Abstract
Identification of essential genes by construction of conditional knockouts with inducible promoters allows the identification of essential genes and creation of conditional growth (CG) mutants that are then available as genetic tools for further studies. We used large-scale transposon delivery of the rhamnose-inducible promoter, PrhaB followed by robotic screening of rhamnose-dependent growth to construct a genomic library of 106 Burkholderia cenocepacia CG mutants. Transposon insertions were found where PrhaB was in the same orientation of widely conserved, well-characterized essential genes as well as genes with no previous records of essentiality in other microorganisms. Using previously reported global gene-expression analyses, we demonstrate that PrhaB can achieve the wide dynamic range of expression levels required for essential genes when the promoter is delivered randomly and mutants with rhamnose-dependent growth are selected. We also show specific detection of the target of an antibiotic, novobiocin, by enhanced sensitivity of the corresponding CG mutant (PrhaB controlling gyrB expression) within the library. Modulation of gene expression to achieve 30–60% of wild-type growth created conditions for specific hypersensitivity demonstrating the value of the CG mutant library for chemogenomic experiments. In summary, CG mutants can be obtained on a large scale by random delivery of a tightly regulated inducible promoter into the bacterial chromosome followed by a simple screening for the CG phenotype, without previous information on gene essentiality.
Keywords: Burkholderia cepacia complex, chemical genetics, essential genome, growth inhibition, inducible gene expression, large scale bacterial genetics
Introduction
The Burkholderia cepacia complex (referred to here as Bcc) is a group of closely related gram-negative bacteria that are widely distributed in natural and man-made environments (Vandamme and Dawyndt 2011). Burkholderia species, including the Bcc, are of great interest because of their large multipartite genomes, their great metabolic versatility, and the wide array of ecological niches they occupy (Sousa et al. 2011). Also of interest is the “dual personality” of the Bcc as these environmental bacteria, which were initially considered harmless, are now known to cause human infections (Chiarini et al. 2006). While strains can be exploited for biocontrol, bioremediation, and plant growth promotion purposes, safety issues arise regarding human infections, as many Bcc strains have emerged as antibiotic-resistant, opportunistic pathogens in patients with cystic fibrosis, chronic granulomatous disease, and other medical conditions associated with a compromised immune system (Mahenthiralingam et al. 2005; Valvano et al. 2005; Loutet and Valvano 2010).
Bacterial genes that are required for growth in rich, undefined media are regarded as essential and hence their encoded products are potential targets of new antibiotics (Brown and Wright 2005). Essential genes have been identified on a genomic scale by high-density transposon knockout mutagenesis (Hutchison et al. 1999; Akerley et al. 2002; Gerdes et al. 2003; Jacobs et al. 2003; Sassetti et al. 2003) or systematic gene-by-gene inactivation (Thanassi et al. 2002; Kang et al. 2004) where genes for which mutants could not be recovered are assumed to be essential. However, identification of essential genes by construction of conditional knockouts with inducible promoters adds the value of obtaining conditional growth (CG) mutants that are then available for further studies (Judson and Mekalanos 2000; DeVito et al. 2002; Forsyth et al. 2002). A number of inducible promoters have been used to express essential genes through the construction of CG mutants (Wong and Akerley 2003; Carroll et al. 2005), with the Escherichia coli arabinose-inducible promoter (PBAD, ParaB) being one of the best characterized (Guzman et al. 1995; Judson and Mekalanos 2000). A great challenge is, however, to achieve genomic representation of essential genes with conditional mutagenesis, probably because of the different range of required expression levels. Promoters with very low uninduced expression levels are necessary to obtain mutants with a CG phenotype. Yet, highly induced expression levels may be necessary for highly expressed essential genes. Promoters that are inducible to such high levels may show uninduced levels of essential gene expression that are tolerable to bacterial growth (Bugrysheva et al. 2011).
The E. coli rhamnose-inducible promoter (PrhaB) is controlled by a cascade of two transcriptional regulators and is more tightly regulated than ParaB (Haldimann et al. 1998). We previously demonstrated that PrhaB is suitable for tightly regulated gene expression in the Bcc clinical isolate B. cenocepacia K56-2, and that essential genes can be identified by transposon-based delivery of PrhaB throughout the bacterial chromosome followed by screening for absence or growth without rhamnose (Cardona et al. 2006).
In this work, we asked whether saturation of a genome with PrhaB allows identification of essential genes at the genomic level with representation of such genes in a library of CG mutants. Using a large-scale mutagenesis approach and robotic screening of more than 200,000 transposon mutants for rhamnose-dependent growth, we constructed a library of CG mutants (CG mutant library) and analyzed the contribution of promoter expression levels and gene redundancy in the identification of essential genes. We demonstrate that screening for CG in one condition identifies CG mutants of similar CG phenotypes, which makes them suitable for chemogenomic experiments.
Experimental Procedures
Bacterial strains and growth conditions
Bacterial strains and plasmids are listed in Table 1, and the identified CG mutants are listed in Table S1. All mutants were made in a B. cenocepacia K56-2 background and were grown in Luria Bertani (LB) media at 37°C, supplemented as required with different concentrations of rhamnose, 100 or 50 μg/mL trimethoprim (Tp) for B. cenocepacia or E. coli, respectively, 50 μg/mL gentamicin and 40 μg/mL kanamycin (Km). All chemicals were purchased from Sigma Chemical Co., St. Louis, MO unless otherwise indicated. To prepare standardized glycerol stocks, overnight cultures were washed twice with LB, adjusted to a final OD600nm of 0.2 in LB 20% glycerol, and aliquoted into polymerase chain reaction (PCR) tubes for storage at −70°C. In assays involving bacterial growth, cultures were diluted to give a theoretical final OD600nm of 0.001, and arranged in 96-well format. Plates were sealed with parafilm and incubated at 37°C with shaking at 200 rpm in a New Brunswick Scientific E24 shaking incubator (Edison, NJ). OD600nm readings were taken using BioTek Synergy 2 plate reader (Winooski, VT).
Table 1.
Bacterial strains and plasmids
| Features | Source | |
|---|---|---|
| Strains | ||
| Burkholderia cenocepacia K56-2 (LMG18863) | ET12 linage, CF isolate | Mahenthiralingam et al. (2000) |
| B. cenocepacia gyrB | Promoter replacement of gyrB PgyrB:: pRB6 | This study |
| Escherichia coli SY327 | araD Δ(lac pro) argE (Am) recA56 Rifr nalA λ pir | Miller and Mekalanos (1988) |
| E. coli DH5α | F−, ϕ 80 lacZΔM15 endA1 recA1 hsdR17(rK−mK+)supE44 thi−1 ΔgyrA96 (ΔlacZYA-argF)U169 relA1 | Invitrogen |
| Plasmids | ||
| pRK2013 | oricolE1, RK2 derivative, Kmr mob+ tra+ | Figurski and Helinski (1979) |
| pRB-rham | pSCrhaboutgfp derivative (Cardona et al. 2006), ori R6K, rhaR rhaS PrhaB e-gfp | This study |
| pSC200 | pGpΩTp derivative (Flannagan et al. 2007), ori R6K, rhaR rhaS PrhaB e-gfp | Ortega et al. (2007) |
| pRB6 | pSC200 containing gyrB 5′end | This study |
Molecular biology techniques
DNA ligase and restriction enzymes (New England Biolabs, Whitby, ON, Canada) were used as recommended by the manufacturers. Escherichia coli SY327 cells were transformed using the Z-competent buffer kit protocol (Zymo Research, Irvine, CA). Conjugation into B. cenocepacia K56-2 was accomplished by triparental mating (Craig et al. 1989) with E. coli DH5α carrying the helper plasmid pRK2013 (Figurski and Helinski 1979). DNA was amplified using a PTC-221 DNA engine (MJ Research, Waltham, MA) or an Eppendorf Mastercycler ep gradient S thermal cycler with either Taq DNA polymerase (Qiagen, Hilden, Germany) or Phusion High-Fidelity PCR Kit (New England Biolabs). Amplification conditions were optimized for each primer pair. PCR products and plasmids were purified using QIAquick PCR Purification Kit (Qiagen) and QIAprep Spin Miniprep Kit (Qiagen), respectively. DNA sequencing was performed by The Center for Applied Genomics (TCAG) at The Hospital for Sick Children, Toronto, Ontario.
Vector constructions
pRB-rham (Fig. 1A) is a derivative of pSCrhaBoutgfp (Cardona et al. 2006), in which the pMB1 origin of replication was replaced with that of the R6K plasmid (ori R6K) to avoid possible reversion to a replicative plasmid in B. cenocepacia. ori R6K-dependent replication has the absolute requirement of the Pir protein (Stalker et al. 1979), and thus plasmids bearing this replication origin do not replicate in bacteria not harboring pir.
Figure 1.
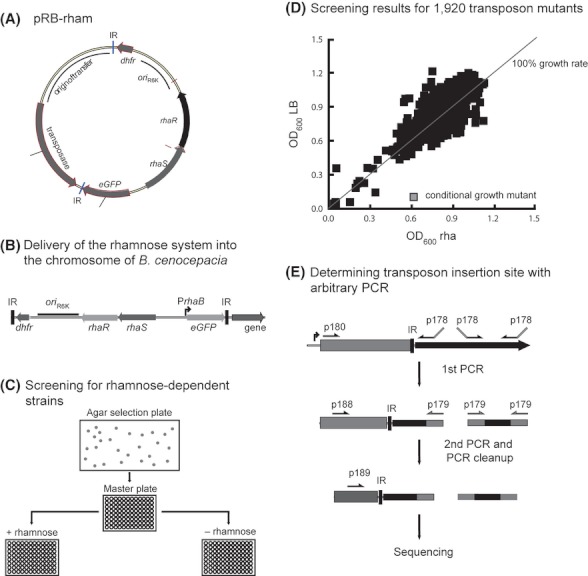
Construction of a Burkholderia cenocepacia CG mutant library. (A) Transposon vector pBR-rham is a derivative of pSCrhaBoutgfp (Cardona et al. 2006). See Experimental Procedures for details on plasmid construction. (B) Trimethoprim resistance provided by the dhfr cassette was used to select for the transconjugants containing an outward-facing rhamnose-inducible promoter PrhaB. (C) The transconjugants were robotically picked into 96- or 384-well master plates before being robotically replicated into 96- or 384-well secondary plates containing LB with and without rhamnose. (D) The OD600nm of the plates were read after 16 h and mutants showing at least 50% less growth in the absence of rhamnose were included in the library. (E) The insertion sites of the mutants were primarily determined using arbitrary primed PCR to preferentially amplify the transposon–genome junction and sequenced using a transposon-specific primer.
To construct pRB-rham, pTnMod-RTp (Dennis and Zylstra 1998) and pSCrhaBoutgfp (Cardona et al. 2006) were digested with SpeI/KpnI and the ori R6K, and dhfr cassette from pTnMod-RTp was ligated to the backbone of pSCrhaBoutgfp. To construct plasmid pRB6, a 300-bp DNA fragment containing the 5′ end of gyrB, flanked by XbaI and NdeI restriction sites, was cloned into pSC200 immediately downstream from the rhamnose-inducible promoter. The resulting plasmid was conjugated into B. cenocepacia K56-2 by triparental mating. Integration of pRB6 and replacement of the gyrB natural promoter was confirmed by colony PCR for the ori R6K and PCR amplification of the insertion interface.
CG mutant library construction
pRB-rham was introduced into B. cenocepacia K56-2 via triparental mating (Craig et al. 1989). The exconjugates were selected for by plating onto 500-cm2 QTrays (Genetix, X6023; San Jose, CA) containing LB agar with 0.2% rhamnose and the appropriate antibiotics and incubating for 48 h at 37°C. The resulting colonies were robotically picked using a Genetix QPix2 XT colony picker into master plates containing liquid LB medium with 0.1% rhamnose and Tp100 and were incubated overnight. While initial picking and replicating was performed in 96-well plates (Greiner Bio-One, 655185; Monroe, NC), the majority of the library was produced in 384-well microplates (Greiner Bio-One, 781186). The master plates were robotically replicated into secondary plates containing LB and LB 0.1% rhamnose and incubated overnight. Bacterial growth was estimated by measuring OD600nm of the cultures using a BioTek Synergy 2 plate reader equipped with a BioTek Bio-Stack automated plate stacker, and the ratio of growth without and with rhamnose was calculated for each mutant. Transposon mutants showing at least a 50% decrease in OD600nm in the absence of rhamnose in comparison to OD600nm in the presence of rhamnose were manually rescreened for growth in LB with or without 0.1% rhamnose. Mutants showing at least a 50% decrease in OD600nm, after 16 h of incubation were stored as glycerol stocks.
Determination of transposon insertion sites and orientations
Transposon insertion sites were identified either by arbitrary primed PCR (Das et al. 2005; Miller-Williams et al. 2006) or by self-cloning as previously described (Dennis and Zylstra 1998). For each clone, we first attempted arbitrary PCR. A 1-μL aliquot of overnight culture was used directly as the template for an initial low-stringency PCR reaction using a transposon-specific primer and a degenerate arbitrary primer, which amplifies the transposon–genome junction as well as other random stretches of DNA. The products of this reaction were used as the template for a second PCR reaction using an inner transposon-specific primer and a primer identical to the tail of the degenerate primer to preferentially amplify the transposon–genome junction (Table S2). The products were purified using a QIAquick PCR Purification Kit (Qiagen) and sequenced using a third transposon-specific primer. Approximately 20% of the PCR products did not return a usable chromatogram. To determine whether the unsuccessful sequencing was due to the presence of multiple insertions, we subjected these clones to Southern blot experiments. All clones showed only one restricted DNA fragment that hybridized with the transposon-complementary probe, demonstrating that the clones with a failed sequencing reaction harbored a single transposon insertion (data not shown). For these clones, insertions were determined successfully using the self-cloning procedure. DNA was digested using NotI or NdeI, and Southern blots for the eGFP on the end of the transposon were performed using an iBlot Gel Transfer System (Invitrogen, Carlsbad, CA) and DIG High-Prime DNA Labeling Kit (Roche, Indianapolis, IN). The location of the insertion site was determined using nucleotide BLAST against the genome of B. cenocepacia J2315 from the Burkholderia Genome Database (Winsor et al. 2008). The distance from the insertion to the start site of the downstream open reading frame for insertions into putatively intergenic regions and the start site of the surrounding open reading frame for insertions within putative genes were also calculated.
Comparisons with essential genes in other bacteria
Essential E. coli genes were obtained from PEC (Profiling of E. coli Chromosome) (Hashimoto et al. 2005), and from Liberati et al. (2006) for Pseudomonas aeruginosa. Orthologs of B. cenocepacia J2315 in E. coli MG1655 and P. aeruginosa PAO1 were found using reciprocal best-hit protein BLAST (Altschul et al. 1990) on the annotated open reading frames with an Expect cutoff of 10. Escherichia coli microarray data were obtained from E-MEXP-3461 (Prieto et al. 2011), RNA-seq data from (Yi et al. 2011), and B. cenocepacia J2315 microarray data from (Bazzini et al. 2011). The expression levels of essential and nonessential genes were compared using the Mann–Whitney sum-rank test (Lehmann 1998) assuming that the test-statistic U is normally distributed given the large sample size. This statistical test assumes neither that the expression is normally distributed across genes nor that there is a linear relationship between the values and the underlying mRNA levels.
Functional characterization of genes in putatively essential operons
As the rhamnose-inducible promoter will control the expression of all downstream genes in the same transcriptional unit, all downstream genes in the same putative operon were included in our analysis. Genes were included if either OperonDB (confidence level of 50 or more) (Pertea et al. 2009) or DOOR (Database of Prokaryotic Operons) (Dam et al. 2007) placed them in the same putative operon.
Enhanced sensitivity assay
To calculate rhamnose concentrations that produced 30–60% of wild-type growth, rhamnose dose–response curves of each mutant were run as follows: mutants were grown for 22 h in LB with a rhamnose gradient of 0–0.16%. The resulting OD600nm readings were then fitted to the Hill equation using GraphPad prism (GraphPad Software, Inc., La Jolla, CA). To develop the enhanced sensitivity assay, all the mutants were grown in 96-well-format plates with 200 μL of LB medium containing rhamnose concentrations required to achieve between 30% and 60% of wild-type growth without the addition of antibiotics. Novobiocin (1 μg/mL) or chloramphenicol (2 μg/mL) was added as required. Plates were incubated for 22 h at 37°C with shaking. For each rhamnose concentration, fold reduction was measured as OD600nm without antibiotic/OD600nm with antibiotic, and mutant sensitivity was defined as log10 of the fold reduction in growth due to the antibiotic.
Results
Building a CG mutant library in B. cenocepacia K56-2
We previously defined essential operons as transcriptional units containing at least one essential gene (Cardona et al. 2006). However, the total number of essential genes and their organization into transcriptional units in the large genome of B. cenocepacia is unknown. To estimate the number of B. cenocepacia essential operons expected to hit to achieve genome coverage, we first analyzed 14 bacterial genomes with experimental data on gene essentiality and compared the number of essential genes versus genome size (Fig. S1). No correlation was found between the size of the essential genome with the overall number of genes with most of the bacterial essential genomes ranging from 300 to 700 genes (Gerdes et al. 2006; Langridge et al. 2009; Christen et al. 2011; Griffin et al. 2011). To estimate the number of operons that may contain essential genes in a model organism, we analyzed the E. coli genome in silico, and based on the 306 experimentally confirmed essential genes and a computational estimate of operons (Zhang et al. 2006), we found 180 E. coli operons that include one or more essential genes (data not shown). Thus, if B. cenocepacia contained 300–700 essential genes as in other bacterial genomes, with a similar distribution to that of E. coli, approximately 200–400 essential operons would be expected.
The construction of the B. cenocepacia CG mutant library is outlined in Figure 1. The previously developed method for delivering PrhaB by transposon mutagenesis and selection by replica plating on LB agar with and without rhamnose (Cardona et al. 2006) was modified to achieve high-throughput levels. After successful transposon mutagenesis and robotic screening in LB liquid media in the presence and absence of rhamnose, transposon mutants with at most 50% growth in the absence of rhamnose were isolated and rescreened before inclusion in the CG library. We reasoned that the permissive 50% cutoff would allow the inclusion of CG mutants with slow growth in the absence of rhamnose due to either low levels of residual expression or remaining essential gene products from growth of the parental culture in rhamnose. This decision also means that the library would include mutants for genes that are important, but not absolutely required for growth. By screening 200,000 transposon mutants with this single condition, 134 CG mutants were isolated and their growth phenotypes confirmed, representing a hit frequency of approximately 1/1500. However, the CG phenotype of 19 of these mutants could not be observed when the original glycerol stocks were further used, leaving 115 CG mutants in the library.
We identified the location and orientation of the delivered PrhaB by arbitrary primed PCR (Das et al. 2005; Miller-Williams et al. 2006) or self-cloning (Dennis and Zylstra 1998) and by aligning the obtained DNA sequences with the genome sequence of the clonally related strain J2315 (Holden et al. 2009). All K56-2 DNA sequences could be matched to equivalent genomic regions in J2315. However, we found that a 57-kb duplication on chromosome 1 of J2315 does not exist in the K56-2 genome. Insertion sites in some mutants were mapped to the 3′ end of lepA1/lepA2, near the middle of the 57-kb duplication on chromosome 1 of J2315. As our system works by usurping the native promoter to control expression of downstream genes, it should be impossible to observe a CG defect from an insertion into only one copy of large identical repeats. After confirming the CG phenotypes in clones that had transposon insertions in the K56-2 putative duplicated region, we verified that the duplication was not present in the genome of K56-2 by PCR amplification of the two duplication-genome interfaces (data not shown). Clones with insertions in this site were the most common ones found with up to 10 clones recovered toward the completion of the screening procedure. Despite the permissive conditions for inclusion into the library the vast majority of the mutants (101 of 115) showed less than 35% of wild-type growth over 22 h in the absence of rhamnose (Fig. 2). Eighty-two mutants in the library showed less than 20% of wild-type growth in the absence of rhamnose, with 42 of these showing less than 5% of wild-type growth over 22 h.
Figure 2.
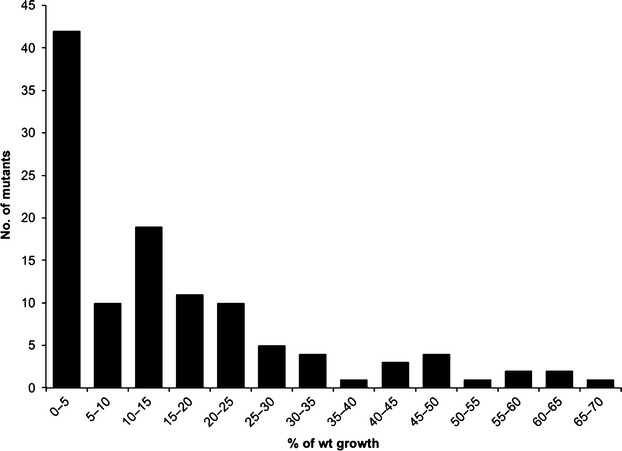
Histogram of growth for 115 CG mutants in the absence of rhamnose. Burkholderia cenocepacia K56-2 (wild-type) and CG mutants were grown in LB without rhamnose and OD600nm was measured after 22 h. Percentage of wild-type growth was defined as the growth of CG mutants relative to that of B. cenocepacia K56-2. Bars represent the total number of CG mutants with percent of wild-type growth within the range indicated by flanking numbers.
Functional characterization of essential operons
Of the 115 CG mutants in the library, 106 were successfully sequenced and had insertion sites in the same orientation as interrupted and/or adjacent downstream genes (Fig. 3, Table S1). As PrhaB controls the expression of downstream genes in the same operon, insertion sites identified 50 unique putative essential operons (Fig. 3). These operons contained 179 genes, which we organized into functional groups using the COG (Cluster of Orthologous Genes) (Tatusov et al. 2003) and GO (Gene Ontology) (Ashburner et al. 2000) annotations from the Burkholderia Genome Database (Winsor et al. 2008). Genes involved in core metabolic functions such as energy production, cell envelope biosynthesis, and DNA replication were overrepresented compared with the entire genome (Fig. 4). Conversely, genes involved in transcription, which are peripheral to growth in the permissive conditions tested, were underrepresented. Similarly, genes involved in carbohydrate metabolism were also underrepresented. This is expected given the range of nutrients available in rich media such as LB. Genes of unknown function were also underrepresented, but still comprised the second largest category after energy production, indicating the large number of uncharacterized genes involved in core B. cenocepacia processes.
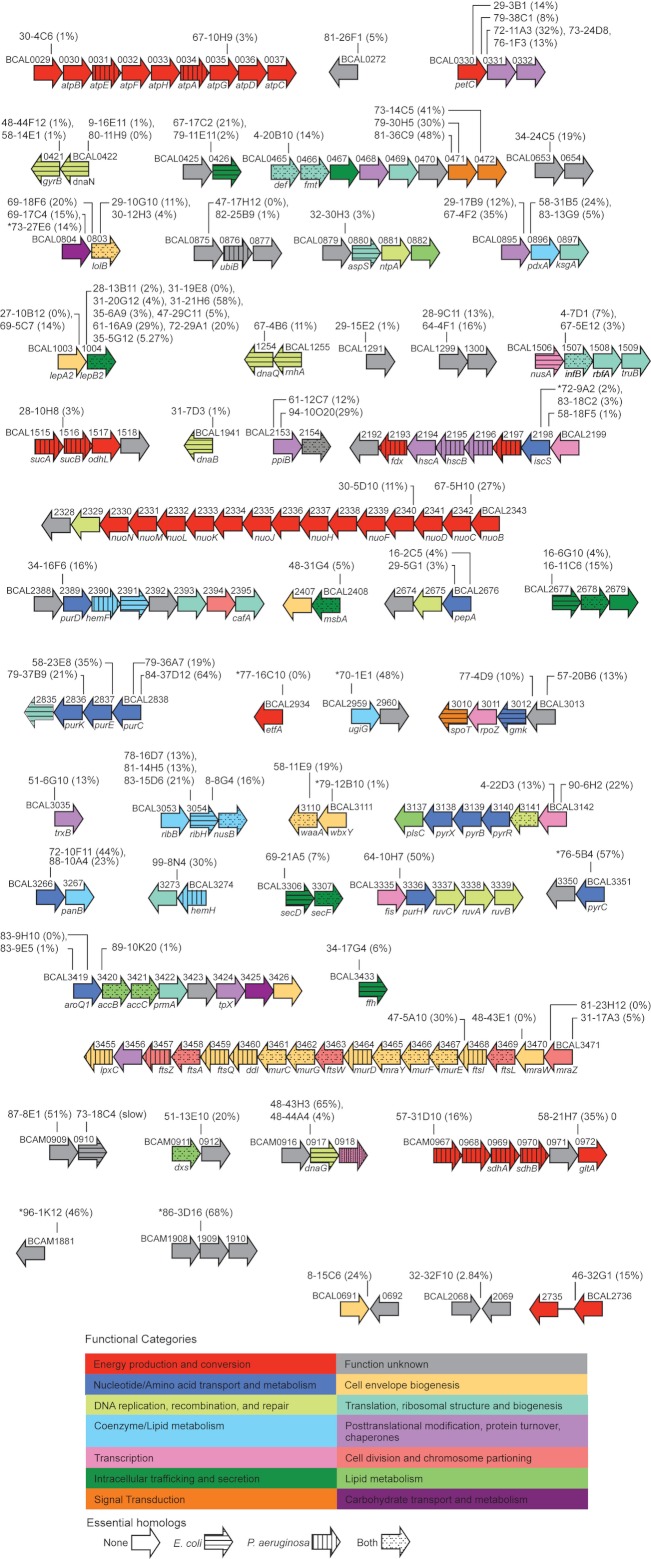
Figure 3.
The 50 putative essential operons identified in Burkholderia cenocepacia 56-2. Each block represents a putative essential operon, and each arrow represents a gene. Operons include genes downstream from the mutant insertion sites according to OperonDB or DOOR. Genes are ordered according to the locus names of the B. cenocepacia J2315 genome. For each mutant, the strain name and the approximate location of the PrhaB are indicated by vertical lines. Exact location of insertion sites are listed in Table S1. Mutants where the location of the transposon insertion site could not be determined at the nucleotide level are indicated with an asterisk. The percentage of wild-type growth in the absence of rhamnose is found between brackets beside the strain name. Genes are color coded according to putative function and black patterns indicate essentiality of homologs in Escherichia coli (horizontal), Pseudomonas aeruginosa (vertical), or both (spots). Three mutant strains, 46-32G1, 8-15C6, and 32-32F10, whose insertion sites and conditional-growth phenotypes are confirmed but no downstream genes appear to be part of the same operon, are shown separately at the end.
Figure 4.
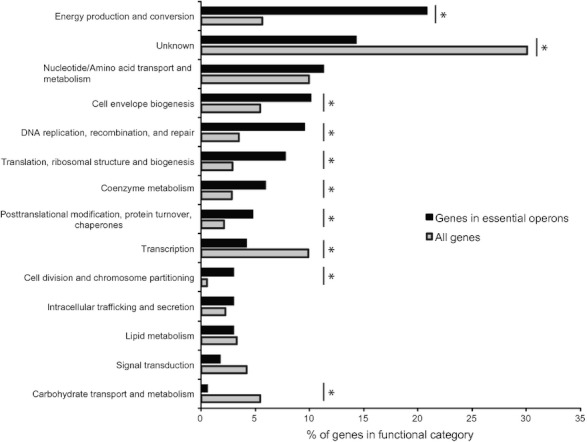
Functional categories of genes in putative essential operons. Putative gene function is based on the GO (Gene Ontology) and COG (Cluster of Orthologous Genes) annotations. For each functional category, Pearson's chi-squared test was used to determine whether the occurrence of the category in the entire genome differs statistically from its occurrence in the putative essential genes identified in this study. A star indicates a P-value of less than 0.05.
A BLASTp reciprocal best hit (RBH) revealed 239 B. cenocepacia J2315 genes with essential orthologs in E. coli (Baba et al. 2008) and 249 with essential orthologs in P. aeruginosa PAO1 (Jacobs et al. 2003), of which 131 were essential in both (data not shown). Of the 179 genes found in essential operons in our study, 63 had essential orthologs in either E. coli or P. aeruginosa, of which 25 were essential in both (Fig. 3, Tables S1 and S3). The identical genomic duplication in B. cenocepacia J2315 caused genes in P. aeruginosa or E. coli that matched to a B. cenocepacia J2315 gene in the duplicated region to return two identical “best hits.” As we determined that the aforementioned duplication is not present in the B. cenocepacia K56-2 strain used for the experiments, the genes downstream of transposon insertions found in the region by our study were manually matched against E. coli and P. aeruginosa and added as orthologs to Table S3. The identification of clones with transposon insertions upstream of genes that are orthologous to essential genes related to cell division (ftsA, ftsZ, ftsW, etc.), and peptidoglycan biosynthesis and assembly (murD, mraF, mraY, etc.) shows that our method of large-scale screening for CG mutants does discover operons containing essential genes.
Of the 179 genes found in this study, 117 do not have essential orthologs in E. coli or P. aeruginosa. Of these, 66 are located in operons with at least one essential ortholog in the aforementioned genomes, for example, BCAL1508 and BCAL1509 (Fig. 3). The remaining 51 genes were organized in 19 operons that did not match to any essential orthologs in either P. aeruginosa or E. coli. We reasoned that if these genes were essential in B. cenocepacia, they might be conserved at least across closely related species. We therefore examined the distribution of these open reading frames across the Burkholderia genus using Burkholderia Ortholog groups from the Burkholderia Genome Database (Winsor et al. 2008). Of the 51 genes, 38 were present in all the sequenced genomes of Burkholderia species and B. cenocepacia strains (Table S4). This is in agreement with the assumption that gene conservation among Burkholderia genomes is related to essentiality (Juhas et al. 2012). However, a few poorly conserved genes that were found in our study also suggest that species or strain-specific requirements for essentiality are also possible. The functions of many of these genes can be inferred from similar genes in other species, but the reasons why they may be required for growth by B. cenocepacia K56-2 remains elusive. Seventy-one mutants had insertions inside of putative coding sequences (Figs. 3 and S2). The essentiality of the downstream genes could then be conditional to the absence of the product encoded by the disrupted gene.
Analyzing the rate at which new essential operons were discovered
Progress toward the identification of new essential operons was monitored by plotting the number of unique operons discovered against the total number of CG mutants sequenced (Fig. 5). We assumed that as more mutants were sequenced, the proportion of new operons was expected to fall at a rate proportional to the fraction of unique operons already discovered. In addition, the proportion of new operons could also depend on whether all essential operons have the same probability of being discovered. We ran computer simulations of randomly chosen essential operons out of a pool of 200 using either an equal chance of picking every operon or applying the frequency distribution of the experimentally identified essential operons to the entire pool. The actual rate at which new operons were being discovered fell below the theoretical predictions (Fig. 5). When we reduced the theoretical number of essential operons to 70, the simulations matched the experimental results. This means that either the probability of finding certain essential genes with our methodology is unequal or B. cenocepacia has a significantly lower number of essential genes than other bacterial genomes. We then analyzed two scenarios that could explain the rate at which new essential operons were being found assuming that B. cenocepacia has the same number of essential genes as other bacterial species: limited expression range of the promoter and biased transposon insertion. Alternatively, we analyzed gene redundancy in B. cenocepacia as a factor that would render fewer essential genes than expected due to gene duplication.
Figure 5.
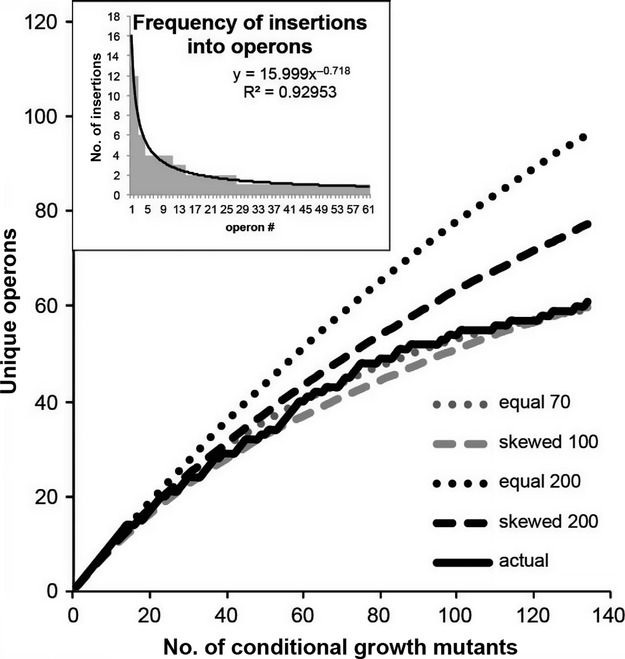
The rate at which new operons were discovered. The rate at which unique operons were discovered experimentally (actual) is compared with four simulations averaging 100 trials of 200, 100, or 70 essential operons, assuming either that every essential operon is equally likely to be detected (equal) or that the observed frequency distribution applies to all essential operons (skewed). The inset shows the frequency at which essential operons were discovered experimentally.
Global analysis of essential gene expression levels
It is reasonable to expect that essential genes require strong promoters as they tend to be highly expressed (Dotsch et al. 2010). In previous attempts to modulate essential gene expression via inducible promoters, achieving a CG phenotype has been complicated by the requirement for the chosen system to allow high levels of expression required by essential genes while simultaneously providing tight regulation (Xu et al. 2010). We previously demonstrated that the PrhaB is tightly regulated and thus can be used to identify essential genes (Cardona et al. 2006). However, we did not rule out the possibility of missing highly expressed essential genes due to a narrow dynamic range of expression levels driven by PrhaB. To examine whether our procedure was excluding highly expressed essential genes, we examined whether global gene expression analysis can show that essential genes are on average more highly expressed than nonessential genes, whether the operons identified by our study showed a similar bias in expression, and finally, whether there was a correlation between frequency at which essential operons were recovered and the level of operon expression.
RNA-Seq measures gene expression by sequencing single molecules of mRNA and is thought to provide the most accurate and unbiased absolute quantitation of transcription on a genomic level (Fu et al. 2009). Therefore, we used previously published RNA-Seq data to examine whether essential genes are more likely to be highly expressed in E. coli (Prieto et al. 2011). Our analysis showed that E. coli essential genes are more highly expressed than nonessential genes in cultures harvested during exponential growth (Fig. 6A), after heat-shock treatment (Fig. 6A), and anaerobic growth (data not shown), with only stationary-phase cells showing no statistically significant difference between essential and nonessential genes (data not shown). As there are no RNA-Seq data available for B. cenocepacia J2315 grown in LB, we looked at whether the normalized fluorescence from cDNA microarrays would show similar differences. As microarrays rely on hybridization of labeled cDNA, the fluorescent intensity for any probe depends not only on the number of transcripts, but also on the hybridization efficiency and the possibility of off-target hybridization to other transcripts (Fu et al. 2009). These biases make comparisons of expression levels between different genes based on differences in microarray fluorescence questionable. As we are interested in the difference in expression between essential and nonessential genes in general, these biases should be equally present in both classes of genes. Therefore, we hypothesized that microarrays could substitute for RNA-Seq for our purposes. We confirmed that previously published cDNA microarray data (DeVito et al. 2002) showed similar differences in expression between essential and nonessential E. coli genes (Fig. 6B), confirming that there is a bias toward higher levels of expression by essential genes and that this difference can be observed using cDNA microarrays. To determine whether the operons in the CG mutant library were similarly biased toward highly expressed genes, we repeated this analysis using previously published cDNA microarray data for B. cenocepacia J2315 grown in LB (Bazzini et al. 2011). As the expression of genes within operons is highly correlated, only the first genes in the identified operons were included in the analysis. There was a significant bias toward high expression of genes from putative essential operons (Fig. 6C). In addition, only 43 genes not represented in our CG mutant library showed higher expression than the most highly expressed operons identified experimentally. Of these, 15 genes in five putative operons have essential orthologs in E. coli or P. aeruginosa, and all had fluorescence within 2.1-fold of that of the experimentally identified genes. These data suggest the PrhaB can drive the levels of expression needed by the vast majority of essential genes.
Figure 6.
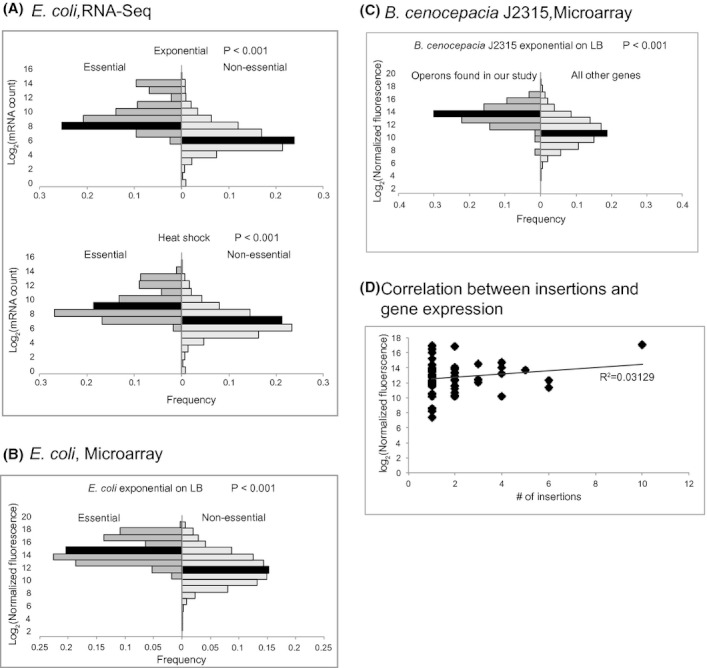
The distribution of gene expression in putatively essential and nonessential genes. The black bars contain the median level of gene expression for each class. The distribution of expression between essential and nonessential genes for each species/condition/methodology was compared using a Mann–Whitney test. Median values, U statistic, and number (n) of essential and nonessential genes are as follows. (A) Exponentially growing cells (median mRNA transcripts: 364.7 essential, 51.8 nonessential, U = 893,584, P < 0.001), heat-shock treatment (median mRNA transcripts: 294 essential, 69.3 nonessential, U = 786,268, P < 0.001) nnonessential = 4038, nessential = 280; (B) cDNA microarray for exponentially growing cells (median fluorescence: 16,700.09 essential, 3328.65 nonessential, U = 701,538, nessential = 280, nnonessential = 3868, P < 0.001); (C) cDNA microarray for Burkholderia cenocepacia J2315 (median fluorescence: 10,448.6 operons in library, 1864.7 other genes, U = 346,475, nessential = 6875, nnonessential = 63, P < 0.001); (D) There is no correlation between levels of gene expression and the frequency at which insertions into an operon were recovered.
We next asked if the higher frequency of finding insertions upstream of certain operons was related to hotspots of the Tn5-mini transposon. These hotspots have been associated with negatively supercoiled regions (Lodge and Berg 1990) that could be created upstream of highly transcribed genes (Rovinskiy et al. 2012). We then analyzed the relationship between the frequency of recovering CG mutants of certain operons and transcription levels. However, we were unable to find a correlation between a high number of CG mutants recovered for essential operons and high level of expression for those operons (Fig. 6D). Taken together, neither the range of expression levels of the PrhaB promoter nor hotspots due to the presence of highly expressed genes can completely explain the observed rate at which new essential operons were discovered.
Analysis of gene redundancy in B. cenocepacia
To explain the lower than expected rate at which we were discovering essential operons (Fig. 5), we reasoned that a higher number of genes encoding for essential proteins have to be duplicated in comparison with other essential genomes. To address a possible gene-redundancy effect, we first analyzed the criteria for two genes to be considered duplicated. While two exact copies of a gene are undoubtedly duplicates, duplicated genes that diverged after the duplication event may only share a certain DNA sequence similarity over a partially alignable region. To analyze the presence of duplicate genes in B. cenocepacia, considering not only identical copies but also duplicate genes that may have further differentiated, we estimated the presence of such genes at different stringency cutoff levels. Similar to other studies of gene duplication (Gu et al. 2002), two parameters were considered for defining stringency: DNA sequence identity and the proportion of alignable sequence over a gene. We compared gene duplications of B. cenocepacia with those of P. aeruginosa and E. coli at all levels of stringency. For example, two genes were considered duplicates with a stringency cutoff of 40 if they shared a DNA sequence identity and percent of alignable sequence equal or higher than 40%. The B. cenocepacia genome showed higher gene duplication than the P. aeruginosa and E. coli genomes at all stringency cutoff levels, but the proportion of duplicates varied greatly with stringency cutoff (Fig. 7). When exact copies were considered (stringency cutoff of 100), approximately 2% and 1% of the genes came out as duplicates for B. cenocepacia and E. coli, respectively (Fig. 7, inset). This twofold difference was also observed at a stringency cutoff of 60, where the percentage of duplicates increased to 7.6% and 3.5% in B. cenocepacia and E. coli, respectively. Thus, if the same proportion of duplicated genes observed in whole genomes is observed in essential genomes, then gene redundancy could explain the previously observed lower than expected hit frequency.
Figure 7.
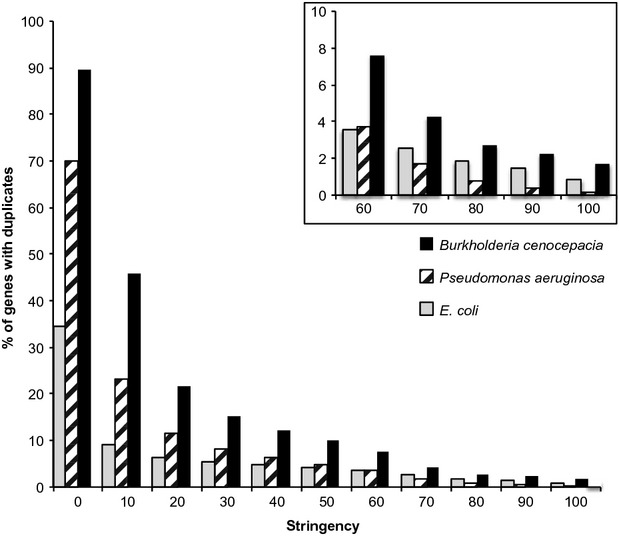
Genetic duplication in Burkholderia cenocepacia J2315, Pseudomonas aeruginosa PA01, and Escherichia coli K12 as a function of DNA sequence similarity. For each strain, the genome was downloaded from the Genome directory of NCBI and a BLAST database was built containing all annotated coding regions. For each gene, similar genes within the same genome were identified using blastn with an expect cutoff of 0.1. Stringency cutoff was defined by percent coverage in the gene alignment and % sequence identity. A stringency cutoff of 40%, for example, means that both percent coverage and sequence identity between two genes are equal or higher than 40%. A gene was included in the same paralogous group when percent coverage and percent identity with at least one member of the group satisfied the stringency cutoff. The inset shows a scaled-up figure of the 60–100 stringency cutoff. Note that paralogous group denotes genes that meet the required conditions for inclusion within the group without any reference to gene history or gene evolution.
CG mutants demonstrate selective hypersensitivity at low rhamnose concentrations
If a small molecule with antibacterial activity exerts its effect by binding and inhibiting an essential protein, then underexpressing this essential gene should cause cells to become more sensitive to that small molecule (DeVito et al. 2002; Donald et al. 2009). This hypersensitivity should allow growth inhibitors to be matched to their specific molecular targets. We then reasoned that only a CG mutant of the gyrB gene should show enhanced sensitivity to the antibiotic novobiocin and other CG mutants should not. Novobiocin exerts its inhibitory action by binding the GyrB subunit of DNA gyrase (Lewis et al. 1996). Using 12 different concentrations of rhamnose and a novobiocin concentration that inhibits 30% of wild-type growth (IC30), we established that a B. cenocepacia gyrB was hypersensitive at rhamnose concentrations that produced less than 60% of wild-type growth (data not shown). Next, we tested the sensitivity to novobiocin of the CG mutant library at various rhamnose concentrations. We reasoned that while mutants under too little stress may not show any hypersensitivity, severely stressed cells could be generally hypersensitive to growth inhibitors. Reducing growth to 30% or less of wild-type growth caused hypersensitivity and high variability across experiments for most of the CG mutants (data not shown). We then screened 25 randomly chosen mutants at rhamnose concentrations that produced 30–60% of wild-type growth (Fig. S3). Chloramphenicol, which inhibits protein synthesis by binding to the ribosome (Wilson 2011), was used as a negative control at its IC30. Only mutants with transposon insertions upstream of gyrB were hypersensitive to novobiocin (Fig. S3A), and none of the CG mutants were hypersensitive to chloramphenicol (Fig. S3B).
Discussion
In this work, we demonstrate that random transposon mutagenesis with an outward-facing promoter PrhaB followed by screening for a rhamnose-dependent CG phenotype can identify putative essential operons at a large scale while simultaneously constructing CG mutants. The recovery of genes with essential orthologs in other species known to be involved in essential cellular processes (e.g., ftsZ) shows that this methodology is capable of recovering mutants of truly essential genes on a large scale. While secondary site mutations that produce a rhamnose-conditional phenotype cannot be ruled out, we consider this possibility to be unlikely. First, rhamnose is not used as a carbon source in B. cenocepacia (data not shown), and it is only known to be part of the O-antigen, a nonessential component of the lipopolysaccharide (Ortega et al. 2005, 2007). Second, we recovered independent transposon mutants that contain the rhamnose promoter controlling the same gene or gene cluster. Finally, independent CG mutants of gyrB were constructed showing the same phenotype as recovered by transposon mutagenesis.
If the number of essential genes in B. cenocepacia essential genome is not substantially different from what was found in other genomes, our study could identify 20–35% of the B. cenocepacia essential operons using a single inducible promoter and single screening condition. A possible explanation for the lower than expected hit frequency could be that the B. cenocepacia genome is not uniformly available to the transposon system used. The Tn5-mini transposon has been shown to insert into a highly degenerate consensus sequence (Shevchenko et al. 2002) allowing for almost unbiased insertions based on sequence. However, hotspots are common and are associated with local DNA topology with highly transcribed negatively supercoiled regions being more favorable (Lodge and Berg 1990). This suggests that operons with high levels of expression should be more accessible to the transposon, and insertions in these operons should be recovered more frequently. However, among the recovered CG mutants, there was no correlation between the frequency of insertions into an operon and the operon's level of expression (Fig. 6D). Conversely, the presence of DNA regions that may never be targeted by a transposon should be considered. Previously, two independent studies performed in P. aeruginosa PAO1 and PA14 used two different transposons, a Tn5–based system and a mariner transposon, respectively, to identify putative essential genes (Jacobs et al. 2003; Liberati et al. 2006). Approximately half of the 678 PA14/PAO1 orthologs not hit with Tn5 were disrupted by the mariner-based transposon in the PA14 library. These 343 genes account for 6.7% of P. aeruginosa PAO1/PA14 orthologs, suggesting that only 6.7% of PAO1 genome was missed by the Tn5 system due to cold spots. Then, a similar distribution of cold spots in B. cenocepacia could not account for the rate of essential operon discovery in our study.
The promoters for essential genes differ from those of nonessential genes by having higher levels of expression and lower levels of noise (Silander et al. 2012). In a previous study, genome-wide promoter replacement of essential genes in Staphylococcus aureus was attempted by site-directed delivery of tetracycline-inducible promoters with different expression levels. Of 150 essential genes that were targeted with three different promoter variants in more than 400 different constructs, only 64 essential genes were found to have a CG phenotype (Xu et al. 2010). An analysis of the expression levels of the B. cenocepacia K56-2 recovered operons showed that the most highly expressed genes in B. cenocepacia J2315 were included and that only five predicted essential operons had higher levels of expression than the most highly expressed operons recovered by our study. This suggests that the rhamnose-inducible promoter is capable of driving the expression required by the vast majority of B. cenocepacia essential genes when the promoter is delivered randomly and CG mutants are isolated by their conditional phenotype.
The presence of duplicated essential genes in B. cenocepacia could lower the hit frequency of our methodology. We predicted that for this effect to account for the lower than expected frequency of finding essential operons, gene duplication in B. cenocepacia should be higher than in E. coli. Our analysis of B. cenocepacia J2315 in comparison with E. coli and P. aeruginosa showed that gene redundancy in B. cenocepacia indeed seems to be twice as high. However, essential genes tend to have fewer paralogs on average than nonessential genes (Deng et al. 2011). Therefore, it is unknown if the observed gene redundancy in B. cenocepacia also occurs among essential genes.
In summary, the reasons for the modest success in promoter replacement of essential genes are currently unknown and could be due to effects of the selection conditions or loss of necessary regulatory regions that account for the genomic organization of essential operons in regulons. The frequency at which CG mutants of different essential operons are recovered could also depend on the length of the region into which the transposon can insert and still produce a phenotype (Christen et al. 2011). Promoter replacement of essential genes that occur in clusters or have very short promoters may require high-throughput approaches such as those performed with high-density transposon mutagenesis and next-generation sequencing technology (Gawronski et al. 2009; Langridge et al. 2009).
This study also shows that the B. cenocepacia library of CG mutants can be used to screen for the targets of specific growth inhibitors. However, one limitation of this methodology is the inability to detect the target of antibiotics that inhibit growth by forming toxic complexes, rather than by target inactivation. During an antisense RNA study (Xu et al. 2010), an S. aureus gyrA mutant was not hypersensitive to ciprofloxacin, an observation we were able to confirm with a B. cenocepacia CG mutant of gyrA (data not shown). Identifying the target of a small molecule with growth-inhibitory characteristics using the developed assay requires screening the whole library against each compound of interest. As the library is sensitive over a broad range of rhamnose concentrations and sublethal concentrations of novobiocin, we predict that it will be possible to screen pools of CG mutants with broadly similar rhamnose sensitivities. This will allow for the simultaneous screening of many targets against small molecules with growth-inhibitory properties.
Acknowledgments
This study was supported by operating grants from the Manitoba Health and Research Council (MHRC), the Canadian Institutes of Health Research (CIHR) – Regional Partnership Program (RPP), a Natural Sciences and Engineering Research Council (NSERC) discovery grant program, the Paul Thorlakson Foundation Fund, and the University of Manitoba University Research Grants Program (URGP). R. A. M. B. was supported by an MHRC Graduate Studentship and an NSERC Postgraduate Scholarship. We thank Alicia Ling, Samira Atoui, Jesse Franklin, and Jacqueline Donogh for technical assistance and Mazdak Khajehpour for informative discussions in regard to data analysis.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. The number of essential genes as a function of genome size. The overall number of essential genes in 14 bacterial genomes is plotted against the total number of genes in the same genome. Data on gene essentiality for all genomes with the exception of data noted with asterisks were collected from Gerdes et al. (2006); gene essentiality of Caulobacter crescentus, Mycobacterium tuberculosis, and Salmonella typhi were obtained from Christen et al. (2011), Griffin et al. (2011), and Langridge et al. (2009), respectively. Mutants were generated by random or targeted transposon mutagenesis, and mutants were propagated. Mutant lack of survival was considered criteria for defining the interrupted gene as essential. Gray and white squares indicate that the data were obtained from propagating the mutants within a population or clonally, respectively. Mge, Mycoplasma genitalium; Hin, Haemophilus influenzae Rd; Hpy, Helicobacter pylori G27; Sau, Staphylococcus aureus RN4220; Mtu, M. tuberculosis H37Rv; Eco, Escherichia coli; Sty, S. typhi; Ccr, C. crescentus; Bsu, Bacillus subtilis; PAO1, Pseudomonas aeruginosa PAO1; PA14, P. aeruginosa PA14.
Figure S2. Transposon insertions relative to the start of relevant coding sequence. (A) Histogram of the distance from insertions into putative intergenic regions to the putative start codon of the downstream gene measured in base pairs. (B) Histogram of the distance from insertions inside of putative coding sequences to the start codon of the surrounding gene measured as a percentage of total gene length.
Figure S3. Conditional growth mutants show selective hypersensitivity. Mutants were grown in rhamnose concentration gradients estimated to produce more than 30% of wild-type growth and challenged with either novobiocin or chloramphenicol at the IC30 of the wild-type. Circles represent CG mutants of the direct target of novobiocin, gyrB. Green crosses represent a CG mutant of the electron transfer flavoprotein gene (etfA). Black crosses correspond to nonsensitive mutants. Mutants of gyrB show hypersensitivity to novobiocin when grown in rhamnose concentrations producing 30–60% of wild-type growth but not when grown in rhamnose concentrations producing 80–100% of wild-type growth. A CG mutant of etfA shows intermediate hypersensitivity. None of the mutants showed hypersensitivity to chloramphenicol. Error bars represent 1 standard deviation calculated from two biological replicates.
Table S1. Mutants found in this study.
Table S2. Primers.
Table S3. Genes found in this study with essential orthologs in Escherichia coli or Pseudomonas aeruginosa.
Table S4. Burkholderia species-specific putative essential operons.
References
- Akerley BJ, Rubin EJ, Novick VL, Amaya K, Judson N, Mekalanos JJ. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA. 2002;99:966–971. doi: 10.1073/pnas.012602299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Huan HC, Datsenko K, Wanner BL, Mori H. The applications of systematic in-frame, single-gene knockout mutant collection of Escherichia coli K-12. Methods Mol. Biol. 2008;416:183–194. doi: 10.1007/978-1-59745-321-9_12. [DOI] [PubMed] [Google Scholar]
- Bazzini S, Udine C, Sass A, Pasca MR, Longo F, Emiliani G, et al. Deciphering the role of RND efflux transporters in Burkholderia cenocepacia. PLoS ONE. 2011;6:e18902. doi: 10.1371/journal.pone.0018902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown ED, Wright GD. New targets and screening approaches in antimicrobial drug discovery. Chem. Rev. 2005;105:759–774. doi: 10.1021/cr030116o. [DOI] [PubMed] [Google Scholar]
- Bugrysheva JV, Froehlich BJ, Freiberg JA, Scott JR. The histone-like protein Hlp is essential for growth of Streptococcus pyogenes: comparison of genetic approaches to study essential genes. Appl. Environ. Microbiol. 2011;77:4422–4428. doi: 10.1128/AEM.00554-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona ST, Mueller C, Valvano MA. Identification of essential operons in Burkholderia cenocepacia with a rhamnose inducible promoter. Appl. Environ. Microbiol. 2006;72:2547–2555. doi: 10.1128/AEM.72.4.2547-2555.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll P, Muttucumaru DG, Parish T. Use of a tetracycline-inducible system for conditional expression in Mycobacterium tuberculosis and Mycobacterium smegmatis. Appl. Environ. Microbiol. 2005;71:3077–3084. doi: 10.1128/AEM.71.6.3077-3084.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarini L, Bevivino A, Dalmastri C, Tabacchioni S, Visca P. Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends Microbiol. 2006;14:277–286. doi: 10.1016/j.tim.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Christen B, Abeliuk E, Collier JM, Kalogeraki VS, Passarelli B, Coller JA, et al. The essential genome of a bacterium. Mol. Syst. Biol. 2011;7:528. doi: 10.1038/msb.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig FF, Coote JG, Parton R, Freer JH, Gilmour NJ. A plasmid which can be transferred between Escherichia coli and Pasteurella haemolytica by electroporation and conjugation. J. Gen. Microbiol. 1989;135:2885–2890. doi: 10.1099/00221287-135-11-2885. [DOI] [PubMed] [Google Scholar]
- Dam P, Olman V, Harris K, Su Z, Xu Y. Operon prediction using both genome-specific and general genomic information. Nucleic Acids Res. 2007;35:288–298. doi: 10.1093/nar/gkl1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Noe JC, Paik S, Kitten T. An improved arbitrary primed PCR method for rapid characterization of transposon insertion sites. J. Microbiol. Methods. 2005;63:89–94. doi: 10.1016/j.mimet.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Deng J, Deng L, Su S, Zhang M, Lin X, Wei L, et al. Investigating the predictability of essential genes across distantly related organisms using an integrative approach. Nucleic Acids Res. 2011;39:795–807. doi: 10.1093/nar/gkq784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis JJ, Zylstra GJ. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 1998;64:2710–2715. doi: 10.1128/aem.64.7.2710-2715.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVito JA, Mills JA, Liu VG, Agarwal A, Sizemore CF, Yao Z, et al. An array of target-specific screening strains for antibacterial discovery. Nat. Biotechnol. 2002;20:478–483. doi: 10.1038/nbt0502-478. [DOI] [PubMed] [Google Scholar]
- Donald RGK, Skwish S, Forsyth RA, Anderson JW, Zhong T, Burns C, et al. A Staphylococcus aureus fitness test platform for mechanism-based profiling of antibacterial compounds. Chem. Biol. 2009;16:826–836. doi: 10.1016/j.chembiol.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Dotsch A, Klawonn F, Jarek M, Scharfe M, Blocker H, Haussler S. Evolutionary conservation of essential and highly expressed genes in Pseudomonas aeruginosa. BMC Genomics. 2010;11:234. doi: 10.1186/1471-2164-11-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannagan RS, Aubert D, Kooi C, Sokol PA, Valvano MA. Burkholderia cenocepacia requires a periplasmic HtrA protease for growth under thermal and osmotic stress and for survival in vivo. Infect. Immun. 2007;75:1679–1689. doi: 10.1128/IAI.01581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, et al. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 2002;43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- Fu X, Fu N, Guo S, Yan Z, Xu Y, Hu H, et al. Estimating accuracy of RNA-seq and microarrays with proteomics. BMC Genomics. 2009;10:161. doi: 10.1186/1471-2164-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc. Natl. Acad. Sci. USA. 2009;106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes SY, Scholle MD, Campbell JW, Balazsi G, Ravasz E, Daugherty MD, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 2003;185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes S, Edwards R, Kubal M, Fonstein M, Stevens R, Osterman A. Essential genes on metabolic maps. Curr. Opin. Biotechnol. 2006;17:448–456. doi: 10.1016/j.copbio.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Cavalcanti A, Chen FC, Bouman P, Li WH. Extent of gene duplication in the genomes of drosophila, nematode, and yeast. Mol. Biol. Evol. 2002;19:256–262. doi: 10.1093/oxfordjournals.molbev.a004079. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldimann A, Daniels LL, Wanner BL. Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon. J. Bacteriol. 1998;180:1277–1286. doi: 10.1128/jb.180.5.1277-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Ichimura T, Mizoguchi H, Tanaka K, Fujimitsu K, Keyamura K, et al. Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol. Microbiol. 2005;55:137–149. doi: 10.1111/j.1365-2958.2004.04386.x. [DOI] [PubMed] [Google Scholar]
- Holden MT, Seth-Smith HM, Crossman LC, Sebaihia M, Bentley SD, Cerdeno-Tarraga AM, et al. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol. 2009;191:261–277. doi: 10.1128/JB.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, et al. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson N, Mekalanos JJ. TnAraOut, a transposon-based approach to identify and characterize essential bacterial genes. Nat. Biotechnol. 2000;18:740–745. doi: 10.1038/77305. [DOI] [PubMed] [Google Scholar]
- Juhas M, Stark M, Lumjiaktase C, von Mering P, Crook DW, Valvano MA, et al. High confidence prediction of essential genes in Burkholderia cenocepacia. PLoS ONE. 2012;7:e40064. doi: 10.1371/journal.pone.0040064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Durfee T, Glasner JD, Qiu Y, Frisch D, Winterberg KM, et al. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 2004;186:4921–4930. doi: 10.1128/JB.186.15.4921-4930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, et al. Simultaneous assay of every Salmonella typhi gene using one million transposon mutants. Genome Res. 2009;19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann EL. Nonparametrics: statistical methods based on ranks. Upper Saddle River, NJ: Prentice Hall; 1998. [Google Scholar]
- Lewis RJ, Tsai FT, Wigley DB. Molecular mechanisms of drug inhibition of DNA gyrase. BioEssays. 1996;18:661–671. doi: 10.1002/bies.950180810. [DOI] [PubMed] [Google Scholar]
- Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge JK, Berg DE. Mutations that affect Tn5 insertion into pBR322: importance of local DNA supercoiling. J. Bacteriol. 1990;172:5956–5960. doi: 10.1128/jb.172.10.5956-5960.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loutet SA, Valvano MA. A decade of Burkholderia cenocepacia virulence determinant research. Infect. Immun. 2010;78:4088–4100. doi: 10.1128/IAI.00212-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam E, Coenye T, Chung JW, Speert DP, Govan JR, Taylor P, et al. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 2000;38:910–913. doi: 10.1128/jcm.38.2.910-913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam E, Urban TA, Goldberg JB. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 2005;3:144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Williams M, Loewen PC, Oresnik IJ. Isolation of salt-sensitive mutants of Sinorhizobium meliloti strain Rm1021. Microbiology. 2006;152:2049–2059. doi: 10.1099/mic.0.28937-0. [DOI] [PubMed] [Google Scholar]
- Ortega X, Hunt TA, Loutet S, Vinion-Dubiel AD, Datta A, Choudhury B, et al. Reconstitution of O-specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis. J. Bacteriol. 2005;187:1324–1333. doi: 10.1128/JB.187.4.1324-1333.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega XP, Cardona ST, Brown AR, Loutet SA, Flannagan RS, Campopiano DJ, et al. A putative gene cluster for aminoarabinose biosynthesis is essential for Burkholderia cenocepacia viability. J. Bacteriol. 2007;189:3639–3644. doi: 10.1128/JB.00153-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Ayanbule K, Smedinghoff M, Salzberg SL. OperonDB: a comprehensive database of predicted operons in microbial genomes. Nucleic Acids Res. 2009;37:D479–D482. doi: 10.1093/nar/gkn784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto AI, Kahramanoglou C, Ali RM, Fraser GM, Seshasayee AS, Luscombe NM. Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12. Nucleic Acids Res. 2011;40:3524–3537. doi: 10.1093/nar/gkr1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovinskiy N, Agbleke AA, Chesnokova O, Pang Z, Higgins NP. Rates of gyrase supercoiling and transcription elongation control supercoil density in a bacterial chromosome. PLoS Genet. 2012;8:e1002845. doi: 10.1371/journal.pgen.1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Shevchenko Y, Bouffard GG, Butterfield YS, Blakesley RW, Hartley JL, Young AC, et al. Systematic sequencing of cDNA clones using the transposon Tn5. Nucleic Acids Res. 2002;30:2469–2477. doi: 10.1093/nar/30.11.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silander OK, Nikolic N, Zaslaver A, Bren A, Kikoin I, Alon U, et al. A genome-wide analysis of promoter-mediated phenotypic noise in Escherichia coli. PLoS Genet. 2012;8:e1002443. doi: 10.1371/journal.pgen.1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa SA, Ramos CG, Leitao JH. Burkholderia cepacia complex: emerging multihost pathogens equipped with a wide range of virulence factors and determinants. Int. J. Microbiol. 2011;2011:607575. doi: 10.1155/2011/607575. Epub 2010 Aug 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalker DM, Kolter R, Helinski DR. Nucleotide sequence of the region of an origin of replication of the antibiotic resistance plasmid R6K. Proc. Natl. Acad. Sci. USA. 1979;76:1150–1154. doi: 10.1073/pnas.76.3.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanassi JA, Hartman-Neumann SL, Dougherty TJ, Dougherty BA, Pucci MJ. Identification of 113 conserved essential genes using a high-throughput gene disruption system in Streptococcus pneumoniae. Nucleic Acids Res. 2002;30:3152–3162. doi: 10.1093/nar/gkf418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvano MA, Keith KE, Cardona ST. Survival and persistence of opportunistic Burkholderia species in host cells. Curr. Opin. Microbiol. 2005;8:99–105. doi: 10.1016/j.mib.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Vandamme P, Dawyndt P. Classification and identification of the Burkholderia cepacia complex: past, present and future. Syst. Appl. Microbiol. 2011;34:87–95. doi: 10.1016/j.syapm.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Wilson DN. On the specificity of antibiotics targeting the large ribosomal subunit. Ann. NY Acad. Sci. 2011;1241:1–16. doi: 10.1111/j.1749-6632.2011.06192.x. [DOI] [PubMed] [Google Scholar]
- Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD, Brinkman FS. The Burkholderia Genome Database: facilitating flexible queries and comparative analyses. Bioinformatics. 2008;24:2803–2804. doi: 10.1093/bioinformatics/btn524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SM, Akerley BJ. Inducible expression system and marker-linked mutagenesis approach for functional genomics of Haemophilus influenzae. Gene. 2003;316:177–186. doi: 10.1016/s0378-1119(03)00762-5. [DOI] [PubMed] [Google Scholar]
- Xu HH, Trawick JD, Haselbeck RJ, Forsyth RA, Yamamoto RT, Archer R, et al. Staphylococcus aureus TargetArray: comprehensive differential essential gene expression as a mechanistic tool to profile antibacterials. Antimicrob. Agents Chemother. 2010;54:3659–3670. doi: 10.1128/AAC.00308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Cho YJ, Won S, Lee JE, Jin Yu H, Kim S, et al. Duplex-specific nuclease efficiently removes rRNA for prokaryotic RNA-seq. Nucleic Acids Res. 2011;39:e140. doi: 10.1093/nar/gkr617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GQ, Cao ZW, Luo QM, Cai YD, Li YX. Operon prediction based on SVM. Comput. Biol. Chem. 2006;30:233–240. doi: 10.1016/j.compbiolchem.2006.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.