Abstract
Replication of the mammalian mitochondrial DNA (mtDNA) is dependent on the minimal replisome, consisting of the heterotrimeric mtDNA polymerase (POLG), the hexameric DNA helicase TWINKLE and the tetrameric single-stranded DNA-binding protein (mtSSB). TWINKLE has been shown to unwind DNA during the replication process and many disease-causing mutations have been mapped to its gene. Patients carrying Twinkle mutations develop multiple deletions of mtDNA, deficient respiratory chain function and neuromuscular symptoms. Despite its importance in human disease, it has been unclear whether TWINKLE is the only replicative DNA helicase in mammalian mitochondria. Furthermore, a substantial portion of mtDNA replication events is prematurely terminated at the end of mitochondrial control region (D-loop) and it is unknown whether TWINKLE also has a role in this abortive replication. Here, we present a conditional mouse knockout for Twinkle and demonstrate that TWINKLE is essential for mouse embryonic development and thus is the only replicative DNA helicase in mammalian mitochondria. Conditional knockout of Twinkle results in severe and rapid mtDNA depletion in heart and skeletal muscle. No replication intermediates or deleted mtDNA molecules are observed after Twinkle knockout, suggesting that TWINKLE once loaded is very processive. We also demonstrate that TWINKLE is essential for nascent H-strand synthesis in the D-loop, thus showing that there is no separate DNA helicase responsible for replication of this region. Our data thus suggest that the relative levels of abortive D-loop synthesis versus complete mtDNA replication are regulated and may provide a mechanism to control progression to complete mtDNA replication.
INTRODUCTION
Mammalian mtDNA only encodes 13 proteins, but these are nevertheless essential for cell viability as they are crucial components of the oxidative phosphorylation system, located in the inner mitochondrial membrane (1). It is, therefore, not surprising that defects in mtDNA expression and maintenance are associated with numerous mitochondrial diseases, age-associated diseases and the ageing process in humans (2,3). All of the proteins required for mtDNA maintenance and expression are encoded by the nuclear genome and have to be imported into mitochondria after synthesis in the cytosol. Therefore, it is clear that a coordinated action between the two cellular genomes is required to regulate oxidative phosphorylation capacity in response to physiological demand and disease states (3).
The mitochondrial genome is a compact, double-stranded, circular molecule of ∼16.5 kb in mammals. As a consequence of a strand-bias in nucleotide base content, the two strands of mtDNA show different sedimentation in alkaline cesium chloride gradients. Consistent with this difference, the two strands are designated as the heavy (H) and the light (L) strand (4). The mtDNA sequence only contains one longer, non-coding region, the control region, which contains the promoters for transcription of the L and H strands (LSP and HSP) and the origin of replication of the H-strand (OH) (4). The origin of replication of the L-strand (OL) is located elsewhere in a cluster of five Transfer RNA (tRNA) genes (5,6). The exact mechanism of mtDNA replication is still under intense debate. The strand displacement model proposes that mtDNA is replicated asymmetrically and that replication of the H-strand starts from the OH and proceeds unidirectionally until almost two-thirds of the leading strand is replicated. When the leading strand replication unwinds, the region where OL is located, L-strand replication is initiated from the OL (7). A recent study has provided genetic and biochemical support for a critical role of OL in mtDNA replication (5,6). A second, strand-coupled model for mtDNA replication is based on the results from two-dimensional agarose electrophoresis analysis of mtDNA replication intermediates. Finally, a third model, ribonucleotide incorporation throughout the lagging strand, has been proposed for mtDNA replication. This model claims that RNA intermediates are laid down throughout the lagging strand and that these RNA species later on are replaced by DNA (8–10).
Many replication events initiated at OH are prematurely terminated at the end of the mtDNA control region at the so-called termination associated sequences (TAS) (11), which also acts as termination sites for H-strand transcription (12). This prematurely terminated replication leads to the formation of the characteristic triple-stranded DNA structure, the D-loop, in which a 650 bp single-stranded nascent H-strand DNA molecule is hybridized to its parental strand. Mechanisms that lead to D-loop formation and its function in mammalian cells are still unknown, although a role in nucleoid organization has been proposed (13). In addition, it is possible that the D-loop has a regulatory role in providing a switch between abortive and genome length mtDNA replication (14).
Mitochondria contain their own enzymatic machinery that is dedicated to mtDNA replication. Basic components of this apparatus have been characterized in vitro (15); however, also other factors, besides the already identified minimal replisome, must be important to complete mtDNA replication. The minimal replisome is composed of the heterotrimeric mtDNA polymerase (POLG), the hexameric replicative helicase TWINKLE and the tetrameric mitochondrial single-stranded binding protein (mtSSB) (5,15). In addition, the complete replisome requires a primase to form the primers for initiation of DNA synthesis and this role is likely served by the mitochondrial RNA polymerase (POLRMT) (5).
TWINKLE is the mitochondrial replicative DNA helicase that can catalyze 5′ to 3′ NTP-dependent unwinding of the mtDNA duplex in vitro (16). This protein was first identified in a screen for mutations in patients with multiple mtDNA deletions. The carboxy-terminal domain of TWINKLE is homologous to the helicase domain of the T7 phage gp4 protein (17). However, the gp4 protein also has a primase activity in its amino-terminal region that has been lost in the metazoan TWINKLE protein (18). Another distinctive domain within TWINKLE is the linker region that is important for multimerization. More than 30 mutations causing disease in humans have been mapped to the Twinkle gene, most of them affecting sequences encoding the linker or the carboxy-terminal domains. Patients carrying such mutations have been reported to display deletions or depletion of mtDNA leading to respiratory chain deficiency and neuromuscular symptoms. All of the dominant TWINKLE mutations cause autosomal dominant progressive external ophthalmoplegia (adPEO) with exercise intolerance and muscle weakness, most severely affecting ocular muscles (14,19). The effects of some of these TWINKLE mutations have been studied in vitro using purified recombinant proteins (20–23). In an in vivo approach, a transgenic mouse model expressing a mutant variant carrying a duplication of 13 amino acids in the TWINKLE linker region was shown to accumulate multiple DNA deletions, the so-called ‘deletor mice’ (24). Recently, mutant TWINKLE (d-mtDNA helicase) variants expressed in the flies were shown to result in severe depletion of mtDNA and lethality (25). Consistently, depletion of TWINKLE in cell lines leads to decreased mtDNA copy number (26,27). We decided to further characterize the in vivo function of TWINKLE by generating and analyzing conditional Twinkle knockout mice. We show here that TWINKLE is essential for mouse embryonic development, and thus is the only replicative DNA helicase in mammalian mitochondria. A loss of TWINKLE in heart and skeletal muscle leads to drastic mtDNA depletion. Importantly, we demonstrate that TWINKLE is necessary for DNA unwinding during both abortive (D-loop) and complete, genomic length, mtDNA replication.
RESULTS
TWINKLE is essential for embryonic development in the mouse
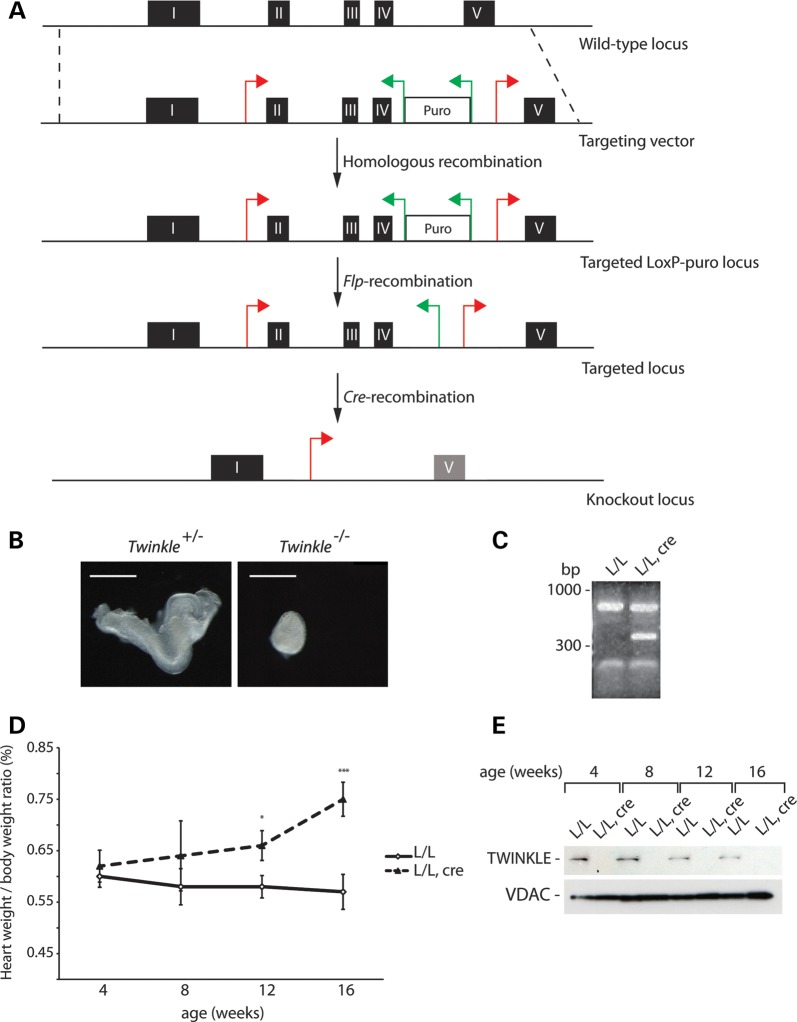
In order to assess the in vivo role of TWINKLE, we generated a mouse with a conditional knockout allele of the Twinkle gene (Fig. 1A). After targeting of the gene in embryonic stem (ES) cells, the mutated locus was transmitted through the mouse germline resulting in heterozygous Twinkle+/loxP-pur mice. The puromycin resistance cassette was removed by mating of Twinkle+/loxP-pur mice with transgenic mice ubiquitously expressing the Flp-recombinase, thus generating mice heterozygous for a loxP-flanked Twinkle allele. Heterozygous Twinkle knockout mice were obtained (Twinkle+/− mice) after mating Twinkle+/loxP mice to mice ubiquitously expressing cre-recombinase (β-actin-cre mice). An intercross of heterozygous Twinkle+/− animals gave no viable homozygous knockout pups, whereas all other genotypes were represented at the expected Mendelian ratios (genotyped pups n = 65; Twinkle+/−n = 41; Twinkle+/+ n = 24). We dissected embryos at embryonic day (E) 8.5 (n = 23) derived from Twinkle+/− intercrosses and found 26% embryos (n = 6) to have a mutant appearance and all of them were homozygous knockouts (Twinkle−/−) (Fig. 1B). The remaining normally appearing embryos were either Twinkle+/+ (n = 4) or Twinkle+/− (n = 13), thus showing that loss of TWINKLE causes embryonic lethality at ∼E8.5. Other mouse mutants lacking expression of critical genes controlling mtDNA maintenance or expression, e.g. PolgA (28), Tfam (29), Mterf3 (30) and Tfb1m (31), also die at ∼E8.5, showing that embryos with disrupted mtDNA expression as a general rule survive until shortly after implantation.
Figure 1.
Conditional knockout of Twinkle. (A) The targeting strategy for the conditional disruption of the Twinkle gene. (B) Morphology of heterozygous (Twinkle+/−) and homozygous knockout (Twinkle−/−) embryos at E8.5. Scale bar, 0.5 mm. (C) RT–PCR analysis of Twinkle transcripts from wild-type (L/L) and tissue-specific knockout mice (L/L, cre). (D) Heart-to-body weight ratio of control (L/L) and tissue-specific knockout mice (L/L, cre) at indicated time points. Error bars represent SEM; *P<0.05, **P<0.01, ***P<0.001, Student's t-test. (E) Western blot analysis of TWINKLE protein levels from wild-type (L/L) and tissue-specific knockout mice (L/L, cre) at different ages. VDAC levels were used as a loading control.
The loss of TWINKLE causes mitochondrial dysfunction
Tissue-specific (heart and skeletal muscle) Twinkle knockout mice were generated by mating of Twinkle+/loxP mice with mice expressing cre-recombinase under the control of the creatinine kinase promoter (Ckmm-cre). The tissue-specific knockout mice (TwinkleloxP/loxP, +/Ckmm-cre) die prematurely at the age of 19 weeks. Using RT–PCR, we could verify the absence of mRNAs containing sequences corresponding to exons II–IV of Twinkle in the tissue-specific knockout hearts (Fig. 1C). The presence of wild-type transcripts in knockout hearts is likely explained by the presence of other cell types that are unaffected by the cardiomyocyte-specific knockout (Fig. 1C). From 8 weeks of age, the tissue-specific knockout mice began to show signs of progressive heart enlargement (Fig. 1D) consistent with mitochondrial cardiomyopathy. Western blot analysis to assess the steady-state levels of TWINKLE protein showed the absence of the TWINKLE protein in knockout hearts from an early age (Fig. 1E).
Twinkle tissue-specific knockout mice have drastic depletion of mtDNA
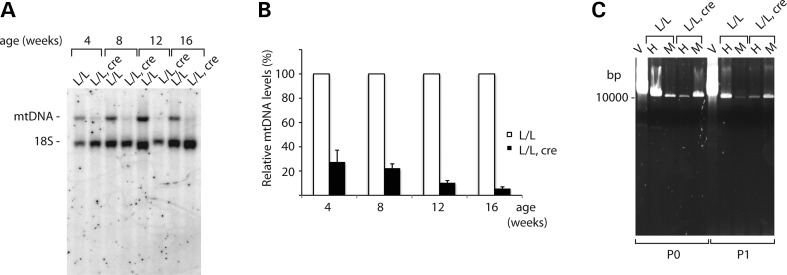
Southern blot analysis of mtDNA levels of control (TwinkleloxP/loxP) and tissue-specific knockout mice (TwinkleloxP/loxP, +/Ckmm-cre) revealed that there is a significant reduction in mtDNA levels in the mutant heart tissue (Fig. 2A). This depletion is evident from a young age (<30% mtDNA left at 4 weeks of age) and becomes increasingly pronounced with increasing age (6% mtDNA left at 16 weeks of age) (Fig. 2B). Although it has been reported that mutations in Twinkle lead to mtDNA deletions (20,24,32), no rearranged or deleted mtDNA was detectable in mouse hearts lacking TWINKLE by Southern blot analysis (Fig. 2A). Moreover, only full-length mtDNA was amplified by a long-extension PCR of DNA isolated from hearts and skeletal muscles of the mutant animals (Fig. 2C).
Figure 2.
MtDNA depletion in TWINKLE knockout mice. (A) Southern blot analysis of mtDNA levels from wild-type (L/L) and tissue-specific knockout mice (L/L, cre) at different ages. The nuclear 18S rRNA gene was used as a loading control. (B) Southern blot quantification: ratio of the mtDNA signal to 18S DNA loading control from four independent experiments. Error bars represent the SEM. (C) Long-extension PCR of heart (H) skeletal muscle (M) isolated total DNA from 12-week-old wild-type (L/L) and tissue-specific knockout mice (L/L, cre) using primers P0 and P1. A vector containing whole mtDNA was used as a control (V).
TWINKLE is required for the replication of the D-loop region
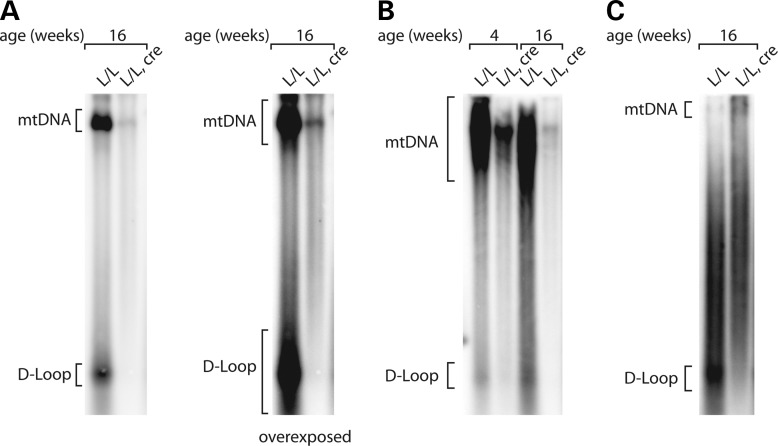
Next, we performed Southern blot analyses to determine whether TWINKLE is required for unwinding of the control region to synthesize the nascent D-loop strand. The severe depletion of mtDNA in Twinkle knockout hearts made it impossible to directly compare absolute levels of D-loop DNA. The D-loop was barely detectable (4 weeks of age) or absent (16 weeks of age) in the knockout samples, even after prolonged exposure of Southern blots (Fig. 3A and B). To overcome this problem, excess mtDNA was loaded in the lane containing Twinkle knockout sample, but virtually no signal corresponding to the D-loop strand was detected, thus showing that TWINKLE is also essential for the replication of the D-loop region (Fig. 3C).
Figure 3.
Severely impaired D-loop formation in Twinkle knockout mice. (A) Southern blot analysis of the absolute D-loop levels of wild-type (L/L) and tissue-specific knockout mice (L/L, cre) at 16 weeks of age. The same amount of total DNA was loaded in both lanes and the same blot is shown at two different exposures. (B) Southern blot analysis of the absolute D-loop levels of wild-type (L/L) and tissue- specific knockout mice (L/L, cre) at 4 and 16 weeks of age. The same amount of total DNA was loaded in each lane. (C) Southern blot analysis of the 7S DNA levels of wild-type (L/L) and tissue-specific knockout mice (L/L, cre) at 16 weeks of age. To allow relative comparisons, the amount of loaded mtDNA from knockout sample (L/L, cre) was approximately four times higher than the amount from wild-type samples (L/L, cre).
Steady-state levels of mitochondrial transcripts are decreased in tissue-specific Twinkle knockout mice
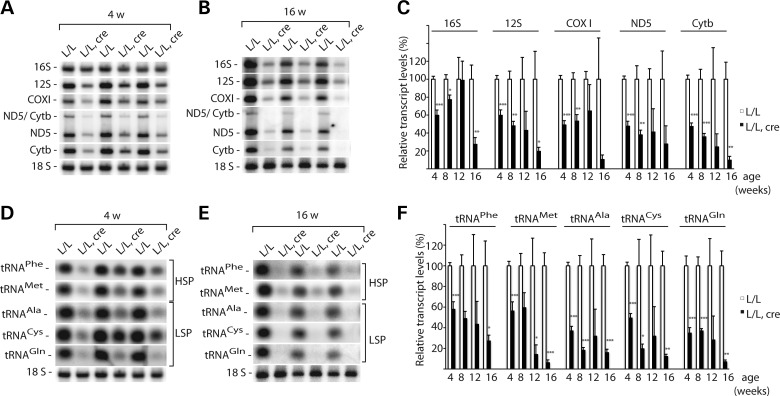
To study the steady-state levels of mitochondrial transcripts at different ages, we performed northern blot analyses. We investigated mtDNA-encoded mRNAs and rRNAs transcribed from the H-strand and tRNAs transcribed from both the H- and L- strands and found decreased levels of all of these transcripts (Fig. 4A–F). The decrease became more pronounced at higher ages in the mice with heart-specific Twinkle knockout (Fig. 4A–F).
Figure 4.
Steady-state levels of mitochondrial transcripts in the absence of TWINKLE. (A) Northern blot analysis of mRNAs and rRNAs isolated from the heart tissue of control (L/L) and knockout (L/L, cre) mice at 4 weeks of age. The nuclear 18S rRNA was used as a loading control. (B) Northern blot analysis of mRNAs and rRNAs isolated from the heart tissue of control (L/L) and knockout (L/L, cre) mice at 16 weeks of age. The nuclear 18S rRNA was used as a loading control. (C) Quantification of steady-state levels of the mRNA and rRNA transcripts from wild-type (L/L) and knockout (L/L, cre) mice at different ages (age 4 weeks, n = 5 pairs of animals; 8 weeks, n = 3; 12 weeks, n = 4; 16 weeks, n = 3;). Error bars represent SEM; *P < 0.05, **P < 0.01, ***P < 0.001, Student's t-test. (D) Northern blot analysis of tRNAs isolated from the heart tissue of control (L/L) and knockout (L/L, cre) mice at 4 weeks of age. The nuclear 18S rRNA was used as a loading control. (E) Northern blot analysis of tRNAs isolated from the heart tissue of control (L/L) and knockout (L/L, cre) mice at 16 weeks of age. The nuclear 18S rRNA was used as a loading control. The autoradiographs shown in panels A and D, as well as those shown in panels B and E, originate from the same gel. Therefore, the 18S loading control is identical in those panels. (F) Quantification of steady-state levels of tRNAs from wild-type (L/L) and knockout (L/L, cre) mice at different ages (4 weeks, n = 5 pairs of animals; 8 weeks, n = 3; 12 weeks, n = 4; 16 weeks, n = 3). Error bars represent SEM; *P < 0.05, **P < 0.01, ***P < 0.001, Student's t-test.
Deficient respiratory chain assembly of Twinkle tissue-specific knockout mice
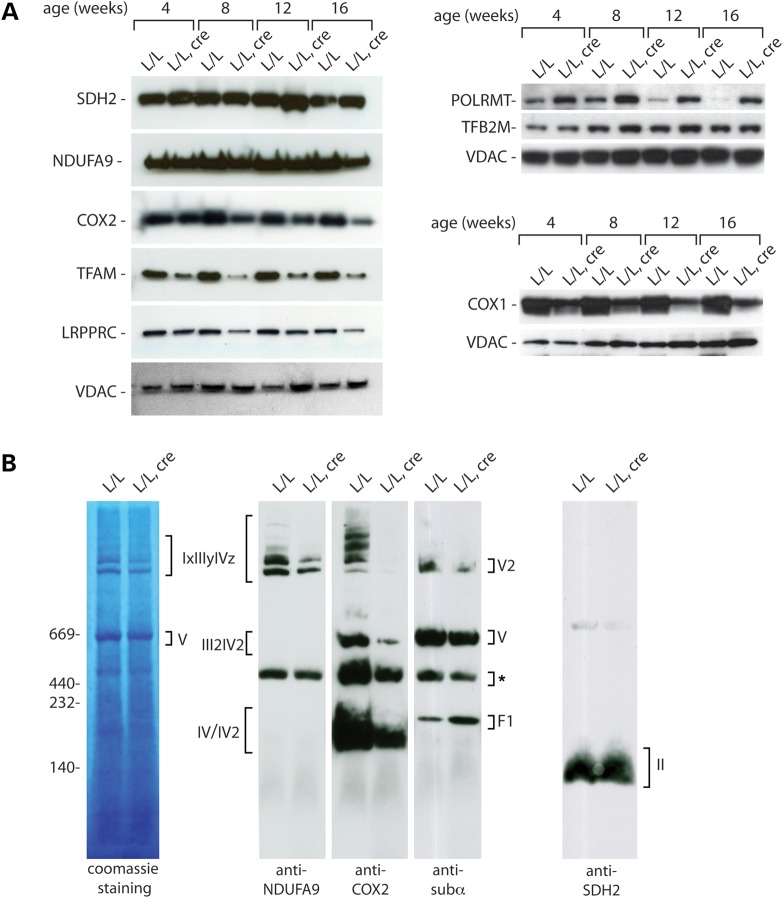
As a consequence of the severe mtDNA depletion and the concomitant decrease of mtDNA-encoded transcripts, the levels of the mtDNA-encoded COX1 and COX2 proteins were decreased in mitochondria from heart tissues of the tissue-specific Twinkle knockout animals (Fig. 5A). This decrease was obvious already at early time points and became even more pronounced with increasing age. Levels of NDUFA9, a nuclear-encoded subunit of complex I, remained stable with only a small decrease visible at 16 weeks of age, the last time point analyzed. The NDUFA9 decrease is likely secondary to the absence of the seven mtDNA-encoded subunits that are essential for the assembly of complex I. Complex II consists of exclusively nuclear-encoded subunits and consequently the steady-state levels of SDH2 were not affected at any age. We also examined the levels of mitochondrial transcription factor A (TFAM), that in addition to its role in transcription initiation also fully coats and packages mtDNA into nucleoids (33–35). Knockout of Tfam results in a loss of mtDNA (29) and overexpression of TFAM leads to an increase of mtDNA copy number (36), whereas decrease of the mtDNA copy number by other means, e.g. due to pharmacological inhibition of mtDNA replication, leads to a secondary decrease of TFAM protein levels (37). We observed a gradual depletion of TFAM protein levels in Twinkle knockout hearts (Fig. 5A), consistent with the idea that TFAM levels do not only control mtDNA levels but also secondarily decrease in response to mtDNA depletion. The leucine-rich pentatricopeptide repeat containing (LRPPRC) protein regulates steady-state levels of mtDNA-encoded transcripts (38,39) and has also been shown to promote polyadenylation of mtDNA-encoded mRNAs and to co-ordinate translation of mitochondrial transcripts (38,40). The Twinkle knockout hearts have reduced levels of mitochondrial transcripts and this leads to a secondary reduction of LRPPRC (Fig. 5A), likely because of reciprocal inter-dependency between LRPPRC protein levels and levels of mtDNA-encoded mRNAs. In contrast to the decreased levels of TFAM and LRPPRC proteins, steady-state levels of the mitochondrial RNA polymerase, POLRMT were significantly increased in Twinkle knockout tissues. Given the fact that POLRMT has been shown to generate the primers for mtDNA replication in vitro (5,41), in addition to its role in mtDNA gene expression, we propose that this upregulation might be a compensatory response to mtDNA depletion. Steady-state levels of mitochondrial transcription factor TFB2M were not significantly affected by the absence of TWINKLE (Fig. 5A). Consistent with the reduction of mtDNA expression in Twinkle knockout hearts, the levels of respiratory complexes and their super assemblies containing mtDNA-encoded subunits (complexes I, III, IV and V) were decreased in comparison with the exclusively nucleus-encoded complex II (Fig. 5B). Furthermore, western blot analysis of blue native PAGE separated respiratory chain complexes revealed the presence of a subcomplex of complex V (Fig. 5B), likely representing the F1 portion of the ATP synthase. Such a decreased assembly of complex V has previously been observed in other mouse knockouts with impaired mtDNA expression (30,31,38,42).
Figure 5.
Loss of TWINKLE leads to reduced respiratory chain assembly. (A) Western blot analysis of the steady-state levels of various mitochondrial proteins from isolated heart mitochondria of control (L/L) and knockout (L/L, cre) mice at different ages. (B) BN–PAGE analysis of mitochondria isolated from TWINKLE knockout (L/L, cre) hearts and controls (L/L) at 10 weeks of age. OXPHOS complexes were detected with subunit-specific antibodies or Coomassie Brilliant Blue staining. *-unspecific bands.
TWINKLE overexpression leads to elevated mtDNA levels
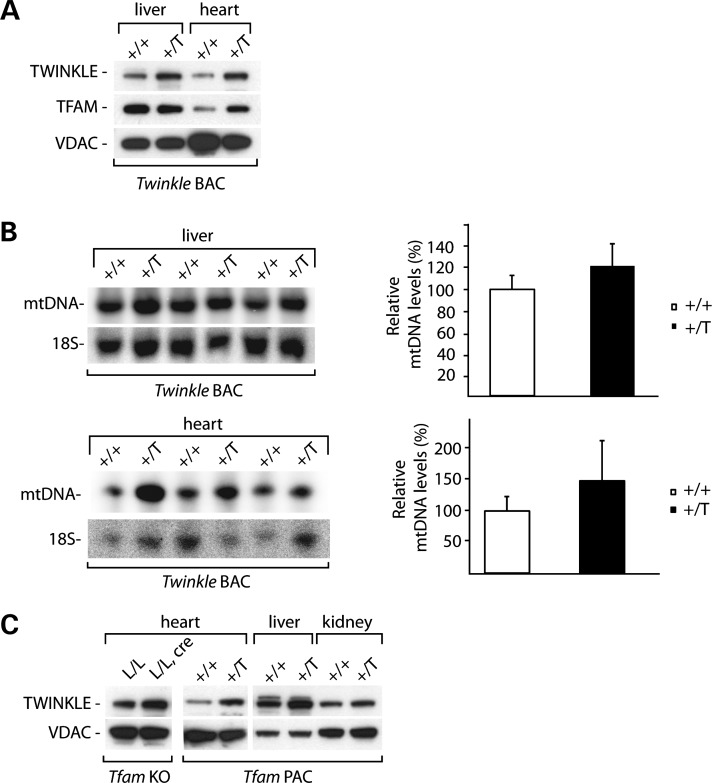
Subsequently, we examined the effect of TWINKLE overexpression on the levels of mtDNA. For this purpose, we generated BAC transgenic mice by pronuclear injection of BAC clones containing a large genomic fragment harboring the mouse Twinkle gene (also known as Peo1). No detrimental effects of Twinkle overexpression were noted as transgenic mice were apparently normal. We performed western blot analyses of mitochondria isolated from liver and heart tissues of wild-type and Twinkle BAC transgenic mice and found that TWINKLE levels were increased approximately two to three times in the overexpressing mice (Fig. 6A). In addition, TFAM levels were obviously upregulated in heart mitochondria of the transgenic mice (Fig. 6A). The TFAM increase strongly suggests upregulation of mtDNA levels and this prediction was confirmed by Southern blot analyses of mtDNA levels (Fig. 6B). The overall mtDNA increase was ∼20–50%. A more drastic effect of Twinkle overexpression on the mtDNA copy number (approximately three times) has been described in mice expressing Twinkle under the strong β-actin promoter (27,43). In contrast, the BAC transgenic overexpression strategy we used here generates a more moderate increase of TWINKLE expression as the endogenous regulatory elements were used for overexpression.
Figure 6.
TWINKLE overexpression increases mtDNA in BAC transgenic mice. (A) Western blot analysis of the steady-state levels of various mitochondrial proteins from isolated liver and heart mitochondria of wild-type (+/+) and transgenic (+/T) mice. (B) Southern blot analysis and quantification of mtDNA levels in total DNA extracts from liver and heart from wild-type (+/+) and transgenic (+/T) mice. The nuclear 18S rRNA gene was used as a loading control. Error bars represent SEM. (C) Western blot analysis of the steady-state levels of TWINKLE protein from isolated heart, liver and kidney mitochondria from TFAM knockout mice (L/L, cre) and their controls (L/L) and from Tfam PAC transgenic animals (+/T) and their controls (+/+). VDAC levels were used as a loading control.
Given the observations that Twinkle knockout leads to mtDNA depletion and that TWINKLE overexpression leads to an increased mtDNA copy number, we investigated whether there is a general connection between the levels of TWINKLE protein and mtDNA. To this end, we analyzed TWINKLE levels in TFAM conditional knockout (44) and TFAM overexpressing mice that display mtDNA copy number decrease and increase, respectively (36). Mice with general TFAM overexpression showed increased steady-state levels of TWINKLE in all analyzed tissues, consistent with the idea that the mtDNA copy number increase is linked to increased mtDNA replication (Fig. 6C). However, heart-specific Tfam knockout mice also showed upregulation of TWINKLE levels, suggesting that mtDNA replication is activated as a futile compensatory response to counteract the profound mtDNA depletion (Fig. 6C).
DISCUSSION
In this study, we have investigated the in vivo function of the mtDNA helicase TWINKLE by using a conditional knockout mouse model. Previous experiments have provided substantial knowledge about TWINKLE involvement in the mtDNA replication process (20–23), but it has remained unclear whether TWINKLE is the only replicative DNA helicase in mammalian mitochondria and if the synthesis of the nascent D-loop strands also depends on TWINKLE. We demonstrate here that TWINKLE is essential for mtDNA replication and that no other helicase can substitute in its absence. Mouse embryos lacking TWINKLE die in mid-gestation because of severe mtDNA depletion. To further study the role of Twinkle, we also disrupted its gene in the mouse heart and found a progressive, severe mtDNA depletion leading to severely impaired mtDNA expression and profound respiratory chain deficiency.
In contrast to the mutated forms of TWINKLE causing adPEO in humans or an mtDNA deletor phenotype in mice, a loss of TWINKLE does not cause accumulation of mtDNA deletions in tissue-specific knockout mice. AdPEO is a late-onset mitochondrial disease and the accumulation of deleted mtDNA molecules varies widely in different post-mitotic tissues collected from affected patients (19). Similarly, in the transgenic mtDNA deletor mouse model, mtDNA deletions were only detectable after 18 months of age (24), suggesting that deletions may be a consequence of age-associated decline of DNA repair mechanisms (32). Moreover, the absence of mtDNA deletions in our model is in line with observations in human cells expressing dominant Twinkle mutations (32) and transgenic flies over-expressing mutant d-mtDNA helicase versions (25). In the mouse model, we describe here, we successfully abolished TWINKLE protein expression, whereas other models and human patients have phenotypes caused by dominant negative effects of a mutant TWINKLE protein expressed against a background of wild-type TWINKLE. Our results, therefore, suggest that the wild-type TWINKLE protein, once it is loaded on an mtDNA template, is able to support a complete round of replication without stalling and creating deletions, even if the intramitochondrial TWINKLE levels are very low. This result is also in good agreement with previous in vitro studies that propose that TWINKLE at the mtDNA replication fork is very processive in the presence of POLG and mtSSB (15,45).
Our results confirm that TWINKLE is involved in mtDNA copy number regulation in mammals (27,43), because the mtDNA copy number is directly determined by the TWINKLE protein levels in the various mouse models we present here. Regulation of mammalian mtDNA replication most likely involves molecular events controlling the initiation of leading-strand synthesis in the control region. Premature termination of transcription from the light strand promoter provides the primers necessary for initiation of H-strand replication at OH. Furthermore, yet unknown mechanisms that determine whether mtDNA replication will stop at the end of the D-loop to provide a nascent H-strand (7S DNA) or whether it should proceed around the genome are likely also important for regulation of mtDNA replication. Given its importance for mtDNA replication, it is formally possible that TWINKLE might play an active regulatory role in mtDNA replication. Consistent with this possibility, it has been shown that the maxicircle mtDNA copy number in trypanosomes is regulated by proteolysis of the mitochondrial replicative DNA helicase (46).
To assess the role played by TWINKLE during the replication of the D-loop region, we examined the levels of D-loop intermediates in Twinkle knockout mouse hearts. The amount of nascent H-strands in the D-loop region was severely decreased or even undetectable in hearts lacking TWINKLE protein. In contrast, another mouse strain overexpressing TFAM has increased steady-state levels of TWINKLE. This latter mouse model has previously been shown to display upregulation of the absolute levels of nascent D-loop strands, but it maintains a constant ratio between nascent D-loop strands and mtDNA (36). These findings suggest a requirement for TWINKLE in the replication of the D-loop region. It is possible that the fate of the replication machinery at the end of the control region is dependent on the interaction between TWINKLE and an unknown antihelicase protein. Such an antihelicase activity at the end of the D-loop might be crucial for disengagement of the replication machinery. It is possible that overexpression of TWINKLE will overcome the blocking action of such an antihelicase, thus allowing increased complete mtDNA replication and increased mtDNA copy number. Consistent with this possible scenario, a protein with antihelicase function, mtDBP, has been identified in sea urchins (47). Moreover, in bovine mitochondria, an unidentified 48 kD protein has been shown to bind to the TAS sequence located at the 3′ end of the D-loop region (48). Future studies will be needed to identify this potential TAS-binding protein and to show whether it has antihelicase activity or if a completely different regulatory mechanism is operating.
In summary, we demonstrate here that TWINKLE has a critical and essential role in mtDNA replication as knockout of its gene causes severe mtDNA depletion and embryonic lethality in mid-gestation. TWINKLE is thus likely the only replicative DNA helicase in mammalian mitochondria. Furthermore, TWINKLE is necessary for both abortive replication to produce nascent D-loop strands and complete replication of the mtDNA molecule. Moreover, mtDNA copy number regulation in vivo is directly dependent on the TWINKLE protein levels, which, in turn, affect mtDNA replication. These findings point towards a model where TWINKLE has a critical role in the regulation of mtDNA replication.
MATERIAL AND METHODS
Generation of TWINKLE knockout mice
The targeting vector for disruption of Twinkle in ES cells (derived from C57BL/6J mice) was generated using the BAC clones from the C57BL/6J RPCI-23 BAC by Taconic Artemis. To generate conditional knockout Twinkle mice, exons II, III and IV of the Twinkle locus were flanked by loxP sites. The positive selection marker (PuroR) was flanked by F3 sites and inserted into intron IV. The puromycin resistance cassette was removed by mating of Twinkle+/loxP-pur mice with transgenic mice ubiquitously expressing Flp-recombinase. The resulting Twinkle+/loxP mice were mated with mice ubiquitously expressing cre-recombinase to generate heterozygous knockout Twinkle+/− mice (Fig. 1A).
Tissue-specific disruption of Twinkle
Tissue (heart and skeletal muscle)-specific knockout mice were generated as described previously (30,31,38,42). Basically, TwinkleloxP/loxP mice were crossed with transgenic mice expressing cre-recombinase under the control of the muscle creatinine kinase promoter (Ckmm-cre). Double heterozygous mice (Twinkle+/loxP, +/Ckmm-cre) were mated to homozygous TwinkleloxP/loxP mice to generate tissue-specific knockout (TwinkleloxP/loxP, +/Ckmm-cre) and control (TwinkleloxP/loxP) mice.
Generation and genotyping of BAC transgenic mice
A BAC clone containing Twinkle was identified by Clone Finder in the National Center for Biotechnology Information database. A 218 kb clone (RP23-34C7) with 95 and 67 kb flanking sequences upstream and downstream of Twinkle was obtained from Children's Hospital Oakland-BAC-PAC Resources. We used a BAC recombination technology (49) to alter the coding sequence of Twinkle, so that a synonymous change was created to introduce an XhoI restriction enzyme site in exon I. The modified BAC clone was prepared with CsCl2 gradient centrifugation and microinjected into pronuclei of one-cell stage mouse embryos. The presence of the BAC transgene DNA was detected with a transgene-specific PCR and restriction enzyme digestion.
RT–PCR
For the reverse transcription experiments, RNA was isolated using the total RNA isolation kit (ToTALLY, Ambion) and used for complementary DNA (cDNA) synthesis with the High Capacity cDNA reverse transcription kit (Applied Biosystems). Primers used for the PCR were Tw-a: ATGCTCTCTGGTGAG; Tw-b: CCATCAAAGCGATTCTTG.
Long-extension PCR
Mouse mtDNA was amplified from 2 to 20 ng of total DNA or control plasmid containing whole mtDNA molecule (pAM1) with the primers P0 (Fw:ggttcgtttgttcaacga ttaaagtcctacgtg, Rev:aggtgatgtttttggtaaacaggcggggt) or P1 (Fw:cctactagcaattatcccca, Rev:catagtggggtatctaatcccag) using LA Taq polymerase (TAKARA, Japan).
Isolation of mitochondria and BN-PAGE
Mitochondria were isolated from mouse tissue using differential centrifugation as previously described (31). For blue native electrophoresis, 100 µg mitochondria were solubilized in solubilization buffer: 1% (w/v) digitonin (Calbiochem), 20 mM Tris, pH 7.4, 0.1 mM EDTA, 50 mM NaCl, 10% (v/v) glycerol. Following 15 min of incubation on ice, non-solubilized material was removed by centrifugation and the supernatant was mixed with loading dye (5% (w/v) Coomassie Brilliant Blue G-250 (Serva), 100 mM Tris pH 7, 500 mM 6-aminocaproic acid). Samples were resolved on 4–10% (w/v) acrylamide gradient BN-PAGE gels (50). BN gels were further subjected to western blot analysis or Coomassie Brilliant Blue R staining as indicated.
Southern blot analysis
Phenol–chloroform extraction was used to isolate total DNA from tissues. DNA, 3–20 µg, was digested with SacI or SphI endonuclease, fragments were separated by agarose gel electrophoresis, transferred to nitrocellulose membranes (Hybond-N+ membranes, GE Healthcare) and hybridization was carried out with α-32P-dCTP-labeled probes for detection of total mtDNA (pAM1), the D-loop or nuclear DNA (18S rDNA) as loading control. For the D-loop, Southern samples were heated for 10 min at 100°C prior loading.
Northern blot analysis
RNA, 2 µg, was isolated using the ToTALLY RNA Total RNA isolation kit (Ambion), separated in formaldehyde agarose gels and transferred to Hybond-N+ membranes (GE Healthcare) by northern blotting. DNA probes, α-32P-dCTP-labeled, were used for visualization of mRNA and rRNA levels. Different tRNAs were detected using specific oligonucleotides labeled with γ-32P-ATP.
Western blot analysis
Proteins were separated by SDS–PAGE or BN–PAGE and then transferred to polyvinylidene difluoride membranes. Immunodetection was performed according to the standard techniques using enhanced chemiluminescence (Immun-Star HRP Luminol/Enhancer Bio Rad). Rabbit polyclonal antisera were used for detection of TFAM, COX2 and LRPPRC proteins (29,38). Rabbit polyclonal antisera against recombinant mouse TWINKLE, POLRMT and TFB2M proteins were generated by Agrisera. TWINKLE and TFB2M antisera were subsequently affinity purified. Monoclonal antibodies specific for NDUFA9 (complex I) and SDHA (70 kD subunit of complex II) were obtained from Invitrogen. F1α (complex V) monoclonal antibody was produced by Abcam. VDAC and COX1 monoclonal antibodies were purchased from Mitoscience.
FUNDING
This work was supported by a European Research Council Advanced Investigator grant to N.G.L. and by Swedish Research Council, Swedish Cancer Foundation and European Research Council Starting Independent Investigator grants to M.F. Funding to pay the Open Access publication charges for this article was provided by Max-Planck Society.
ACKNOWLEDGEMENTS
We acknowledge Emily Hoberg for technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Park C.B., Larsson N.-G. Mitochondrial DNA mutations in disease and aging. J. Cell Biol. 2011;193:809–818. doi: 10.1083/jcb.201010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larsson N.-G. Somatic mitochondrial DNA mutations in mammalian aging. Annu. Rev. Biochem. 2010;79:683–706. doi: 10.1146/annurev-biochem-060408-093701. [DOI] [PubMed] [Google Scholar]
- 3.Nunnari J., Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falkenberg M., Larsson N.-G., Gustafsson C.M. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 5.Fuste J.M., Wanrooij S., Jemt E., Granycome C.E., Cluett T.J., Shi Y., Atanassova N., Holt I.J., Gustafsson C.M., Falkenberg M. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol. Cell. 2010;37:67–78. doi: 10.1016/j.molcel.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 6.Wanrooij S., Miralles Fusté J., Stewart J.B., Wanrooij P.H., Samuelsson T., Larsson N.-G., Gustafsson C.M., Falkenberg M. In vivo mutagenesis reveals that OriL is essential for mitochondrial DNA replication. EMBO Rep. 2012 doi: 10.1038/embor.2012.161. doi:10.1038/embor.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clayton D.A. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 8.Yasukawa T., Reyes A., Cluett T.J., Yang M.-Y., Bowmaker M., Jacobs H.T., Holt I.J. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25:5358–5371. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pohjoismäki J.L.O., Holmes J.B., Wood S.R., Yang M.-Y., Yasukawa T., Reyes A., Bailey L.J., Cluett T.J., Goffart S., Willcox S., et al. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J. Mol. Biol. 2010;397:1144–1155. doi: 10.1016/j.jmb.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holt I.J., Lorimer H.E., Jacobs H.T. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100:515–524. doi: 10.1016/s0092-8674(00)80688-1. [DOI] [PubMed] [Google Scholar]
- 11.ter Schegget J., Flavell R.A., Borst P. DNA synthesis by isolated mitochondria. 3. Characterization of D-loop DNA, a novel intermediate in mtDNA synthesis. Biochim. Biophys. Acta. 1971;254:1–14. [PubMed] [Google Scholar]
- 12.Freyer C., Park C.B., Ekstrand M.I., Shi Y., Khvorostova J., Wibom R., Falkenberg M., Gustafsson C.M., Larsson N.-G. Maintenance of respiratory chain function in mouse hearts with severely impaired mtDNA transcription. Nucleic Acids Res. 2010;38:6577–6588. doi: 10.1093/nar/gkq527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holt I.J., He J., Mao C.-C., Boyd-Kirkup J.D., Martinsson P., Sembongi H., Reyes A., Spelbrink J.N. Mammalian mitochondrial nucleoids: organizing an independently minded genome. Mitochondrion. 2007;7:311–321. doi: 10.1016/j.mito.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Wanrooij S., Falkenberg M. The human mitochondrial replication fork in health and disease. Biochim. Biophys. Acta. 2010;1797:1378–1388. doi: 10.1016/j.bbabio.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 15.Korhonen J.A., Pham X.H., Pellegrini M., Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korhonen J.A., Gaspari M., Falkenberg M. TWINKLE Has 5′ -> 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J. Biol. Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 17.Spelbrink J.N., Li F.Y., Tiranti V., Nikali K., Yuan Q.P., Tariq M., Wanrooij S., Garrido N., Comi G., Morandi L., et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 18.Shutt T.E., Gray M.W. Twinkle, the mitochondrial replicative DNA helicase, is widespread in the eukaryotic radiation and may also be the mitochondrial DNA primase in most eukaryotes. J. Mol. Evol. 2006;62:588–599. doi: 10.1007/s00239-005-0162-8. [DOI] [PubMed] [Google Scholar]
- 19.Suomalainen A., Majander A., Wallin M., Setälä K., Kontula K., Leinonen H., Salmi T., Paetau A., Haltia M., Valanne L., et al. Autosomal dominant progressive external ophthalmoplegia with multiple deletions of mtDNA: clinical, biochemical, and molecular genetic features of the 10q-linked disease. Neurology. 1997;48:1244–1253. doi: 10.1212/wnl.48.5.1244. [DOI] [PubMed] [Google Scholar]
- 20.Wanrooij S., Goffart S., Pohjoismäki J.L.O., Yasukawa T., Spelbrink J.N. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goffart S., Spelbrink H. Inducible expression in human cells, purification, and in vitro assays for the mitochondrial DNA helicase Twinkle. Methods Mol. Biol. 2009;554:103–119. doi: 10.1007/978-1-59745-521-3_7. [DOI] [PubMed] [Google Scholar]
- 22.Matsushima Y., Farr C.L., Fan L., Kaguni L.S. Physiological and biochemical defects in carboxyl-terminal mutants of mitochondrial DNA helicase. J. Biol. Chem. 2008;283:23964–23971. doi: 10.1074/jbc.M803674200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korhonen J.A., Pande V., Holmlund T., Farge G., Pham X.H., Nilsson L., Falkenberg M. Structure-function defects of the TWINKLE linker region in progressive external ophthalmoplegia. J. Mol. Biol. 2008;377:691–705. doi: 10.1016/j.jmb.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 24.Tyynismaa H., Mjosund K.P., Wanrooij S., Lappalainen I., Ylikallio E., Jalanko A., Spelbrink J.N., Paetau A., Suomalainen A. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc. Natl Acad. Sci. USA. 2005;102:17687–17692. doi: 10.1073/pnas.0505551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez-Martinez A., Calleja M., Peralta S., Matsushima Y., Hernandez-Sierra R., Whitworth A.J., Kaguni L.S., Garesse R. Modeling pathogenic mutations of human twinkle in Drosophila suggests an apoptosis role in response to mitochondrial defects. PLoS ONE. 2012;7:e43954. doi: 10.1371/journal.pone.0043954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsushima Y., Kaguni L.S. Differential phenotypes of active site and human autosomal dominant progressive external ophthalmoplegia mutations in drosophila mitochondrial DNA helicase expressed in Schneider cells. J. Biol. Chem. 2007;282:9436–9444. doi: 10.1074/jbc.M610550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyynismaa H., Sembongi H., Bokori-Brown M., Granycome C., Ashley N., Poulton J., Jalanko A., Spelbrink J.N., Holt I.J., Suomalainen A. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 2004;13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 28.Hance N., Ekstrand M.I., Trifunovic A. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum. Mol. Genet. 2005;14:1775–1783. doi: 10.1093/hmg/ddi184. [DOI] [PubMed] [Google Scholar]
- 29.Larsson N.G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G.S., Clayton D.A. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 30.Park C.B., Asin-Cayuela J., Cámara Y., Shi Y., Pellegrini M., Gaspari M., Wibom R., Hultenby K., Erdjument-Bromage H., Tempst P., et al. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 31.Metodiev M.D., Lesko N., Park C.B., Cámara Y., Shi Y., Wibom R., Hultenby K., Gustafsson C.M., Larsson N.-G. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Goffart S., Cooper H.M., Tyynismaa H., Wanrooij S., Suomalainen A., Spelbrink J.N. Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum. Mol. Genet. 2009;18:328–340. doi: 10.1093/hmg/ddn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kukat C., Wurm C.A., Spåhr H., Falkenberg M., Larsson N.-G., Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl Acad. Sci. USA. 2011;108:13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaufman B.A., Durisic N., Mativetsky J.M., Costantino S., Hancock M.A., Grutter P., Shoubridge E.A. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ngo H.B., Kaiser J.T., Chan D.C. The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA. Nat. Struct. Mol. Biol. 2011;18:1290–1296. doi: 10.1038/nsmb.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ekstrand M.I., Falkenberg M., Rantanen A., Park C.B., Gaspari M., Hultenby K., Rustin P., Gustafsson C.M., Larsson N.-G. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum. Mol. Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- 37.Larsson N.G., Oldfors A., Holme E., Clayton D.A. Low levels of mitochondrial transcription factor A in mitochondrial DNA depletion. Biochem. Biophys. Res. Commun. 1994;200:1374–1381. doi: 10.1006/bbrc.1994.1603. [DOI] [PubMed] [Google Scholar]
- 38.Ruzzenente B., Metodiev M.D., Wredenberg A., Bratic A., Park C.B., Cámara Y., Milenkovic D., Zickermann V., Wibom R., Hultenby K., et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2011;31:443–456. doi: 10.1038/emboj.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sasarman F., Brunel-Guitton C., Antonicka H., Wai T., Shoubridge E.A. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol. Biol. Cell. 2010;21:1315–1323. doi: 10.1091/mbc.E10-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bratic A., Wredenberg A., Grönke S., Stewart J.B., Mourier A., Ruzzenente B., Kukat C., Wibom R., Habermann B., Partridge L., et al. The bicoid stability factor controls polyadenylation and expression of specific mitochondrial mRNAs in Drosophila melanogaster. PLoS Genet. 2011;7:e1002324. doi: 10.1371/journal.pgen.1002324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wanrooij S., Fuste J.M., Farge G., Shi Y., Gustafsson C.M., Falkenberg M. Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc. Natl Acad. Sci. USA. 2008;105:11122–11127. doi: 10.1073/pnas.0805399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cámara Y., Asin-Cayuela J., Park C.B., Metodiev M.D., Shi Y., Ruzzenente B., Kukat C., Habermann B., Wibom R., Hultenby K., et al. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 43.Ylikallio E., Tyynismaa H., Tsutsui H., Ide T., Suomalainen A. High mitochondrial DNA copy number has detrimental effects in mice. Hum. Mol. Genet. 2010;19:2695–2705. doi: 10.1093/hmg/ddq163. [DOI] [PubMed] [Google Scholar]
- 44.Wang J., Wilhelmsson H., Graff C., Li H., Oldfors A., Rustin P., Brüning J.C., Kahn C.R., Clayton D.A., Barsh G.S., et al. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat. Genet. 1999;21:133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 45.Jemt E., Farge G., Bäckström S., Holmlund T., Gustafsson C.M., Falkenberg M. The mitochondrial DNA helicase TWINKLE can assemble on a closed circular template and support initiation of DNA synthesis. Nucleic Acids Res. 2011;39:9238–9249. doi: 10.1093/nar/gkr653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu B., Wang J., Yaffe N., Lindsay M.E., Zhao Z., Zick A., Shlomai J., Englund P.T. Trypanosomes have six mitochondrial DNA helicases with one controlling kinetoplast maxicircle replication. Mol. Cell. 2009;35:490–501. doi: 10.1016/j.molcel.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polosa P.L., Deceglie S., Roberti M., Gadaleta M.N., Cantatore P. Contrahelicase activity of the mitochondrial transcription termination factor mtDBP. Nucleic Acids Res. 2005;33:3812–3820. doi: 10.1093/nar/gki693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doda J.N., Wright C.T., Clayton D.A. Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proc. Natl Acad. Sci. USA. 1981;78:6116–6120. doi: 10.1073/pnas.78.10.6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee E.-C., Yu D., Martinez de Velasco J., Tessarollo L., Swing D.A., Court D.L., Jenkins N.A., Copeland N.G. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 50.Schägger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]