differential expression of two 5S RNA genes. Cell. 1985 Oct;42(3):759–767. doi: 10.1016/0092-8674(85)90272-7. [DOI] [PubMed] [Google Scholar]
- Brown D. D. The role of stable complexes that repress and activate eucaryotic genes. Cell. 1984 Jun;37(2):359–365. doi: 10.1016/0092-8674(84)90366-0. [DOI] [PubMed] [Google Scholar]
- Brown R. S., Sander C., Argos P. The primary structure of transcription factor TFIIIA has 12 consecutive repeats. FEBS Lett. 1985 Jul 8;186(2):271–274. doi: 10.1016/0014-5793(85)80723-7. [DOI] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Fairall L., Rhodes D., Klug A. Mapping of the sites of protection on a 5 S RNA gene by the Xenopus transcription factor IIIA. A model for the interaction. J Mol Biol. 1986 Dec 5;192(3):577–591. doi: 10.1016/0022-2836(86)90278-0. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguère V., Hollenberg S. M., Rosenfeld M. G., Evans R. M. Functional domains of the human glucocorticoid receptor. Cell. 1986 Aug 29;46(5):645–652. doi: 10.1016/0092-8674(86)90339-9. [DOI] [PubMed] [Google Scholar]
- Ginsberg A. M., King B. O., Roeder R. G. Xenopus 5S gene transcription factor, TFIIIA: characterization of a cDNA clone and measurement of RNA levels throughout development. Cell. 1984 Dec;39(3 Pt 2):479–489. doi: 10.1016/0092-8674(84)90455-0. [DOI] [PubMed] [Google Scholar]
- Green S., Chambon P. Oestradiol induction of a glucocorticoid-responsive gene by a chimaeric receptor. Nature. 1987 Jan 1;325(6099):75–78. doi: 10.1038/325075a0. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanas J. S., Hazuda D. J., Bogenhagen D. F., Wu F. Y., Wu C. W. Xenopus transcription factor A requires zinc for binding to the 5 S RNA gene. J Biol Chem. 1983 Dec 10;258(23):14120–14125. [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984 Sep 25;12(18):7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar A. B., Martin P. L., Roeder R. G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 Nov 18;222(4625):740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- Losa R., Brown D. D. A bacteriophage RNA polymerase transcribes in vitro through a nucleosome core without displacing it. Cell. 1987 Aug 28;50(5):801–808. doi: 10.1016/0092-8674(87)90338-2. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConkey G. A., Bogenhagen D. F. Transition mutations within the Xenopus borealis somatic 5S RNA gene can have independent effects on transcription and TFIIIA binding. Mol Cell Biol. 1987 Jan;7(1):486–494. doi: 10.1128/mcb.7.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S., Tjian R. Transcriptional selectivity of viral genes in mammalian cells. Cell. 1986 Sep 12;46(6):795–805. doi: 10.1016/0092-8674(86)90061-9. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R., Brown D. D. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieler T., Appel B., Oei S. L., Mentzel H., Erdmann V. A. Point mutational analysis of the Xenopus laevis 5S gene promoter. EMBO J. 1985 Jul;4(7):1847–1853. doi: 10.1002/j.1460-2075.1985.tb03859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieler T., Hamm J., Roeder R. G. The 5S gene internal control region is composed of three distinct sequence elements, organized as two functional domains with variable spacing. Cell. 1987 Jan 16;48(1):91–100. doi: 10.1016/0092-8674(87)90359-x. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. An underlying repeat in some transcriptional control sequences corresponding to half a double helical turn of DNA. Cell. 1986 Jul 4;46(1):123–132. doi: 10.1016/0092-8674(86)90866-4. [DOI] [PubMed] [Google Scholar]
- Rusconi S., Yamamoto K. R. Functional dissection of the hormone and DNA binding activities of the glucocorticoid receptor. EMBO J. 1987 May;6(5):1309–1315. doi: 10.1002/j.1460-2075.1987.tb02369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryrie I. J., Gallagher A. The yeast mitochondrial ATPase complex. Subunit composition and evidence for a latent protease contaminant. Biochim Biophys Acta. 1979 Jan 11;545(1):1–14. doi: 10.1016/0005-2728(79)90108-7. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D., Engelke D., Ng S. Y., Shastry B. S., Roeder R. G. The binding of a transcription factor to deletion mutants of a 5S ribosomal RNA gene. Cell. 1981 Mar;23(3):665–669. doi: 10.1016/0092-8674(81)90429-3. [DOI] [PubMed] [Google Scholar]
- Schuh R., Aicher W., Gaul U., Côté S., Preiss A., Maier D., Seifert E., Nauber U., Schröder C., Kemler R. A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Krüppel, a Drosophila segmentation gene. Cell. 1986 Dec 26;47(6):1025–1032. doi: 10.1016/0092-8674(86)90817-2. [DOI] [PubMed] [Google Scholar]
- Segall J., Matsui T., Roeder R. G. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980 Dec 25;255(24):11986–11991. [PubMed] [Google Scholar]
- Smith D. R., Jackson I. J., Brown D. D. Domains of the positive transcription factor specific for the Xenopus 5S RNA gene. Cell. 1984 Jun;37(2):645–652. doi: 10.1016/0092-8674(84)90396-9. [DOI] [PubMed] [Google Scholar]
- Taylor W., Jackson I. J., Siegel N., Kumar A., Brown D. D. The developmental expression of the gene for TFIIIA in Xenopus laevis. Nucleic Acids Res. 1986 Aug 11;14(15):6185–6195. doi: 10.1093/nar/14.15.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso J. Y., Van Den Berg D. J., Korn L. J. Structure of the gene for Xenopus transcription factor TFIIIA. Nucleic Acids Res. 1986 Mar 11;14(5):2187–2200. doi: 10.1093/nar/14.5.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A., Churchill M. E., Kam L. Hydroxyl radical footprinting: a high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 1987;155:537–558. doi: 10.1016/0076-6879(87)55035-2. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P., Jordan E., Brown D. D. A bacteriophage RNA polymerase transcribes through a Xenopus 5S RNA gene transcription complex without disrupting it. Cell. 1986 Feb 14;44(3):381–389. doi: 10.1016/0092-8674(86)90459-9. [DOI] [PubMed] [Google Scholar]








