Abstract
With white blood cell count emerging as an important risk factor for chronic inflammatory diseases, genetic associations of differential leukocyte types, specifically monocyte count, are providing novel candidate genes and pathways to further investigate. Circulating monocytes play a critical role in vascular diseases such as in the formation of atherosclerotic plaque. We performed a joint and ancestry-stratified genome-wide association analyses to identify variants specifically associated with monocyte count in 11 014 subjects in the electronic Medical Records and Genomics Network. In the joint and European ancestry samples, we identified novel associations in the chromosome 16 interferon regulatory factor 8 (IRF8) gene (P-value = 2.78×10(−16), β = −0.22). Other monocyte associations include novel missense variants in the chemokine-binding protein 2 (CCBP2) gene (P-value = 1.88×10(−7), β = 0.30) and a region of replication found in ribophorin I (RPN1) (P-value = 2.63×10(−16), β = −0.23) on chromosome 3. The CCBP2 and RPN1 region is located near GATA binding protein2 gene that has been previously shown to be associated with coronary heart disease. On chromosome 9, we found a novel association in the prostaglandin reductase 1 gene (P-value = 2.29×10(−7), β = 0.16), which is downstream from lysophosphatidic acid receptor 1. This region has previously been shown to be associated with monocyte count. We also replicated monocyte associations of genome-wide significance (P-value = 5.68×10(−17), β = −0.23) at the integrin, alpha 4 gene on chromosome 2. The novel IRF8 results and further replications provide supporting evidence of genetic regions associated with monocyte count.
INTRODUCTION
The white blood cells (WBCs) are key regulators of the immune system. The differential leukocyte types are reported as a proportion of the total WBC count and have different immunological functions. In order of predominance, these types include neutrophils, lymphocytes, monocytes, eosinophils and basophils. Whereas environmental influences (infection, allergen exposure, medications, etc.) are likely responsible for the majority of intra-individual variability in acute differential counts, inter-individual differences in chronic ‘resting state’ differential counts are known to be heritable traits (1,2). Each cell type matures from hematopoietic stem cells, and the control of the homeostasis of these cell populations continues to be a focus of intense research (3).
Monocytes and some dendritic cells arise from the differentiation of a common precursor, and monocytes themselves can differentiate into dendritic cells under proper conditions. Most monocytes are destined to mature into macrophages. These cells have broad roles in inflammation, autoimmunity and protection from infection and cancer. Understanding the genetic mechanisms influencing monocyte homeostasis could provide insight into the etiology of a variety of disease states. Whereas total WBC has been analyzed for genetic associations several times (4–6), monocyte count has generally been studied less frequently.
Several loci have been reported to be associated with the monocytes. Integrin, alpha 4 (ITGA4), has been associated in multiple cohorts (1,2,7). ITGA4 encodes a receptor for fibronectin in leukocyte cell lines. It may also participate in T-cell interactions with target cells (8). The association of monocytes with variants at the intergenic region at 9q31, lysophosphatidic acid receptor 1 (LPAR1), has also been reported by multiple studies (1,7). LPAR1 is a lysophosphatidic acid (LPA) receptor and mediates diverse biologic functions including proliferation, platelet aggregation, smooth muscle contraction, inhibition of neuroblastoma cell differentiation, chemotaxis and tumor cell invasion (8). Monocyte count associations with the MHC region and near syntaxin-binding protein 6 in a Japanese cohort (2) did not replicate in the large meta-analysis across populations of diverse ancestral background, but did for the 8q24 intergenic region (1). Inconsistent monocyte genome-wide association studies (GWAS) results suggest that clarification is needed to identify loci with consistent impact or race-specific impact. There is increasing evidence that large datasets are required to identify biologically relevant common variants. In this study, we performed a pooled and genetically determined ancestry (GDA)-stratified analysis of monocyte count using 11 014 subjects in the electronic Medical Records and Genomics (eMERGE) Network (4). The Network, consisting of five US cohorts, was linked to electronic medical records (EMRs) for collecting phenotypes. Having access to multiple WBC differentials measured over many years for the duration of the EMR allowed for genomic association with chronic ‘resting state’ of the monocyte count. We were able to assess monocyte count distribution for each subject and allow for the removal of outliers that could be due to environmental conditions like an infection. This is a distinct advantage for such phenotypes with large variability due to environmental conditions that could introduce bias or reduce power. Having access to multiple observations over multiple years also presented challenges and opportunities to assess different association methods utilizing repeated measures from observational data.
RESULTS
Descriptive statistics by eMERGE study site
Table 1 contains descriptive statistics of total monocyte count along with the covariates stratified by eMERGE study site and combined. Northwestern University (23.1%) and Vanderbilt University (32.5%) had the highest proportions of participants who self-identified or were observed reported as having African ancestry (AA) that was generally supported by the genetically determined recent ancestry. There was a significant difference (P < 0.0001) in monocyte count by site, with Marshfield Clinic and Mayo Clinic having the highest median values of 8.4 and 8.6%, respectively. Supplementary Material, Figure S3 illustrates the multivariate effects of predictors on monocyte count for site and interquarile ranges for median BMI, median age and eigenvector 1 and eigenvector 2 from the ancestry principal component analysis (PCA). All covariates in the multivariate were significant (P-value < 0.0001) except for eigenvector 1 that was moderately significant (P-value = 0.03).
Table 1.
Summary statistics of demographic data and phenotypes by eMERGE participating site and combined. The number of records per site is listed under site name (n), and the total number of records was 11 014. This table was produced using the Hmisc library in R
| Group Health (n = 1841) | Marshfield (n = 3614) | Mayo Clinic (n = 2653) | Northwestern (n = 1109) | Vanderbilt (n = 1797) | Combined (n = 11 014) | |
|---|---|---|---|---|---|---|
| Sex (M) | 41% (754) | 41% (1492) | 62% (1646) | 43% (473) | 36% (641) | 46% (5006) |
| Median BMI | 27.0 ± 5.1 | 29.5 ± 5.9 | 28.7 ± 5.2 | 30.8 ± 8.3 | 29.9 ± 7.6 | 29.1 ± 6.3 |
| Median age | 74.2 ± 8.0 | 61.2 ± 11.9 | 64.0 ± 9.1 | 52.4 ± 13.6 | 50.0 ± 16.5 | 61.4 ± 14.1 |
| Self-reported race | ||||||
| American Indian or Alaska Native | 0.1% (2) | 0.6% (22) | 0.2% (5) | 0.0% (0) | 0.0% (0) | 0.3% (29) |
| Asian | 2.7% (50) | 0.3% (10) | 0.1% (2) | 0.0% (0) | 0.1% (3) | 0.6% (62) |
| Black or African American | 3.2% (59) | 0.1% (3) | 0.3% (8) | 26.8% (297) | 34.6% (622) | 9.0% (989) |
| Other | 1.2% (22) | 0.4% (13) | 0.4% (10) | 0.0% (0) | 0.1% (2) | 0.4% (47) |
| White | 92.6% (1705) | 98.7% (3566) | 94.7% (2513) | 73.2% (812) | 57.2% (1028) | 87.4% (9624) |
| Unknown | 0.2% (3) | 0.0% (0) | 4.3% (115) | 0.0% (0) | 8.1% (145) | 2.4% (263) |
| GDA of African continent | 2.6% (47) | 0.1% (2) | 0.2% (5) | 23.1% (256) | 32.5% (584) | 8.1% (894) |
| GDA of European continent | 93.1% (1714) | 99.2% (3586) | 98.9% (2625) | 70.7% (784) | 63.4% (1140) | 89.4% (9849) |
| Median monocytes (%) | 7.6 ± 2.4 | 8.4 ± 1.9 | 8.6 ± 1.9 | 7.9 ± 2.2 | 7.1 ± 2.4 | 8.0 ± 2.2 |
x± I represents X ± 1 SD for continuous variables.
Numbers after percentages are frequencies.
Monocyte count association
Novel variant associations
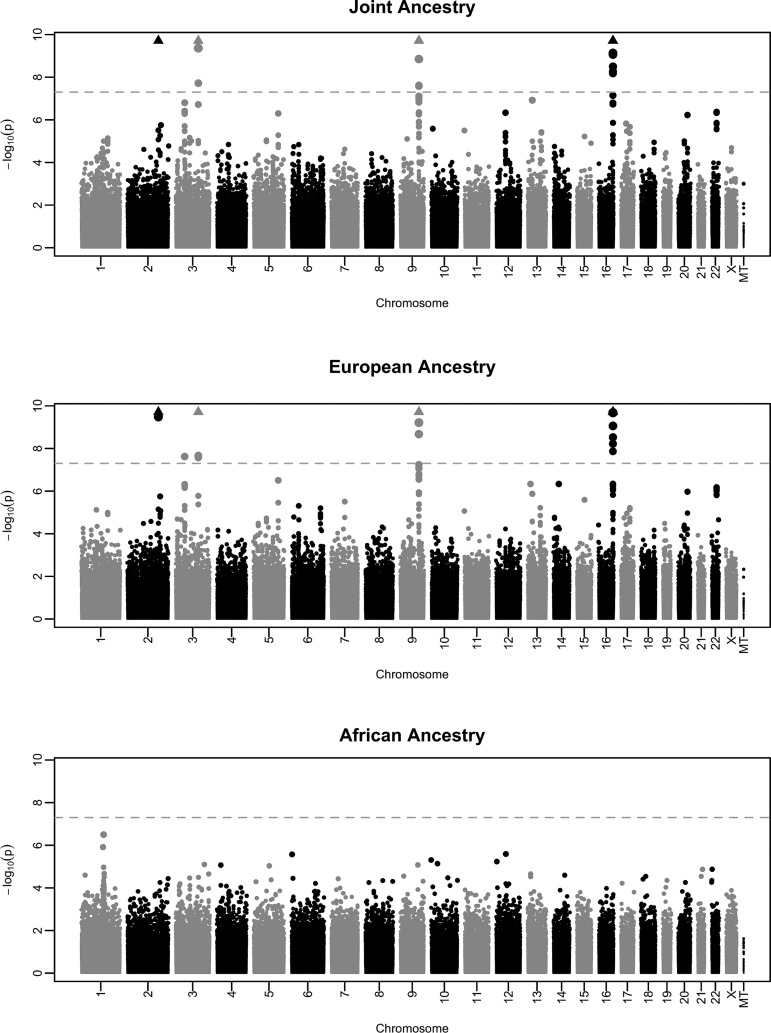
Table 2 outlines variants (both novel and replicated) that reached or approached genome-wide significance for the joint (λ = 1.032), European ancestry (EA) stratified (λ = 1.024) or AA stratified (λ = 1.004) analyses of monocyte count. The Manhattan and QQ plots are illustrated in Figure 1 and Supplementary Material, Figure S1, respectively. To ensure that ancestry-specific admixture was not yielding false positives, we additionally included eigenvectors 1 and 2 to each ancestry-stratified multivariate model that resulted in nominal differences in genomic inflation (λ = 1.021 and λ = 1.006). The Manhattan and QQ plots are illustrated in Supplementary Material, Figures S3 and S4, respectively.
Table 2.
Summary of effects of loci that reached monocyte count genome-wide significance for the joint (n=11 014), EA (n = 9849) and AA (n=887) analyses. Actual numbers for the analyses may vary due to missing phenotype and/or covariate data
| CHR | SNP | A | BP (GRCh37/hg19) | Joint MAF | P-value (β) | EA P-value (β) | AA P-value (β) | Gene | Function |
|---|---|---|---|---|---|---|---|---|---|
| (a) Novel variant associations | |||||||||
| 3 | rs2228467 | G | 42906116 | 0.06 | 1.57×10(−07) (0.30) | 2.39×10(−08) (0.32) | 3.90×10(−01) (−0.39) | CCBP2 | Missense |
| 3 | rs2228468 | C | 42907112 | 0.36 | 5.15×10(−07) (−0.14) | 6.93×10(−07) (−0.15) | 3.87×10(−01) (−0.11) | CCBP2 | Missense |
| 9 | rs2273788 | A | 114348617 | 0.26 | 4.50×10(−07) (0.16) | 1.59×10(−07) (0.17) | 5.62×10(−02) (−0.02) | PTGR1 | Intronic |
| 16 | rs424971 | G | 85946450 | 0.47 | 3.16×10(−16) (−0.22) | 6.32×10(−18) (−0.25) | 5.93×10(−01) (−0.07) | IRF8 | Intronic |
| (b) Replicated variant associations | |||||||||
| 2 | rs2124440 | G | 182328214 | 0.45 | 4.63×10(−17) (−0.22) | 1.35×10(−14) (−0.22) | 5.70×10(−03) (−0.31) | ITGA4 | Intronic |
| 3 | rs2712381 | A | 128338600 | 0.40 | 1.94×10(−16) (−0.23) | 4.52×10(−14) (−0.22) | 1.50×10(−03) (−0.36) | RPN1 | Near 3′ |
BP, base pair; CHR, chromosome; SNP, single nucleotide polymorphism.
Figure 1.
From top to bottom, Manhattan plots of P-values from monocyte count association analyses in the joint, EA and AA groups, respectively.
We identified three separate novel monocyte genetic variant associations, one on chromosome 3 in CCBP2, one on chromosome 16 in the interferon regulatory factor 8 gene (IRF8) and one in PTGR1 (Table 2). Figure 1 illustrates the Manhattan plots for these analyses. QQ plots are shown in Supplementary Material, Figure S1. For the most part, the novel associations identified in the joint analyses were driven by the larger EA group and supported by the results found in the EA-enriched sites (Group Health, Marshfield Clinic and Mayo Clinic; see site specific results, Table 3). In some cases, the AA results were consistent with the European results, but not significant at a genome-wide level.
Table 3.
Summary of effects of loci that reached monocyte count genome-wide significance for the joint analyses, stratified by eMERGE site. Actual numbers for the analyses may vary due to missing phenotype and/or covariate data
| Group Health (n = 2358) | Marshfield (n = 3883) | Mayo Clinic (n = =2809) | Northwestern (n = 1303) | Vanderbilt (n = 3570) | Joint | ||||
|---|---|---|---|---|---|---|---|---|---|
| CHR | SNP | BP (GRCh37/hg19) | GENE | P-value (β) | P-value (β) | P-value (β) | P-value (β) | P-value (β) | P-value (β) |
| 2 | rs2124440 | 182328214 | ITGA4 | 7.94×10(−05) (−0.31) | 2.59×10(−07) (−0.23) | 7.48×10(−05) (−0.21) | 8.75×10(−01) (0.01) | 1.50×10(−02) (−0.19) | 5.68×10(−17) (−0.23) |
| 3 | rs2228467 | 42906116 | CCBP2 | 0.32×10(−01) (0.34) | 2.28×10(−03) (0.26) | 5.10×10(−03) (0.30) | 1.97×10(−01) (0.20) | 7.07×10(−02) (0.32) | 1.88×10(−07) (0.30) |
| 3 | rs2228468 | 42907112 | 6.48×10(−03) (−0.21) | 1.44×10(−05) (−0.19) | 1.28×10(−01) (−0.08) | 3.21×10(−03) (−0.22) | 6.99×10(−01) (−0.03) | 6.39×10(−07) (−0.14) | |
| 3 | rs2712381 | 128338600 | RPN1 | 1.08×10(−03) (−0.26) | 1.01×10(−05) (−0.20) | 7.64×10(−05) (−0.21) | 6.48×10(−01) (0.03) | 1.54×10(−03) (−0.25) | 2.63×10(−16) (−0.23) |
| 9 | rs2273788 | 114348617 | PTGR1 | 1.74×10(−02) (0.21) | 3.02×10(−03) (0.15) | 2.89×10(−02) (0.13) | 9.62×10(−02) (−0.13) | 1.39×10(−01) (0.13) | 3.29×10(−07) (0.16) |
| 16 | rs424971 | 85946450 | IRF8 | 3.71×10(−04) (−0.28) | 5.85×10(−13) (−0.32) | 1.18×10(−02) (−0.13) | 6.64×10(−01) (0.03) | 9.37×10(−02) (0.14) | 2.78×10(−16) (−0.22) |
For the CCBP2 joint analysis, there are multiple variants found within the peak on chromosome 3; we report two based on significance and function (Table 2). Both rs2228467 and rs2228468 are missense variants, V[VAL] → A[ALA] and Y[TYR] → S[SER], respectively. The rs2228467 variant is the most significant [P-value = 1.57×10(−07)] with effect size of β = 0.30 and has a minor allele frequency (MAF) of 0.06 in our joint sample. The minor allele for rs2228467 was associated with a lower monocyte count. Similar effects were seen in the EA group (P-value = 2.39×10(−08); β = 0.32), but not the AA group. The other CCBP2 missense variant (rs2228468; MAF = 0.36) is not in LD with rs2228467 (r2 = 0.04), but this is most likely due to the large difference in MAF between the variants. The joint and EA analyses produced similarly significant results for rs2228468 [P-values = 5.15×10(−07), 6.93×10(−07)] and effect sizes (β = −0.14, β = −0.15) for this variant. Unlike rs2228467, the minor allele for rs2228468 is not associated with a lower monocyte count. Both missense variants in CCBP2 were not significant in our smaller AA group [P-value ≥ 3.87×10(−01)], and the direction of effect was mixed with respect to the joint and EA analyses.
There are also multiple variants in LD found within the peak on chromosome 16 at IRF8, so we report the most significantly associated variant. Again, the joint and EA analyses results for rs424971 (intronic) have similar significance [P-values = 3.16×10(−16), 6.32×10(−18)] and effect size (β = −0.23, β = −0.25). The minor allele for rs424971 (EA MAF = 0.44) is associated with a lower monocyte count. None of the variants in the region were significant in our AA analyses.
Our PTGR1 results suggest a novel monocyte-associated gene of 680 kb from previously reported association to variants in the LPAR1 gene on chromosome 9 (1,7). The EA group again drove the association in our sample. The joint analysis for rs2273788 (intronic) produced marginal genome-wide significance (P-value = 4.50×10(−07), β = 0.16), as did the EA analysis (P-value = 1.59×10(−07), β = 0.17). In both groups, the minor allele for rs424971 was associated with a higher monocyte count. We also included the previously reported LPAR1 variants in the multivariate model to perform conditional analyses (1,7). Both rs7023923 and rs10980800 were separately included in the model, and the significance of rs2273788 was assessed. In general, neither variant significantly changed our rs2273788 association results (P-value = 8.23×10(−07) and 7.92×10(−07), respectively).
Replicated variant associations
We replicated previously reported findings in two regions - ITGA4 on chromosome 2 and RPN1 on chromosome 3 (1,2). The most significant variant in the joint analysis is an intronic single nucleotide polymorphism (SNP) (rs2124440, P-value = 4.63×10(−17), β = −0.22). Subjects with the rs2124440 minor allele on average have lower monocyte count. The EA analysis results reached genome-wide significance (P-value = 1.35×10(−14), β = −0.22), and the smaller AA results were highly consistent (P-value = 5.70×10(−03), β = −0.31). In both ancestry groups, the minor allele for rs2124440 predicted a lower monocyte count. We also replicated an association of RPN1 with a variant (rs2712381) located near the 3′ end. This variant was significant in all the groups, with the joint and EA analysis yielding P-values = 1.94×10(−16), 4.52×10(−14) with β = −0.23, −0.22, respectively. The AA analysis had the same direction of effect (P-value = 1.50×10(−03), β = −0.36). In all ancestry groups, the rs2712381 minor allele predicted a lower monocyte count.
Replication in additional cohorts (CHARGE Hematology Working Group)
We compared our novel and replicated results of variants reaching genome-wide significance with the results from the same variants in a meta-analysis of 19 509 subjects in seven CHARGE cohorts (1). The cohorts include the following studies: (i) The Rotterdam Study (RS), (ii) Framingham Heart Study (FHS), (iii) the NHLBI's Atherosclerosis Risk in Communities (ARIC) Study; (iv) the Age, Gene/Environment Susceptibility—Reykjavik Study (AGES) Study, (iv) Health Aging and Body Composition Study (Health ABC), (vi) the Baltimore Longitudinal Study of Aging (BLSA) and (vii) the Invecchiare in Chianti Study (InChianti). The meta-analyses were performed using the inverse-variance weighted fixed effects model to combine β coefficients and standard errors (SE) (1). The meta-analysis P-values, alleles, βs and SEs are listed in Table 4. We also included adjusted effect (based on strand), if aligned to the eMERGE MAF. For all variants, the directions of effect are the same as we report. The ITGA4 variant (rs2124440) was significant at a genome-wide level in the meta-analysis (P-value = 6.93×10(−14)). The variants in RPN1 (rs2712381), PTGR1 (rs2273788) and IRF8 (rs424971) were significant at levels sufficient for replication, but not at a genome-wide level (P-values = 1.50×10(−02), 3.97×10(−03) and 2.32×10(−05), respectively). Variants for CCBP2 (rs2228467 and rs2228468) did not reach a significance of P-value = 5.00×10(−02).
Table 4.
Replication results from a meta-analysis of 19 509 subjects in seven CHARGE cohorts. Listed from the CHARGE Hematology Working Group are the inverse-variance weighted fixed effects model P-values, alleles, βs and SEs. From the eMERGE analysis, we have included P-values and MAF for reference. Also included are the adjusted effects (based on strand), if aligned to the eMERGE MAF
| CHR | SNP | CHARGE Allele 1 | CHARGE Allele 2 | CHARGE P-value (β;SE) | CHARGE adjusted effect | eMERGE MAF | eMERGE P-value (β) |
|---|---|---|---|---|---|---|---|
| 2 | rs2124440 | A | G | 6.93×10(−14) (0.04; 0.005) | −0.04 | G | 4.63×10(−17) (−0.22) |
| 3 | rs2228467 | T | C | 2.58×10(−01) (−0.02; 0.014) | 0.02 | G | 1.57×10(−07) (0.30) |
| 3 | rs2228468 | A | C | 1.98×10(−01) (0.01; 0.007) | −0.01 | C | 5.15×10(−07) (−0.14) |
| 3 | rs2712381 | A | C | 1.50×10(−02) (−0.02; 0.007) | −0.02 | A | 1.94×10(−16) (−0.23) |
| 9 | rs2273788 | T | C | 3.97×10(−03) (0.02; 0.004) | 0.02 | A | 4.50×10(−07) (0.16) |
| 16 | rs424971 | T | C | 2.32×10(−05) (0.02; 0.005) | −0.02 | G | 3.16×10(−16) (−0.22) |
DISCUSSION
This genome-wide assessment of circulating monocyte count prediction in the eMERGE Network identified novel variants in CCBP2, IRF8 and PTGR1 and validated previous genes reported to be associated with monocyte count—ITGA4 and RPN1 (2,5). The fact we were able to replicate previous associations increases confidence in the phenotypes and, thus, in the currently reported novel associations of CCBP2, IRF8 and PTGR1. PTGR1 lies in a chromosomal region that had showed prior association LPAR1; these results may provide an additional clue to that region's association with monocyte count. The large number of subjects with repeated measures over a number of years through the EMR provided an assessment of an individual's underlying resting state. This was a distinct advantage over other studies, as we had both repeated measures and the ability to remove environmental variation due to factors such as medications or infections. These advantages may have allowed the discovery of the novel findings we report.
Chemokines, such as that coded for by CCBP2, are small molecules that play a role in cell chemotaxis, including the recruitment of effector immune cells to the inflammation site (8). The role of chemokines in the recruitment of macrophages and dendritic cells could obviously affect monocyte development. The direction of effect and the interplay of the two missense variants (rs2228467 and rs228468) will be further explored in future studies.
IRF8 is another excellent candidate given its role with inflammatory signaling. Although common variation in IRF8 has not previously been found to be associated with monocyte count, existing biologic evidence supports this as the causative locus underlying the GWAS signal identified here. IRF8 codes for a transcription factor of the interferon (IFN) regulatory factor (IRF) family (8) that is known to be important in monocyte differentiation (9–11). Coding mutations in IRF8 were recently identified as underlying both an autosomal dominant and an autosomal recessive deficiency of dendritic cells in humans (12), clearly demonstrating the importance of IRF8 in dendritic cell levels. IFNs are also signaling molecules that are released by host cells in the presence of pathogens. This signaling triggers the protective defenses of the immune system to eradicate infected cells that include macrophages and dendritic cells. In addition to regulating the IFN system, the IRF family participates in directing the development of innate and adaptive immunity (13).
We were also able to replicate and/or provide further clarification of monocyte count associations reported by others. Our ITGA4 and RPN1 results provide further evidence that these genes or regions do influence monocyte count. Whereas the LPAR1 gene has previously been reported to be associated with monocyte count, we have reported the association of a nearby gene PTGR1. This gene encodes an enzyme that is involved in the inactivation of the chemotactic factor, leukotriene B4 (8). All variants we presented were replicated in the seven-cohort meta-analysis, except those in CCBP2. Because the novel CCBP2 variants (rs2228467 and rs2228468) did not replicate, despite being an excellent candidate gene, further validation efforts are warranted. Our novel and replicated association results demonstrate genetic variants influencing monocyte count that are not identified for total WBC and may be relevant to WBC differentiation.
MATERIALS AND METHODS
Selection and description of participants
The selection and description of participants are documented in a previous study (4). The eMERGE Network is a consortium of US cohorts linked to EMR data for conducting large-scale, high-throughput genetic research (14). The participating sites included the following: (i) Group Health Cooperative, University of Washington and Fred Hutchinson Cancer Research Center partnership, Seattle, WA, USA, (ii) Marshfield Clinic, Marshfield, WI, USA, (iii) Mayo Clinic, Rochester, MN, USA, (iv) Northwestern University, Chicago, IL, USA and (v) Vanderbilt University, Nashville, TN, USA that also served as the Coordinating Center (15). The Center for Inherited Disease Research (CIDR) at Johns Hopkins University and The Broad Institute of MIT and Harvard served as the genotyping centers for the Network.
Like the previously published total WBC data (4), the differential extraction algorithm for EMRs was developed and validated by Group Health and the University of Washington. As outlined in detail in that paper, we attempted to exclude any visit and/or subject whose values were possibly reflective of something other than resting state differential counts. Subject-level exclusions included indication of HIV or dialysis at any time. Visit-level exclusions are grouped into three categories: (i) any medications (including chemotherapy) that could affect the immune system, (ii) any indication of an infection and (iii) a diagnosis that we deemed a confounder with respect to levels of monocyte count (i.e. a diagnosis of Alzheimer's disease).
Defining GDA
As presented previously (4), to define the genetically determined EA sample, we identified all subjects with values less than three (+3) and greater than negative one (−1) SD from the means of eigenvectors 1 and 2 of self-identified EA, respectively. For genetically determined AA, we identified all subjects with values less than two (+2) and greater than negative one (−1) SD from the mean of eigenvector 1 and less than and greater than one SD (±1) for eigenvector 2 of self-identified AA subjects, respectively. Respective numbers of self-identified and GDA are listed in Table 1. To control for ancestry-specific admixture, PCA was performed within each respective ancestry group (Supplementary Material, Figure S5). This included 12 046 subjects of EA and 1479 of AA.
Technical information
Genotyping
As reported previously (4), most subjects were genotyped on the Illumina Human660W-Quadv1_A (660 W) genotyping platform. A subset of subjects who were self-reported (Northwestern) or observer reported (Vanderbilt University) to have AA was genotyped on the Illumina Human1M-Duo (1 M) genotyping platform. Genotyping calls for both platforms were made at CIDR and Broad using BeadStudio version 3.3.7 and Gentrain version 1.0. Both samples and SNPs were assessed for quality and subsequently filtered from the production data, if thresholds were not met. Cryptic relatedness was assessed for all sites, and pairs at half-sibling level (kinship coefficient θ = k1/4 + k2/2 = 1/8) or higher were randomly broken (by dropping one) before assessing whole-genome association. Subjects identified for filtering at each particular site through the quality control/quality assurance process were subsequently filtered for the entire merged dataset.
Statistics
Analyses of monocyte count as a quantitative phenotype (ordinary least squares) with the given covariates and genotypes were performed in PLINK (16). For subjects with repeated measures, median values for monocytes, BMI and age were utilized in the linear model. For consistency with the WBC count analyses (4), covariates included sex, median BMI, median age, eigenvectors 1 and 2. For the joint analysis, study site was included as a covariate. For the ancestry-stratified analyses, eigenvectors 1 and 2 were dropped from the model. Subsequently, eigenvectors 1 and 2 from each ancestry-specific PCA were added to the model and reanalyzed to ensure that ancestry-specific admixture was not yielding false positives. SNP genotypes were coded as 0, 1 and 2 copies of the minor allele (additive genotypic model). In general, we deemed genome-wide significance at P-value < 5.0×10(−8) that approximates a Bonferroni correction for approximately 1 000 000 independent hypothesis tests (17).
WEB RESOURCES
eMERGE website: http://www.gwas.net
R package Hmisc: http://cran.r-project.org/web/packages/Hmisc/index.html
R package rms: http://cran.r-project.org/web/packages/rms/index.html
R package SNPRelate: http://cran.r-project.org/web/packages/SNPRelate/index.html
SUPPLEMENTARY MATERIAL
FUNDING
eMERGE: This study was supported by the following U01 grants from the National Human Genome Research Institute (NHGRI), a component of the National Institutes of Health (NIH), Bethesda, MD, USA: (i) HG004610, AG06781 (Group Health Cooperative), (ii) HG04599 (Mayo Clinic), (iii) HG004608 (Marshfield Clinic), (iv) HG004609 (Northwestern University), (v) HG004438 (CIDR), (vi) HG004424 (BROAD) and (vii) HG004603 (Vanderbilt University). Additional support was provided by a State of Washington Life Sciences Discovery Fund award to the Northwest Institute of Genetic Medicine (GPJ). CHARGE: This research was made possible by NIA/NIH contract AG000932-02 (2009) Characterization of Normal Genomic Variability. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov). The Age, Gene/Environment Susceptibility Reykjavik Study was funded by NIH contract N01-AG-12100, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association) and the Althingi (the Icelandic Parliament). The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022 and grants R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. The BLSA was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. A portion of that support was through a R&D contract with MedStar Research Institute. The National Heart, Lung, and Blood Institute's Framingham Heart Study is a joint project of the National Institutes of Health and Boston University School of Medicine and was supported by the National Heart, Lung, and Blood Institute's Framingham Heart Study (contract No. N01-HC-25195) and its contract with Affymetrix for genotyping services (contract No. N02-HL-6-4278). Analyses reflect the efforts and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. A portion of this research was conducted using the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. The Health ABC Study was supported in part by the Intramural Research Program of the NIH, National Institute on Aging, NIA contracts N01AG62101, N01AG62103 and N01AG62106. The genome-wide association study was funded by NIA grant 1R01AG032098-01A1 to Wake Forest University Health Sciences, and genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. The InChianti Study was supported as a ‘targeted project’ (ICS 110.1RS97.71) by the Italian Ministry of Health, by the U.S. National Institute on Aging (Contracts N01-AG-916413, N01-AG-821336, 263 MD 9164 13 and 263 MD 821336), and in part by the Intramural Research Program, National Institute on Aging, National Institutes of Health, USA. The GWAS database of the Rotterdam Study was funded through the Netherlands Organization of Scientific Research NWO (nr. 175.010.2005.011, 911.03.012) and the Research Institute for Diseases in the Elderly (RIDE). This study was supported by the Netherlands Genomics Initiative (NGI)/NWO project number 050 060 810 (Netherlands Consortium for Healthy Ageing). The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University, Rotterdam, the Netherlands organization for scientific research (NWO), the Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Netherlands Heart Foundation, the Ministry of Education, Culture, and Science, the Ministry of Health, Welfare, and Sports, the European Commission (DG XII) and the Municipality of Rotterdam. J.F.F. and A.D. were supported by the Netherlands Organization for Scientific Research (NWO, VICI no. 918-76-619). The participation of A.P.R. was supported by National Heart, Lung, and Blood Institute grant R01 HL-071862.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to all the participants in the eMERGE study. The authors thank the staff and participants of the ARIC study for their important contributions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Nalls M.A., Couper D.J., Tanaka T., van Rooij F.J., Chen M.H., Smith A.V., Toniolo D., Zakai N.A., Yang Q., Greinacher A., et al. Multiple loci are associated with white blood cell phenotypes. Plos Genet. 2011;7:e1002113. doi: 10.1371/journal.pgen.1002113. doi:10.1371/journal.pgen.1002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okada Y., Hirota T., Kamatani Y., Takahashi A., Ohmiya H., Kumasaka N., Higasa K., Yamaguchi-Kabata Y., Hosono N., Nalls M.A., et al. Identification of nine novel loci associated with white blood cell subtypes in a Japanese population. Plos Genet. 2011;7:e1002067. doi: 10.1371/journal.pgen.1002067. doi:10.1371/journal.pgen.1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chow A., Brown B.D., Merad M. Studying the mononuclear phagocyte system in the molecular age. Nat. Rev. Immunol. 2011;11:788–798. doi: 10.1038/nri3087. doi:10.1038/nri3087. [DOI] [PubMed] [Google Scholar]
- 4.Crosslin D.R., McDavid A., Weston N., Nelson S.C., Zheng X., Hart E., de Andrade M., Kullo I.J., McCarty C.A., Doheny K.F., et al. Genetic variants associated with the white blood cell count in 13,923 subjects in the eMERGE Network. Hum. Genet. 2012;131:639–652. doi: 10.1007/s00439-011-1103-9. doi:10.1007/s00439-011-1103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nalls M.A., Wilson J.G., Patterson N.J., Tandon A., Zmuda J.M., Huntsman S., Garcia M., Hu D., Li R., Beamer B.A., et al. Admixture mapping of white cell count: genetic locus responsible for lower white blood cell count in the Health ABC and Jackson Heart studies. Am. J. Hum. Genet. 2008;82:81–87. doi: 10.1016/j.ajhg.2007.09.003. doi:10.1016/j.ajhg.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reiner A.P., Lettre G., Nalls M.A., Ganesh S.K., Mathias R., Austin M.A., Dean E., Arepalli S., Britton A., Chen Z., et al. Genome-wide association study of white blood cell count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT) Plos Genet. 2011;7:e1002108. doi: 10.1371/journal.pgen.1002108. doi:10.1371/journal.pgen.1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maugeri N., Powell J.E., Hoen P.A., de Geus E.J., Willemsen G., Kattenberg M., Henders A.K., Wallace L., Penninx B., Hottenga J.J., et al. LPAR1 and ITGA4 regulate peripheral blood monocyte counts. Hum. Mutat. 2011;32:873–876. doi: 10.1002/humu.21536. doi:10.1002/humu.21536. [DOI] [PubMed] [Google Scholar]
- 8.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheller M., Foerster J., Heyworth C.M., Waring J.F., Lohler J., Gilmore G.L., Shadduck R.K., Dexter T.M., Horak I. Altered development and cytokine responses of myeloid progenitors in the absence of transcription factor, interferon consensus sequence binding protein. Blood. 1999;94:3764–3771. [PubMed] [Google Scholar]
- 10.Tamura T., Ozato K. ICSBP/IRF-8: its regulatory roles in the development of myeloid cells. J. Interferon Cytokine Res. 2002;22:145–152. doi: 10.1089/107999002753452755. doi:10.1089/107999002753452755. [DOI] [PubMed] [Google Scholar]
- 11.Tsujimura H., Nagamura-Inoue T., Tamura T., Ozato K. IFN consensus sequence binding protein/IFN regulatory factor-8 guides bone marrow progenitor cells toward the macrophage lineage. J. Immunol. 2002;169:1261–1269. doi: 10.4049/jimmunol.169.3.1261. [DOI] [PubMed] [Google Scholar]
- 12.Hambleton S., Salem S., Bustamante J., Bigley V., Boisson-Dupuis S., Azevedo J., Fortin A., Haniffa M., Ceron-Gutierrez L., Bacon C.M., et al. IRF8 mutations and human dendritic-cell immunodeficiency. N. Engl. J. Med. 2011;365:127–138. doi: 10.1056/NEJMoa1100066. doi:10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozato K., Tailor P., Kubota T. The interferon regulatory factor family in host defense: mechanism of action. J. Biol. Chem. 2007;282:20065–20069. doi: 10.1074/jbc.R700003200. doi:10.1074/jbc.R700003200. [DOI] [PubMed] [Google Scholar]
- 14.McCarty C.A., Chisholm R.L., Chute C.G., Kullo I.J., Jarvik G.P., Larson E.B., Li R., Masys D.R., Ritchie M.D., Roden D.M., et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med. Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. doi:10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roden D.M., Pulley J.M., Basford M.A., Bernard G.R., Clayton E.W., Balser J.R., Masys D.R. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin. Pharmacol. Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. doi:10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnabel R.B., Baumert J., Barbalic M., Dupuis J., Ellinor P.T., Durda P., Dehghan A., Bis J.C., Illig T., Morrison A.C., et al. Duffy antigen receptor for chemokines (Darc) polymorphism regulates circulating concentrations of monocyte chemoattractant protein-1 and other inflammatory mediators. Blood. 2010;115:5289–5299. doi: 10.1182/blood-2009-05-221382. doi:10.1182/blood-2009-05-221382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.