Abstract
Fragile X syndrome (FXS) is a neurodevelopmental disorder caused by the loss-of-function of fragile X mental retardation protein (FMRP). The loss of FMRP function in neurons abolishes its suppression on mGluR1/5-dependent dendritic protein translation, enhancing mGluR1/5-dependent synaptic plasticity and other disease phenotypes in FXS. In this study, we describe a new activation function of FMRP in regulating protein expression in astroglial cells. We found that astroglial glutamate transporter subtype glutamate transporter 1 (GLT1) and glutamate uptake is significantly reduced in the cortex of fmr1−/− mice. Correspondingly, neuronal excitability is also enhanced in acute fmr1−/− (but not in fmr1+/+ control) cortical slices treated with low doses (10 μm) of the GLT1-specific inhibitor dihydrokainate (DHK). Using mismatched astrocyte and neuron co-cultures, we demonstrate that the loss of astroglial (but not neuronal) FMRP particularly reduces neuron-dependent GLT1 expression and glutamate uptake in co-cultures. Interestingly, protein (but not mRNA) expression and the (S)-3,5-dihydroxyphenylglycine-dependent Ca2+ responses of astroglial mGluR5 receptor are also selectively reduced in fmr1−/− astrocytes and brain slices, attenuating neuron-dependent GLT1 expression. Subsequent FMRP immunoprecipitation and QRT–PCR analysis showed that astroglial mGluR5 (but not GLT1) mRNA is associated with FMRP. In summary, our results provide evidence that FMRP positively regulates translational expression of mGluR5 in astroglial cells, and FMRP-dependent down-regulation of mGluR5 underlies GLT1 dysregulation in fmr1−/− astrocytes. The dysregulation of GLT1 and reduced glutamate uptake may potentially contribute to enhanced neuronal excitability observed in the mouse model of FXS.
INTRODUCTION
Fragile X syndrome (FXS) is the most commonly inherited form of autism. The transcriptional silencing of fragile X mental retardation protein (FMRP) expression, as a result of hypermethylation on the abnormally high copy number (>200) of trinucleotide (CGG) repeats at the fmr1 locus (1), directly causes FXS. The loss of FMRP function in FXS patients has been modeled in the fmr1−/− mouse, which recapitulates typical behavioral and synaptic phenotypes of FXS (2), including susceptibility to seizures (3) and abnormal dendritic development (4). Previous biochemical studies have characterized FMRP as an important mRNA-binding protein that negatively regulates protein translation (5,6), especially in post-synaptic neuronal dendrites (7). FMRP-mediated translational regulation plays important roles in proper synaptic connectivity (8) and synaptic plasticity (9,10). In FXS, the loss of FMRP function in neurons abolishes its suppression of mGluR1/5-dependent dendritic protein synthesis, contributing to abnormal mGluR1/5-dependent synaptic plasticity and other disease phenotypes (5,11).
Expression and function of FMRP in glial cells of the nervous system remain largely unexplored. FMRP protein has been detected in oligodendrocyte lineage cells in the developing brain (12). FMRP can directly interact with myelin basic protein (MBP) mRNA in oligodendrocyte cell lines (12), implying a potential role of FMRP in the regulation of MBP expression. FMRP is also present in developing astrocytes in vitro and in vivo (13). FMRP-deficient astrocytes derived from fmr1−/− mice are capable of inducing abnormal dendritic morphology of wild-type (WT) hippocampal neurons in vitro (14,15), suggesting that the selective loss of FMRP in astrocytes may contribute to the pathogenesis of FXS.
In the mammalian central nervous system (CNS), plasma membrane glutamate transporter subtypes glutamate transporter 1 (GLT1) and glutamate aspartate transporter (GLAST) (human EAAT2 and EAAT1) are predominantly and abundantly expressed in astroglial cells, playing critical roles in maintaining glutamate homeostasis (16,17), particularly GLT1/EAAT2 (18). Abundant peri-synaptic astroglial GLT1/GLAST tightly controls extracellular glutamate levels at synapses and effectively modulates neuronal mGluR activation (19,20). Pharmacological or genetic inhibition of GLT1 activity potentiates post-synaptic neuronal mGluR activation (21), while up-regulation of GLT1 expression severely impairs mGluR-dependent long-term depression at rat mossy fiber-CA3 synapses (22). In addition, physiological induction of GLT1 expression in astrocytes is important for normal brain development. Extracellular glutamate levels increase 30 and 90% in GLT1+/− and GLT1−/− mice over WT mice (23), respectively, and severe seizures are observed in GLT1−/− mice as early as P14 (18). In this study, we examined alterations of glutamate transporter expression/function and investigated the underlying mechanisms that contribute to the GLT1 dysregulation in the mouse model of FXS.
RESULTS
Expression of FMRP in developing and mature cortical astrocytes in vitro and in vivo
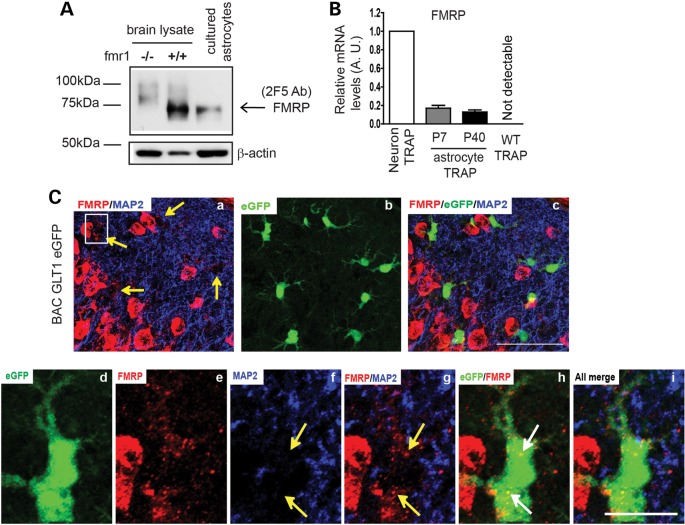
We used two-specific FMRP monoclonal antibodies (2F5 and 7G1) to detect the expression of FMRP in astrocytes in vitro and in vivo (24). The 2F5 and 7G1 antibodies recognize 1–200 or 354–368 amino acid sequences of human FMRP, respectively (24). Both antibodies specifically detect FMRP (71 kDa) in brain lysates from WT (fmr1+/+), but not fmr1−/− mice (Fig. 1A and Supplementary Material, Fig. S1A). Expression of FMRP in cultured primary astrocytes was clearly detected by immunoblotting with 2F5 or 7G1 (Fig. 1A and Supplementary Material, Fig. S1A), consistent with previous observations (13). By employing the translating ribosomal affinity purification (TRAP) approach (25), we isolated ribosome-bound translating mRNA from the cortex of BAC ALDH1L1 TRAP transgenic mice (obtained from the GENSAT project) that selectively express the enhanced green fluorescent protein (eGFP)-L10 fusion ribosome subunit protein in astrocytes of the CNS. Specific activation of the ALDH1L1 promoter in astrocytes has been previously characterized (25,26). We first validated the specificity of translating mRNA transcripts by comparing the relative expression levels of CNS cell-type-specific mRNA transcripts in TRAP samples and total cortical RNA. QRT–PCR results show that glial fibrillary acidic protein (GFAP) and GLT1 mRNA is greatly enriched, while other cell-type-specific mRNAs are minimally detectable or undetectable in TRAP-isolated mRNA (Supplementary Material, Fig. S1B–G), confirming that the TRAP approach selectively isolates mRNA from astrocytes in BAC ALDH1L1 TRAP mice. In addition, TRAP isolation from WT mice resulted in undetectable levels of mRNA by QRT–PCR (Fig. 1B and Supplementary Material, Fig. S1B–D). Subsequent QRT–PCR analysis of FMRP in astrocyte and neuron TRAP samples (from CaMKIIα TRAP transgenic mice, a kind gift of Dr Leon Reijmers, Tufts University) found that translating FMRP mRNA levels in astrocytes are 15–20% of those in neurons, suggesting active translation of the FMRP mRNA in both developing (P7) and mature (P40) astrocytes in vivo (Fig. 1B).
Figure 1.
Expression of FMRP in developing and mature astrocytes in vitro and in vivo. (A) Detection of FMRP protein expression in cultured astrocytes by 2F5 monoclonal antibody. (B) Relative translating FMRP mRNA levels in cortical astrocytes and in neurons in vivo determined by TRAP and QRT–PCR approach. WT TRAP: TRAP procedures from wild-type mice; (C) FMRP immunoreactivity in cortical astrocytes of BAC GLT1 eGFP mice; (a–c) double-immunostaining of MAP2/FMRP on a cortical section of BAC GLT1 eGFP mice; scale bar: 50 μm; (d–i) a magnified view of MAP2/FMRP immunostaining in a single cortical astrocyte of BAC GLT1 eGFP mice. White arrows to highlight positive FMRP signals in astrocytes; yellow arrows to highlight FMRP staining signals not overlap with MAP2 signals. Scale bar: 10 μm.
Immunostaining with the 2F5 antibody also revealed FMRP immunoreactivity in cortical astrocytes (yellow arrows, Fig. 1Ca) in brain sections (P26) from BAC GLT1 eGFP astrocyte reporter mice following antigen retrieval procedures. GLT1 genomic promoter-driven eGFP protein is expressed in astrocyte soma in BAC GLT1 eGFP mice (27), facilitating the identification of astrocytes and detection of the FMRP immunoreactivity in astrocytes in situ (Fig. 1Cc). In astrocytes, FMRP immunoreactivity was found peri-soma or inside the cytoplasm (Fig. 1Ca, d, e and h), and has no overlap with counterstained MAP2 immunoreactivity (Fig. 1Ce–g). In contrast, staining with secondary antibody alone or staining in the negative control BAC GLT1 eGFP X fmr1−/− mouse brain sections gives no positive FMRP staining in astrocytes (Supplementary Material, Fig. S1H). These molecular and immunohistochemical results show that FMRP is expressed in both developing and mature astrocytes in vitro and in vivo.
Transcriptional dysregulation of astroglial GLT1 in fmr1−/− mice
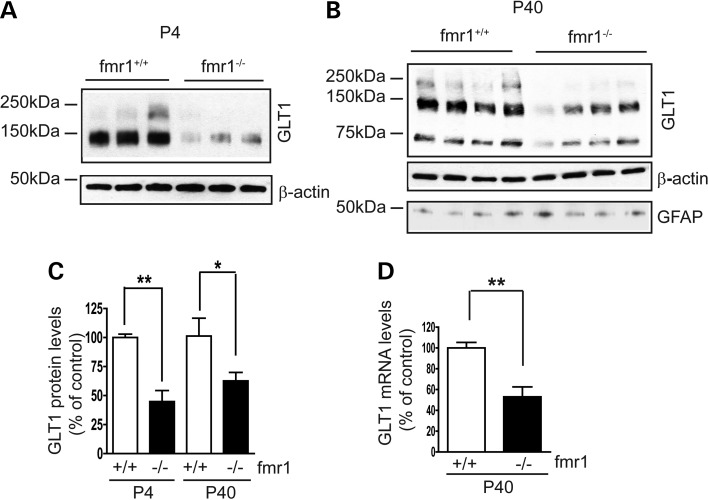
Rapid clearance of extracellular glutamate is one of the most important functions of peri-synaptic astrocytes, and is largely mediated by the astroglial glutamate transporters GLT1 and GLAST (16). In particular, the expression of GLT1 is strongly induced in the CNS during early post-natal development (28,29), becoming the physiologically dominant glutamate transporter for clearance of extracellular glutamate in vivo. In contrast, GLAST is a relatively dominant glutamate transporter in developing astrocytes, and its expression levels undergo only a modest increase during post-natal development. We determined alterations of major glutamate transporter subtypes (GLT1, GLAST and EAAC1) expression levels in the fmr1−/− mouse cortex at multiple developmental stages (as early as P4), since FXS is a developmental disorder. As shown in Figure 2A–C, GLT1 protein levels are significantly reduced in the fmr1−/− mouse cortex compared with that of the WT (fmr1+/+) mice at both P4 (55%) and P40 (40%). This reduction in GLT1 persists into adulthood (P60 and P120, Supplementary Material, Fig. S2A–C). Expression of GLAST was also reduced in the fmr1−/− cortex from P4 to P14 (50 and 30% respectively), but not at P24 or P40 (Supplementary Material, Fig. S2D and E). In contrast, protein levels of neuronal glutamate transporter EAAC1 were not significantly altered in fmr1−/− mice during development (Supplementary Material, Fig. S2F and G). Astrocyte reactivity, indicated by the GFAP expression levels, was unchanged in fmr1−/− mice (Fig. 2B), consistent with the clinical observation that there is no dramatic reactive astrogliosis in FXS patients (30). Significantly reduced (50%) GLT1 mRNA levels, determined by QRT–PCR with GLT1-specific probes, were also found in fmr1−/− mice (Fig. 2D). These results show that GLT1 expression is transcriptionally dysregulated in fmr1−/− mice.
Figure 2.
Transcriptional dysregulation of astroglial glutamate transporter GLT1 expression in the cortex of fmr1−/− mice. A representative immunoblot of GLT1 protein expression in the cortex of fmr1−/− mice at P4 (A) and P40 (B) glutamate transporter (including GLT1) immunoblots often show monomer (62 kDa), dimer (120 kDa) and sometimes multimers (250 kDa), as previously described (16,17). Significantly reduced GLT1 protein (C) and mRNA (D) levels in the cortex of fmr1−/− mice at P4 and P40; n = 5–6 mice/group; All monomer, dimer and multimers (if present) were used for GLT1 quantification. *P < 0.05; **P < 0.01 from Student's t-test.
Reduced glutamate uptake and enhanced neuronal excitability in fmr1−/− mice
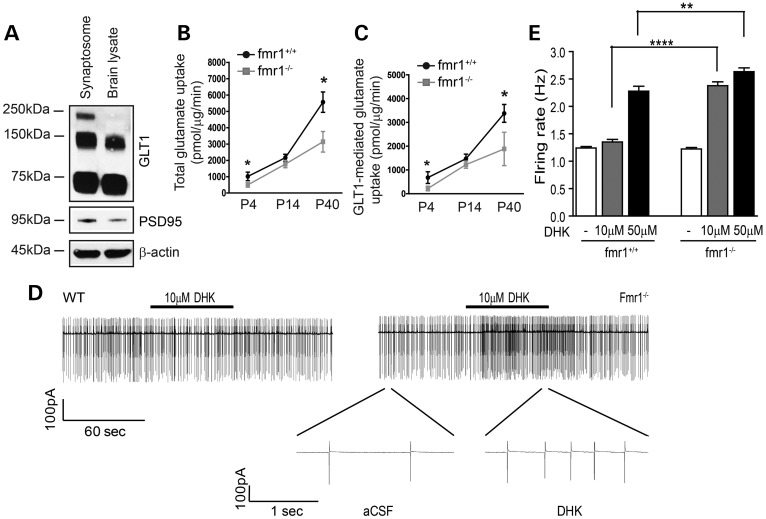
To investigate whether decreased GLT1 (and GLAST) protein leads to reduced clearance of extracellular glutamate, we performed the functional glutamate uptake assay using crude synaptosome preparation procedures (31,32) and [3H] glutamate, with the presence of dihydrokainate (DHK, a GLT1-specific inhibitor, 500 μm) or threo-β-benzyloxyaspartic acid (TBOA) (a pan-glutamate transporter inhibitor, 500 μm). This procedure has been widely and effectively used for determining glutamate transporter activity (16,18). We first confirmed the presence of GLT1 and a known post-synaptic density protein (PSD95) in crude synaptosome preparations by immunoblot (Fig. 3A). Using such preparations, we found that both total and GLT1-mediated glutamate uptake were significantly reduced in the fmr1−/− cortex as early in development as P4 (Fig. 3B and C), consistent with our GLT1/GLAST immunoblot results (Fig. 2A and B; Supplementary Material, Fig. S2D). Glutamate uptake was nearly abolished by adding TBOA (a pan-glutamate transporter inhibitor, 500 μm) to the reaction or by removing Na+ from the reaction buffer (data not shown), confirming that the glutamate uptake measured in our assay was indeed mediated specifically by Na+-dependent glutamate transporters.
Figure 3.
Reduced glutamate uptake and enhanced neuronal excitability in fmr1−/− mice. (A) Immunoblot of GLT1 protein in crude synaptosome preparation from P40 cortex. Total (B) and GLT1-mediated (C) functional glutamate uptake levels in the cortex of fmr1−/− mice during post-natal development n = 6 mice. DHK: 500 μm; TBOA: 500 μm. *P < 0.05; **P < 0.01 from Student's t-test. (D) A representative example of firing rate in a layer 5 somatosensory neocortical neuron in fmr1+/+ (left) control and fmr1−/− (right) cortical slices before and during bath application of GLT1 inhibitor DHK (10 μm). Segment of same trace in fmr1−/− slice is shown in more expanded time scale. (E) Group data showing the effects of DHK on a neuronal firing rate in fmr1+/+ (n = 9) and fmr1−/− (n = 9) cortical slices with 10 and 50 μm DHK application. **P < 0.01, ****P < 0.0001 from one-way ANOVA with the Bonferroni post-test.
To determine whether reduced GLT1 expression and glutamate uptake contributes to enhanced neuronal excitability in fmr1−/− mice, we performed cell-attached voltage-clamp recordings and monitored changes in firing activity of layer 5 somatosensory pyramidal neurons following application of the GLT1-specific inhibitor DHK to acute cortical slices. The application of low dose (10 μm) DHK will partially block GLT1 activity and potentiate the neuronal activation, which helps distinguish differences in extracellular glutamate environment between fmr1+/+ and fmr1−/− mice. Bath application of DHK in fmr1−/− cortical slices resulted in a 2-fold increase (Fig. 3D and E) in the firing rate compared with that in fmr1+/+ slices. Similar difference in neuronal firing between fmr1+/+ and fmr1−/− cortical slices persists in the presence of increased DHK (50 μm) application (Fig. 3E), consistent with the reduced uptake of extracellular glutamate in the fmr1−/− cortex.
Loss of astroglial but not neuronal FMRP is associated with reduced neuron-dependent GLT1 expression in astrocytes
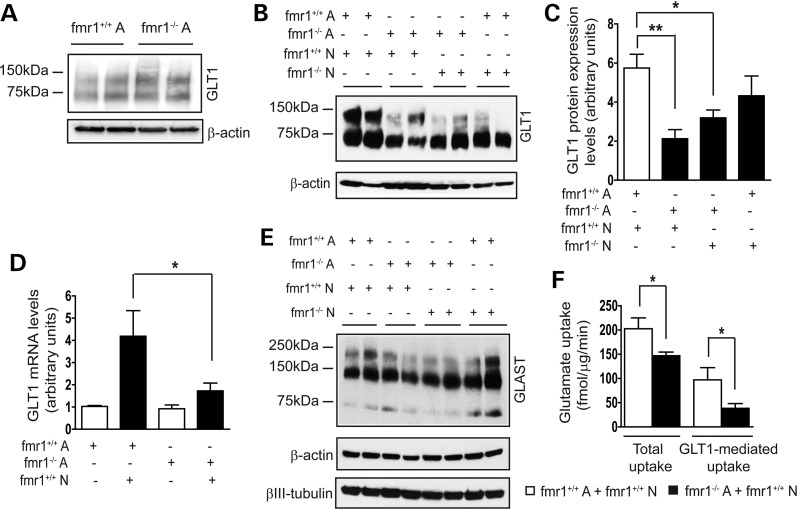
Neuron-dependent GLT1 expression in astrocytes has been previously established as an in vitro model to study GLT1 expression regulation (33,34). Primary astrocytes alone express minimal GLT1 mRNA and protein; however, GLT1 mRNA and protein levels are greatly increased in astrocytes co-cultured with neurons (34,35), mimicking the strong GLT1 induction in astrocytes during early post-natal development (29). In contrast, low levels of GLT1 mRNA are expressed in cultured neurons regardless of the presence of astrocytes (36). To investigate the mechanisms underlying GLT1 transcriptional dysregulation in fmr1−/− mice, we examined GLT1 expression in mismatched astrocyte and neuron co-cultures. We first found that GLT1 protein expression levels in FMRP-deficient astrocytes alone are unchanged comparing with those in WT astrocytes (Fig. 4A). In all co-cultures, we plated similar number of neurons on top of astrocytes, indicated by the equal βIII-tubulin levels (Fig. 4E). Consistent with the previous study (15), no obvious cytotoxicity was observed in any of these co-cultures as evidenced by the LIVE/DEAD cell assay (data not shown).
Figure 4.
Loss of astroglial FMRP attenuates neuron-dependent GLT1 expression in astrocytes. (A) Comparable GLT1 expression levels in cultured fmr1−/− and WT control astrocytes (B) A representative immunoblot of GLT1 protein in mismatched astrocyte and neuron co-cultures from fmr1+/+ or fmr1−/− mice; (A) astrocytes; N, neurons. (C) Differential expression levels of GLT1 protein expression in mismatched astrocyte and neuron co-cultures. n = 4–6 independent co-cultures/group; *P < 0.05; **P < 0.01 from Student's t-test; (D) reduced GLT1 mRNA induction in fmr1−/− astrocyte and wild-type neuron co-cultures; n = 6 independent co-cultures/group; *P < 0.05 from one-way ANOVA with the Bonferroni post-test; (E) a representative immunoblot of GLAST protein in all mismatched co-cultures; (F) total and GLT1-mediated glutamate uptake levels in fmr1−/− astrocyte and WT neuron co-cultures; n = 5 independent co-cultures/group; DHK: 500 μm; TBOA: 500 μm; *P < 0.05 from Student's t-test.
The neuron-dependent up-regulation of GLT1 in astrocytes seen in WT (fmr1+/+) co-cultures was reduced in all other mismatched co-cultures (Fig. 4B and C). In particular, fmr1−/− astrocyte and fmr1+/+ neuron co-cultures have the lowest GLT1 protein levels (65% less than those of WT co-cultures). GLT1 protein levels from fmr1−/− astrocyte and fmr1−/− neuron co-cultures were also significantly reduced by 50% compared with those of WT co-cultures. In contrast, GLT1 protein levels in fmr1+/+ astrocyte and fmr1−/− neuron co-cultures were only slightly reduced (20%) compared with those of WT co-cultures. Although differentially reduced GLT1 protein levels from these mismatched co-cultures suggest that the loss of FMRP in both neurons and astrocytes can contribute to the reduced GLT1 expression, the loss of astroglial FMRP is particularly associated with significantly reduced neuron-dependent GLT1 expression in fmr1−/− astrocytes (65% less than that in WT co-cultures). Consistent with the reduced GLT1 protein levels, 60% less GLT1 mRNA was induced in fmr1−/− astrocyte and fmr1+/+ neuron co-cultures compared with that in WT co-cultures (Fig. 4D). In contrast, we found no significant changes in GLAST protein expression in these mismatched co-cultures (Fig. 4E). We also measured functional glutamate uptake in fmr1−/− astrocyte and WT neuron co-cultures using [3H] glutamate. As shown in Fig. 4F, both total and GLT1-mediated glutamate uptake were significantly reduced (25 and 50%, respectively) compared with that of WT co-cultures, consistent with the reduced GLT1 protein expression in these co-cultures.
Loss of astroglial FMRP selectively reduces mGluR5 signaling in astrocytes
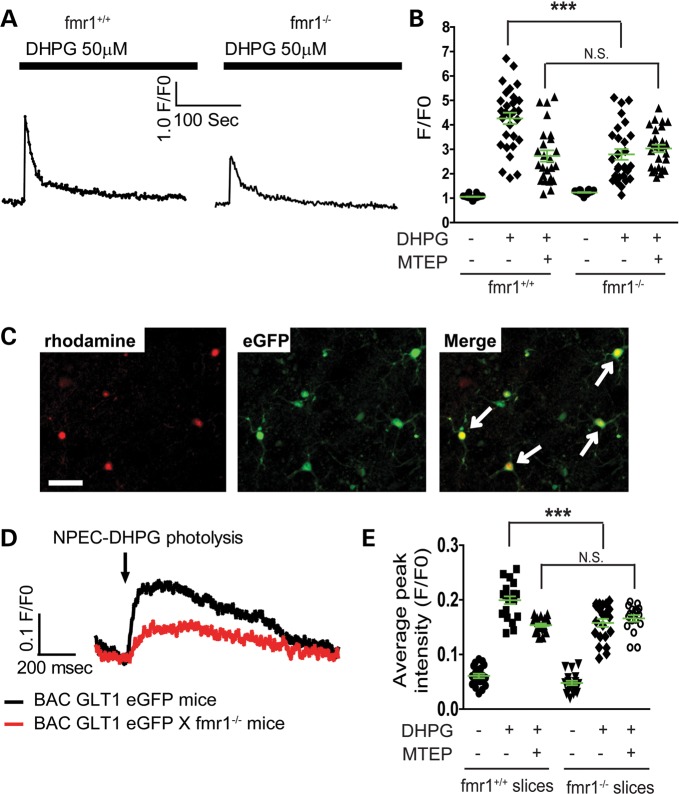
Gq-coupled metabotropic glutamate receptors, especially the group I mGluRs on the astroglial plasma membrane, are central in mediating neuron to astrocyte communication and several important astroglial functions (37). Previous studies have implicated the involvement of mGluR signaling in GLT1 regulation (33). As the fmr1−/− astrocyte and WT neuron co-cultures have the lowest GLT1 expression levels (Fig. 4B and C), we investigated whether the loss of astroglial FMRP particularly alters membrane receptor functions in astrocytes. Primary astrocytes prepared from fmr1+/+ and fmr1−/− mice were treated with (S)-3,5-dihydroxyphenylglycine (DHPG) (50 μm), γ-aminobutyric acid (GABA) (2 μm) and dopamine (20 μm), which are selective agonists of mGluR1/5, GABA and dopamine receptors, respectively. Altered signaling of these receptors in astrocytes was determined by monitoring Ca2+ responses using Ca2+ indicator Fluo4-AM. In WT astrocytes, DHPG treatment increases intracellular Ca2+ (Fig. 5A and B), as a result of the mGluR1/5 activation and production of the second messenger inositol trisphosphate. This DHPG-dependent Ca2+ increase in astrocytes is partially reduced by co-treatment with 3-((2-Methyl-4-thiazolyl)ethynyl)pyridine (MTEP) (50 nm), a mGluR5-specific antagonist (Fig. 5B), confirming that DHPG activates both mGluR1 and mGluR5 receptors in astrocytes. Although CHPG is the selective mGluR5 agonist, CHPG is prepared in DMSO solution, which induces non-specific responses when added to cultured astrocytes. Interestingly, the DHPG-dependent intracellular Ca2+ increase was significantly reduced in fmr1−/− astrocytes (Fig. 5A and B) and no further decrease in the DHPG-stimulated Ca2+ response was observed when fmr1−/− astrocytes were co-treated with MTEP (Fig. 5B), suggesting that mGluR5 signaling is dysfunctional in fmr1−/− astrocytes. In contrast, mGluR1-mediated Ca2+ responses in fmr1+/+ and fmr1−/− astrocytes, determined by the remaining Ca2+ responses in the presence of both DHPG and MTEP (column 3 and 6 in Fig. 5B), are unaltered. GABA and dopamine-induced Ca2+ increases in fmr1−/− astrocytes are also comparable with those observed in WT astrocytes (Supplementary Material, Fig. S3A and B), indicating that the mGluR5 signaling is selectively disrupted in fmr1−/− astrocytes.
Figure 5.
Selective reduction in astroglial mGluR5-dependent Ca2+ responses in fmr1−/− astrocytes. (A) DHPG-induced Ca2+ responses in fmr1−/− and control fmr1+/+ astrocyte; (B) quantitative analysis of mGluR5-dependent Ca2+ responses in fmr1+/+ and fmr1−/− astrocytes; n = 25–27 astrocytes/group. (C) Identification of rhodamine 2-AM loaded astrocytes (highlighted with white arrows) in BAC GLT1 eGFP (or BAC GLT1 eGFP X fmr1−/−) mouse cortical slices. Scale bar: 100 μm. (D) Representative DHPG-induced Ca2+ responses in BAC GLT1 eGFP X fmr1−/− and control BAC GLT1 eGFP cortical slices; (E) quantitative analysis of the mGluR5-dependent Ca2+ response in BAC GLT1 eGFP X fmr1−/− and control BAC GLT1 eGFP cortical slices; n = 17–21 astrocytes from three mice/group, ***P < 0.001 from one-way ANOVA with the Bonferroni post-test; NS, not significant.
We next measured the astroglial mGluR5-dependent Ca2+ response in acute cortical slices from fmr1−/− mice. To identify the cortical astrocytes in slices, we bred BAC GLT1 eGFP reporter mice with fmr1−/− mice. Acute cortical slices were prepared from BAC GLT1 eGFP or BAC GLT1 eGFP X fmr1−/− mice (P15–20) and bulk-loaded with the Ca2+ indicator rhodamine 2-AM. 1-(2-nitrophenyl) ethoxycarbonyl (NPEC)-caged DHPG (38) was bath-applied (100 μm) and flash-photolysed to uncage the DHPG with a UV beam (10 μm diameter for 10 ms) on a single astrocyte identified by eGFP and rhodamine 2-AM fluorescence (Fig. 5C). High speed time-lapse images of astroglial Ca2+ responses were collected during photolysis. In BAC GLT1 eGFP cortical slices, uncaging DHPG elicited robust increases in astrocyte intracellular Ca2+ (Fig. 5D and E). In contrast, uncaging DHPG induced only weak Ca2+ increases in fmr1−/− cortical astrocytes (Fig. 5D and E). Treatment of MTEP and DHPG uncaging induced no further reduction in Ca2+ responses in eGFP+ fmr1−/− cortical astrocytes, whereas the DHPG uncaging induced the Ca2+ response was significantly reduced in BAC GLT1 eGFP cortical astrocytes (Fig. 5E) following MTEP treatment, confirming that mGluR5 signaling becomes dysfunctional in fmr1−/− cortical astrocytes.
FMRP regulates translational expression of mGluR5 in astrocytes
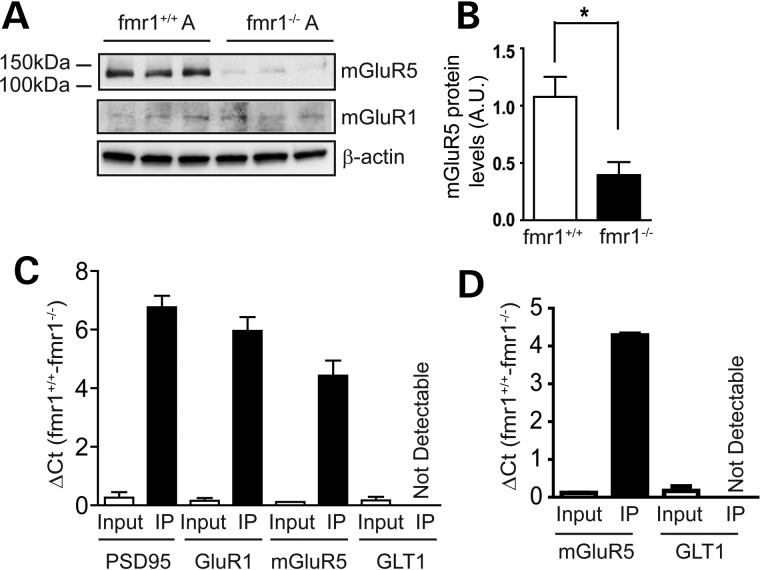
We also determined mGluR5 protein levels in primary fmr1−/− and WT (fmr1+/+) astrocytes. Consistent with the reduced functional response, total mGluR5 (but not mGluR1) protein levels were dramatically reduced in fmr1−/− astrocytes, as determined by immunoblotting (Fig. 6A and B). In contrast, mGluR5 protein expression in fmr1−/− neurons is unchanged (39). Interestingly, mGluR5 mRNA levels were not significantly altered in fmr1−/− astrocytes (Supplementary Material, Fig. S3C). In addition, treatment of MG132, a potent proteasome inhibitor, resulted in comparable increase in mGluR5 protein levels in cultured fmr1+/+ and fmr1−/− astrocytes (Supplementary Material, Fig. S3D and E), suggesting that the mGluR5 protein turn-over rate is not abnormally altered in fmr1−/− astrocytes. These results suggest that FMRP may positively regulate the translation of mGluR5 mRNA in astrocytes. We then determined whether FMRP is directly associated with mGluR5 or GLT1 mRNA in cortical tissue and in astrocyte cultures. QRT–PCR analysis of the mRNA isolated from the FMRP immunoprecipitation (IP) complex revealed abundant mGluR5 (IP, significant ΔCt between fmr1+/+ and fmr1−/− samples, Fig. 6C and D) but not GLT1 mRNA (not detectable, Ct values from IP are in background range) in both cortical and cultured samples. FMRP IP from the fmr1−/− cortex was used as a negative control to determine the background Ct value (≥33) for each mRNA from QRT–PCR. Equal quantity of RNA was inputted for FMRP IP in both fmr1+/+ and fmr1−/− samples (Input, minimal ΔCt between fmr1+/+ to fmr1−/− samples, Fig. 6C and D). We also detected abundant PSD95 and GluR1 mRNA (significant ΔCt between fmr1+/+ and fmr1−/− samples, Fig. 6C and D), two previously identified FMRP targets (39,40), in the IP complex from cortex samples to validate our IP and QRT–PCR procedures. Overall, these biochemical results show that astroglial mGluR5 mRNA is associated with FMRP in astrocytes and mGluR5 protein expression is positively regulated by FMRP in astrocytes.
Figure 6.
FMRP-dependent translational regulation of mGluR5 expression in astrocytes. (A) A representative immunoblot of mGluR5 and mGluR1 in fmr1−/− and fmr1+/+ control astrocytes. (B) Quantification of mGluR5 protein in fmr1−/− and control fmr1+/+ astrocytes; n = 4–5 independent cultures/group; QRT–PCR detection of FMRP-associated mRNA following FMRP immunoprecipitation (IP) from the cortex (C) or cultured astrocytes (D). RNA was analyzed using QRT–PCR following IP. Positive association was shown with significant ΔCt between fmr1+/+ and fmr1−/− samples. Equal quantity of RNA was inputted for IP in both fmr1+/+ and fmr1−/− samples. All Ct values for input are 18–25, depending on different mRNA transcripts. FMRP IP from fmr1−/− mice was used as a negative control to determine the background Ct value (≥33) for each mRNA from QRT–PCR. The Ct value for GLT1 mRNA from FMRP IP is >37 in all samples, though its Ct value in input is 22–25. Results were averaged from two separate IP experiments. *P < 0.05 from Student's t-test;
Loss of astroglial mGluR5 attenuates neuron-dependent GLT1 expression in astrocytes
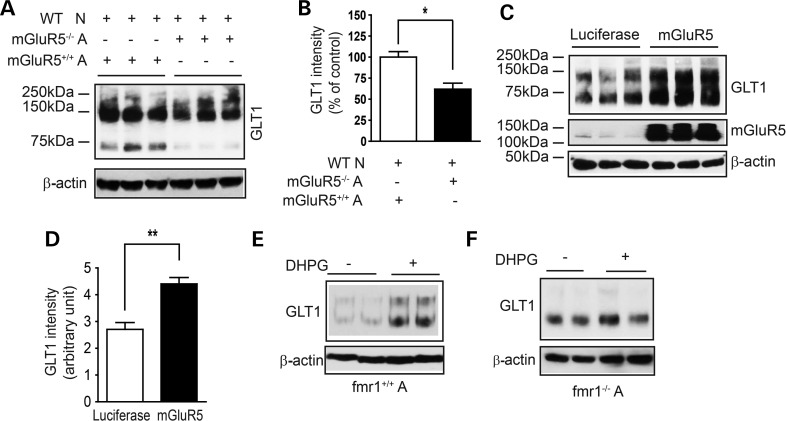
Whether the astroglial mGluR5 receptor directly mediates neuron-dependent GLT1 regulation has not previously been determined. As mGluR5 is expressed in both neurons and astrocytes, pharmacological inhibition of mGluR5 in astrocyte and neuron co-cultures will inhibit mGluR5 in neurons as well. Instead, a selective genetic elimination of astroglial mGluR5 will allow direct examination of its involvement in neuron-dependent GLT1 regulation. Primary mGluR5−/− astrocytes were prepared from the mGluR5−/− mice (a kind gift from Dr Mark Bear, Massachusetts Institute of Technology). Freshly prepared WT neurons were added on top of the confluent mGluR5−/− astrocytes to stimulate GLT1 expression. GLT1 expression levels were then determined following neuronal stimulation, and compared with those in WT (mGluR5+/+) astrocyte and neuron co-cultures. As shown in Figure 7A and B, GLT1 protein levels in mGluR5−/− astrocyte and WT neuron co-cultures are 45% decreased compared with those in the WT co-cultures, indicating that astroglial mGluR5 is important in mediating neuron-dependent GLT1 expression in astrocytes. Whether selective deletion of neuronal mGluR5 alters GLT1 expression in co-cultures will be determined in future studies. GLT1 expression levels were also modestly reduced in the mGluR5−/− mouse cortex (Supplementary Material, Fig. S4A and B). In contrast, GLAST protein expression levels were unchanged in mGluR5−/− astrocyte and WT neuron co-cultures (data not shown). In addition, overexpression of mGluR5 in fmr1−/− astrocytes is sufficient to rescue neuron-dependent GLT1 expression in fmr1−/− astrocyte and WT neuron co-cultures (Fig. 7C and D), confirming the essential and sufficient role of astroglial mGluR5 in mediating neuron-dependent GLT1 expression in astrocytes. Moreover, we examined whether fmr1−/− astrocytes still respond to DHPG treatment to increase GLT1 expression. As shown in Figure 7E, DHPG treatment induces GLT1 up-regulation in WT astrocytes, which is completely abolished by the co-treatment of MTEP (Supplementary Material, Fig. S4C and D). However, DHPG is insufficient to induce GLT1 up-regulation in fmr1−/− astrocytes (Fig. 7F). These results further suggest that the loss of astroglial mGluR5 underlies GLT1 dysregulation in fmr1−/− astrocytes.
Figure 7.
Astroglial mGluR5 contributes to neuron-dependent GLT1 expression in astrocytes. (A) Neuron-induced GLT1 protein expression is attenuated in mGluR5-deficient astrocytes. Wild-type neurons were plated to stimulate GLT1 expression. (B) Quantitative analysis of the GLT1 expression levels in mGluR5-deficient astrocyte and wild-type neuron co-cultures; n = 4 independent co-cultures/group. All monomer, dimer and multimers were used for GLT1 quantification. (C) mGluR5 overexpression in fmr1−/− astrocytes rescues neuron-dependent GLT1 expression. Wild-type neurons were plated to stimulate GLT1 expression. (D) Quantification of GLT1 expression levels in mGluR5-overexpressed fmr1−/− astrocyte and wild-type neuron co-cultures. DHPG (50 μm) treatment significantly increases GLT1 expression in cultured fmr1+/+ (E) but not in fmr1−/− (F) astrocytes; n = 4. *P < 0.05, **P < 0.01 from Student's t-test.
Re-expression of FMRP in fmr1−/− astrocytes rescues mGluR5 expression/function and neuron-dependent GLT1 expression
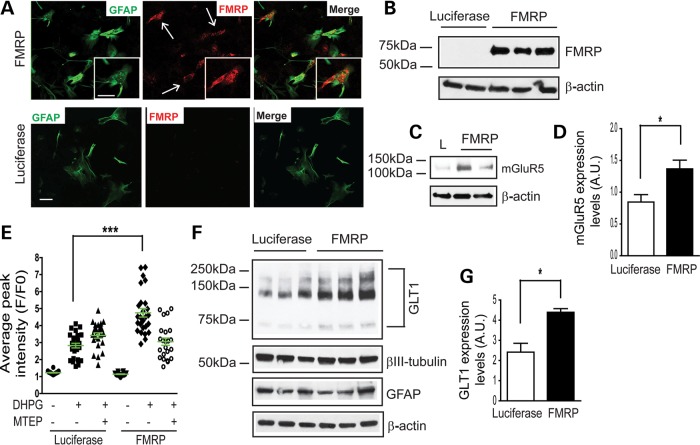
We next tested the rescue effect of FMRP re-expression in cultured fmr1−/− astrocytes on mGluR5 expression/function and neuron-dependent GLT1 expression. FMRP cDNA (41) (a kind gift of Dr Elizabeth Berry-Kravis, Rush University Medical Center) was transiently transfected into cultured fmr1−/− astrocytes, and re-expression of FMRP was confirmed by immunostaining (Fig. 8A) and immunoblotting (Fig. 8B). We achieved 70–80% of FMRP re-expression, indicated by the overlap of FMRP and GFAP immunoreactivity in cultured astrocytes. In contrast, no FMRP immunoreactivity was observed in fmr1−/− astrocytes transfected with luciferase cDNA (negative control). Following the re-expression of FMRP in fmr1−/− astrocytes, mGluR5 protein levels were significantly increased compared with control luciferase-transfected fmr1−/− astrocytes (Fig. 8C and D). In addition, DHPG-induced Ca2+ responses in FMRP transfected fmr1−/− astrocytes were significantly increased compared with those in luciferase-transfected fmr1−/− astrocytes, as determined by Fluo4-AM fluorescence (Fig. 8D). Importantly this DHPG-induced Ca2+ increase was abolished by MTEP, confirming that mGluR5-dependent Ca2+ responses were restored. These results further confirm that astroglial mGluR5 protein expression and function is regulated by astroglial FMRP. We also determined whether FMRP re-expression restores neuron-dependent GLT1 expression in fmr1−/− astrocytes. WT neurons were plated on top of FMRP or luciferase cDNA transfected fmr1−/− astrocytes. As shown in Figure 8E and F, a significant increase in GLT1 expression (45%) was observed with rescued co-cultures relative to controls. This increase almost completely restored the reduced GLT1 expression levels observed in fmr1−/− astrocyte and WT neuron co-cultures, further suggesting that the loss of astroglial FMRP may particularly attenuate neuron-dependent GLT1 expression in fmr1−/− astrocytes.
Figure 8.
Re-expression of FMRP in fmr1−/− astrocytes rescues astroglial mGluR5 expression/function and neuron-dependent GLT1 expression. Re-expression of FMRP in cultured fmr1−/− astrocytes is evidenced by FMRP immunostaining (A) and immunoblot (B). Luciferase (L) cDNA was used as a negative control. A magnified view was shown in the insert image. Scale bar: 50 μm; (C) a representative mGluR5 immunoblot from FMRP re-expressed fmr1−/− astrocytes; (D) quantification of the mGluR5 expression in FMRP re-expressed fmr1−/− astrocytes. n = 4. *P < 0.05 from Student's t-test; (E) DHPG-induced Ca2+ responses in FMRP re-expressed astrocytes n = 24–27 astrocytes/group ***P < 0.001 from one-way ANOVA with the Bonferroni post-test; (F) Neuron-dependent GLT1 expression in FMRP re-expressed astrocyte and wild-type neuron co-cultures; Wild-type neurons were plated 3 days following transfection. Luciferase cDNA transfection serves as a control. (G) Quantitative analysis of GLT1 protein expression levels following FMRP re-expression in co-cultures. n = 6–8 independent transfections/group, *P < 0.05 from Student's t-test.
DISCUSSION
Our studies provide evidence for a new activation function of FMRP in regulating protein expression in astrocytes. The astroglial loss of FMRP in fmr1−/− mice reduces neuron-dependent GLT1 expression in astrocytes, at least partially through the translational down-regulation of astroglial mGluR5. As GLT1 plays critical roles in regulating glutamate homeostasis in the mammalian CNS, its down-regulation and reduced glutamate uptake may shed new light on the roles of astrocytes in the pathogenesis of FXS.
Expression of FMRP in mature astrocytes in vivo
Expression of FMRP has been found in developing astrocytes and oligodendrocytes, but not in the glia of the mature brain (12,13). These previous studies examined FMRP expression in glia cultures, and often performed co-immunostaining of FMRP with a glia-specific marker, i.e. GFAP for astrocytes and MBP for oligodendrocytes, in brain tissue sections. These studies have not found evidence for FMRP expression in mature glia in vivo. However, these previous studies were potentially compromised by the co-immunostaining approach, especially immunostaining of GFAP preferentially labels major branches that are stemmed from astrocyte soma in vivo (42) and FMRP is a cytoplasmic protein. It is, therefore, challenging to co-localize FMRP and GFAP immunoreactivity in mature astrocytes in vivo. In addition, GFAP immunostaining only labels a limited number of cortical astrocytes in non-pathological conditions in the adult brain (43) and is also expressed as a progenitor marker during development and in the subventricular zone in the adult brain (44). We employed astrocyte reporter BAC GLT1 eGFP mice in which the eGFP reporter is highly expressed in the soma of vast majority of cortical astrocytes, driven by the genomic GLT1 promoter that is highly activated in mature astrocytes in vivo (27). This mouse tool greatly facilitates the detection of FMRP immunoreactivity in mature astrocytes in vivo. In addition, we also found sufficient translating FMRP mRNA in both developing and mature cortical astrocytes using TRAP isolation from the BAC ALDH1L1 TRAP mice, providing further evidence that FMRP is expressed in mature cortical astrocytes in vivo.
Activation function of FMRP in regulating protein expression in astrocytes
Although FMRP is known to bind ∼4% of mRNA transcripts (6), whether FMRP regulates protein translation in astrocytes has not previously been explored. The majority of previous studies have suggested that FMRP suppresses protein translation in neurons. However, our current studies provide intriguing evidence that FMRP can positively regulate the translational expression of mGluR5 in astrocytes. Interestingly, mGluR5 protein expression is not altered in cultured fmr1−/− neurons (39). The distinctive role of FMRP in regulating mGluR5 expression in astrocytes not in neurons suggests a cell-type-specific function of FMRP in the mammalian CNS.
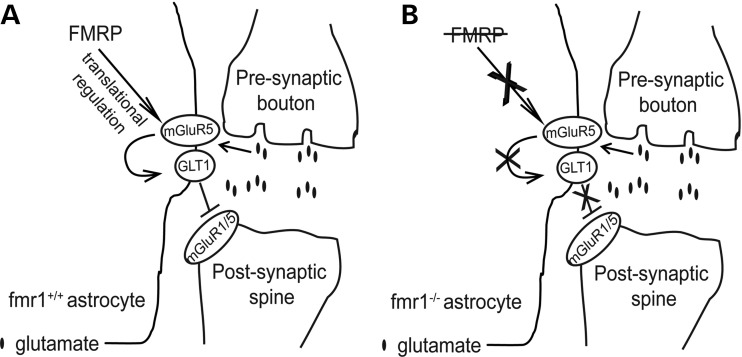
Although GLT1 mRNA has been implicated as one of the FMRP targets in a recent study (45), our results showed that FMRP is not associated with GLT1 mRNA in astrocytes. Notably, low levels of GLT1 mRNA are expressed in cultured cortical neurons and a subset of hippocampal neurons in vivo (36,46), so it is possible that FMRP is associated with GLT1 mRNA in these neurons rather than in astrocytes. In addition, GLT1 protein levels in FMRP-deficient astrocytes alone remain unchanged compared with those in WT astrocytes. Moreover, GLT1 expression is transcriptionally dysregulated with not only reduced protein levels, but also reduced promoter activity and mRNA levels in fmr1−/− mice and co-cultures. These results indicate that it is unlikely that astroglial FMRP directly regulates GLT1 expression at the translational level in astrocytes. Whether astroglial FMRP directly regulates GLT1 transcription remains to be further investigated. Although reduced GLT1 expression is often associated with unhealthy neurons, our results identified the dysfunctional astrocyte pathway, i.e. the reduced mGluR5 signaling resulted from the loss of astroglial (not neuronal) FMRP, may contribute to the dysregulation of neuron-dependent GLT1 expression in fmr1−/− astrocytes (Fig. 9). This offers a new perspective on the pathological mechanisms underlying GLT1 dysregulation in several neurological diseases/disorders. Potential neuronal dysfunctional pathway/signaling involved in GLT1 dysregulation in FXS condition will be investigated in future studies.
Figure 9.
Schematic diagram illustrating the mechanisms underlying the GLT1 dysregulation in fmr1−/− astrocytes (A) FMRP-mediated translational regulation of mGluR5 contributes to neuron-dependent GLT1 expression in fmr1+/+ astrocytes. (B) Translational down-regulation of mGluR5, as a result of the loss of astroglial FMRP, underlies GLT1 dysregulation in fmr1−/− astrocytes.
Dysregulation of astroglial glutamate transporters and synaptic abnormalities in FXS
Active uptake of extracellular glutamate through peri-synaptic astroglial glutamate transporters, especially GLT1, plays important roles in maintaining glutamate homeostasis and limiting the ‘spillout’ and ‘spillover’ of glutamate in the mammalian CNS (19). Manipulation of GLT1 (and GLAST) activity and expression has been shown to significantly modulate synaptic activation, especially mGluR-dependent synaptic activity (19,21,22). Therefore, down-regulation of astroglial glutamate transporters, especially GLT1 in fmr1−/− mice may increase extracellular glutamate levels and induce abnormal mGluR1/5-dependent synaptic activity in FXS. Although it remains uncharacterized whether mGluR1/5 is abnormally activated in FXS, effective reversal of typical FXS phenotypes by inhibiting mGluR5 activation in mouse models of FXS at both developing and adult stages (47,48) implicates potentially enhanced mGluR5 activation in FXS. Indeed, enhanced neuronal excitability has been found in the cortex of fmr1−/− mice (49,50), presumably due to the reduced interneuron inhibition (50). We have also observed enhanced pyramidal neuronal excitability in fmr1−/− (but not fmr1+/+) cortical slices following partial inhibition of GLT1 activity (Fig. 3D and E), suggesting that reduced GLT1 expression directly contribute to enhanced neuronal excitability in pyramidal neurons in the fmr1−/− cortex. This GLT1 (and/or GLAST)-dependent modulation of neuronal (mGluR1/5) activation provides a new astrocyte-mediated mechanism that may contribute to synaptic abnormalities in FXS (Fig. 9). Importantly, the loss of GLT1 (and GLAST) was found as early as P4 in fragile X mice, suggesting that an abnormal expression of astroglial glutamate transporters and reduced glutamate uptake is likely an early causative event for inducing abnormal neuronal activity, not a consequence of abnormal neuronal development in the fmr1−/− mice at a later stage.
Although GLT1 is also known to play a critical role in preventing excitotoxicity (51), the observation that no significant neuronal cell death in FXS patients (30) or fmr1−/− mice suggests that reduction in GLT1 expression in fmr1−/− mice is involved in altering synaptic activity rather than inducing excitotoxicity. Interestingly, elevated extracellular glutamate levels (30% increase) have been observed in GLT1+/− mice where GLT1 expression is reduced by 50% (23), similar to the loss of GLT1 expression observed in fmr1−/− mice in our study (Fig. 2C). Whether typical FXS phenotypes, such as abnormal dendritic spine number/morphology, protein synthesis and increased susceptibility to seizure, are altered in GLT1+/− mice, will be tested in GLT1+/− mice in the future.
MATERIALS AND METHODS
Animals
FVB WT control (fmr1+/+) and fmr1 knockout (fmr1−/−) mice (FVB.129P2(B6) Fmr1-tm1Cgr) were obtained from Jackson Laboratories. Hemizygous BAC GLT1 eGFP mice are a kind gift from Dr Jeffrey Rothstein (Johns Hopkins University); Hemizygous BAC GLT1 eGFP mice were bred with fmr1−/− mice and offsprings (F1) were used as a control for FMRP immunostaining. Hemizygous BAC ALDH1L1 TRAP mice were obtained from the GENSAT project (Rockefeller University). We used only male fmr1−/− mice (or astrocyte cultures from male pups) in all experiments because the fmr1 locus is on the X chromosome and males are more severely affected than females among FXS patients and mouse models (52). Care and treatment of animals in all procedures strictly followed the NIH Guide for the Care and Use of Laboratory Animals and the Guidelines for the Use of Animals in Neuroscience Research and the Tufts University IACUC.
Primary cultures and astrocyte transfection
Cortices of post-natal Day 0–3 (P0–P3) mouse pups were used for astrocyte cultures, as previously described (53) without supplement of cAMP. Cells were plated at a density of 1.0 × 106 cells/ml in 6-well plate or in 100 mm dish. Astrocyte medium is composed of Dulbecco minimum essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Sigma, St Louis, MO, USA) and 1% penicillin/streptomycin. Astrocytes become 90% confluent at 6–7 days in culture and are ready for experiment. For MG132 treatment, MG132 (final conc. 1 μm) was added to the confluent astrocyte cultures in serum-free DMEM for 4 h. For astrocyte and neuron co-cultures, cortical neurons were isolated from embryonic 14–16-day-old mouse brains. The co-culture medium is composed of neurobasal medium, 2% B27 neurobasal supplement, 5% FBS, 2 mm glutamine, 1% of 100x GlutaMAX and 1% penicillin/streptomycin. Freshly prepared neurons (1–2 × 106 cells) were plated on top of astrocytes that had grown on 6-well cell culture plate for 6–7 days. Astrocyte transfection with mGluR5, fmr1 or luciferase cDNA (Promega) was performed using DharmaFECT transfection reagent (Thermo Scientific) as described in the kit. Neurons (1–2 × 106) were added on the transfected astrocytes 72 h following transfection, then the co-cultures were kept for another 6 days.
FMRP immunostaining and confocal imaging
Double-immunostaining of MAP2 and FMRP was performed. Monoclonal antibody 2F5 (Developmental Studies Hybrodoma Bank, IA, USA) was used for FMRP immunostaining with hemizygous BAC GLT1 eGFP or BAC GLT1 eGFP X fmr1−/− mice (negative control). Mice were perfused with 4% paraformaldehyde and cryoprotected with 30% sucrose solution. Thin brain sections (10–20 μm) were prepared using cryostat (Thermo Scienfic) and incubated with 0.01% sodium citrate (pH 6.0) at 85°C for 20 min for antigen retrieval procedure. Brain sections were then incubated with blocking solution [0.01% Triton ×100; 5% goat serum; 1% bovine serum albumin (BSA)] for 30 min followed by overnight incubation with primary antibody (1:1 mAb 2F5) at 4°C. Following overnight incubation brain sections were washed three times with PBS and the FMRP signal was detected with Alexa-555-conjugated goat anti-mouse secondary (1:2000) antibody (Jackson ImmunoResearch, West Grove, PA, USA). MAP2 immunostaining was counterstained following the same procedure. Confocal Z-stack (15 μm) images of eGFP+ astrocytes and FMRP immunoreactivity were captured by the Nikon A1R confocal microscope magnified with 40× (numerical aperture 0.8) and 60× (numerical aperture 1.0) objectives (Nikon Instruments, Inc., Melville, NY, USA).
TRAP mRNA extraction, FMRP immunoprecipitation and quantitative RT–PCR (QRT–PCR)
The ribosome-bound translating mRNA from astrocytes was specifically extracted following eGFP tag IP with BAC ALDH1L1 TRAP transgenic mouse tissue, as previously described (25). Briefly, anti-GFP antibody (HtzGFP-19C8, Monoclonal Antibody Core Facility, Memorial Sloan-Kettering Cancer Center) was coupled to magnetic beads (Dynabeads, Invitrogen) at a concentration of 10 μg/mg. Cortical tissue of BAC ALDH1L1 TRAP mice (P7 or P40) or WT mice (P30) was dissected and dissociated in homogenization buffer using a glass homogenizer. Supernatant was immediately incubated with prepared beads (0.5 mg beads/sample) and gently mixed in a 2 ml eppendorf tube for 1 h at RT. Beads were washed five times with 500 μl of 0.35 m KCL buffer and resuspended in 200 μl of H2O. FMRP IP was performed as described above with 10 μg anti-FMRP antibody (7G1, Developmental Study Hybrodoma Bank, IA, USA) coupled to magnetic beads. Fmr1+/+ control or fmr1−/− mice (P21) cortex and cultured WT primary astrocytes were homogenized and immunoprecipitated with 7G1 antibody. Following IP, RNA was isolated by Trizol reagent and precipitated by isoproponal. Total RNA from the cortex was prepared using an absolutely RNA Miniprep Kit (Stratagene) by following the kit's instruction. Prepared RNA (total or TRAP) was then converted to cDNA using the high archive cDNA synthesis kit (Applied Biosystems, Foster City, CA, USA). QRT–PCR was performed using either TaqMan probes (for GLT1) or specific primers (FMRP) with Syber-Green dye. Primers for FMRP, F: 5′-CATGATGTGAGATTCCCACCAC-3′, R: 5′-GGGATTAACAGATCGTAGACGC-3′. QRT–PCR primers for GFAP, GLT1, CaMKIIα, PLP1, CD11b, PDGFRα, PSD95, GluR1, mGluR5 and β-actin were chosen from the PrimerBank (http://pga.mgh.harvard.edu/primerbank/, 54). β-Actin was used as a control to normalize RNA amount in QRT–PCR.
Preparation of crude synaptosome membrane fractions and glutamate uptake assay
Glutamate uptake was performed with crude synaptosome preparation from the mouse cortex at P4, P14 and P40 using the 0.32 m sucrose centrifugation method (31). After total protein determination, 1 µCi L-3H glutamate and 100 μm non-labeled glutamate were mixed with Na+ uptake buffer (total volume 275 μl) then added into 25 μl of each synaptosome sample in 96-well multiscreen HTS filter plates (Millipore). After 6-min incubation, uptake was terminated by putting into ice bath. Samples were then filtered using the Steriflip vacuum filtration system (Millipore) and washed 6× with ice-cold PBS while continue filtering the samples. Each filtered 96-well membrane was excised out and transferred for scintillation counting. Glutamate uptake from cultures was determined by adding 2 µCi L-3H glutamate (Perkin Elmer) and 100 µm non-labeled glutamate into culture well mixed with Na+ uptake buffer (in millimolar; Tris 5, HEPES 10, NaCl 140, KCl 2.5, CaCl2 1.2, MgCl2 1.2, K2HPO4 1.2, Glucose 10) in total volume of 1 ml. After 6 min incubation at 37°C uptake was terminated by putting into ice bath, buffer was removed and reactions were washed twice with ice-cold Hank's balanced salt solution (HBSS) (Life Technologies, NY, USA). Ice-cold 0.5 ml of 0.1 N NaOH was used to lyse the cells. Aliquots of samples were transferred for scintillation counting (100 μl each with triplicate) and total protein amount was determined by the Bradford assay (Bio-Rad). DHK (500 μm) or dl-TBOA (500 μm) was added into appropriate wells in the glutamate uptake assay. The disintegration per min (DPM) value was normalized by total protein concentration and converted to pmol/μg/min or fmol/μg/min unit.
Immunoblot
Anti-GLT1 (1:5000, rabbit), anti-GLAST (1:500, rabbit) and anti-EAAC1 (1:100, rabbit) were generous gifts from J.D. Rothstein (John Hopkins University, Baltimore) and have been widely used for immunoblot (17). Glutamate transporter immunoblots often show monomer (62 kDa), dimer (120 kDa) and sometimes multimers (250 kDa), as previously described (16,17). All monomer, dimer and multimers (if present) were used for the quantification of transporter expression levels. Anti-FMRP monoclones 2F5 and 7G1 (1:1, Developmental Study Hybrodoma Bank), anti-β-actin (1:1000, Sigma), anti-GFAP (1:1000; Dako) and anti-β-III tubulin (1:1000, R&D system) were purchased. Cerebral cortices from fmr1+/+ or fmr1−/− mice were homogenized. Total protein amount was determined by the Bradford protein assay; 10 μg of cortical lysate or 50 μg of total cell lysate was loaded to 4–15% gradient SDS–PAGE gels. Separated proteins were transferred onto a polyvinylidene difluoride membrane (Bio-Rad) for 1 h. The membrane was blocked with 3% BSA in Tris buffer saline with 0.1% Tween 20 (TBST) then incubated with appropriate primary antibody overnight at 4°C. On following day, membrane was exposed to horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:5000) diluted in TBST. Bands were visualized on CL-XPosureTM film (Thermo Scientific) by ECL Plus chemiluminescent substrate (Thermo Scientific). Different exposure time was used for detecting different proteins.
Cortical slice preparation
For electrophysiological recording, cortical brain slices were prepared from juvenile (P14–28) male fmr1+/+ control or fmr1−/− mice. For measuring the Ca2+ response, cortical brain slices were prepared from P15–20 male BAC GLT1 eGFP or BAC GLT1 eGFP X fmr1−/− mice. Animals were anesthetized with ketamine (110 mg/kg)/xylazine (10 mg/kg) cocktail; the cortex was quickly removed and 300 μm cortical slices were cut using a vibrotome (Leica VT1200, Leica Microsystems) in ice-cold artificial cerebrospinal fluid (aCSF) (in millimolar): KCl 3, NaCl 125, MgCl2 1, NaHCO3 26, NaH2PO4 1.25, glucose 10, CaCl2 2 and 400 μm l-ascorbic acid, with osmolarity at 300–305 mOsm, equilibrated with 95% O2/5% CO2. Slices were incubated at RT until needed.
Calcium imaging
Cultured astrocytes Ca2+ imaging was performed using the Leica TCS SP2 confocal scanning unit. Astrocytes grown on the coverslip were incubated at RT in HBSS (pH 7.3) containing 3 μm Fluo-4 AM and pluronic acid (2.5 μg/ml). Fluorescence images of dye-loaded astrocytes were obtained using a krypton/argon laser at 488 nm and emitted light at >495 nm. Images were acquired every 3 s for 10 min. After 1 min baseline acquisition, DHPG (50 μm) or DHPG with MTEP (50 nm) (Sigma) was applied and images were acquired for another 9 min. Ca2+ imaging from astrocytes in acute cortical slices was accomplished using a Neuro-CCD-SM camera (Redshirt imaging, Decatur, GA, USA) attached to a Olympus BX51 microscope. Slices were incubated with 10 μm rhodomine-2 and pluronic acid (2.5 μg/ml) in aCSF for 1 h in RT then transferred to aCSF solution until needed. Slices were perfused with aCSF containing NPEC-DHPG (100 μm) (38), DNQX (20 μm), CPP (10 μm) and with or without MTEP hydrochloride (50 nm) at a rate of 2 ml/min. Dye-loaded astrocytes were visualized with ×60 water-immersion objective (numerical aperture 1.0) and identified by the eGFP signal. Florescence images were acquired using full-field epi-fluorscent illumination (mercury bulb). An LPSS 355 nm laser was directed at the brain slice and a 10 ms flash of laser light (10 μm diameter) was selectively targeted to a single astrocytic soma to uncage NPEC-DHPG compound. Images were acquired at one image per millisecond for 4 s. No spatial binning or photo-multiplication was used.
Electrophysiology
Tight seal cell-attached recordings (55) from pyramidal neurons in the layer 5 somatosensory cortex were obtained by Multiclamp 700B amplifier filtered at 2 kHz and sampled at 10 kHz with Digidata 1322A (Molecular Devices, Sunnyvale, CA, USA). Pipettes were pulled by model P-97 puller (Shatter instrument, Novato, CA, USA). Pipettes had resistances of 4–6 MΩ. A pClamp 9.2 (Molecular Devices) was used for data acquisition and storage. Pipettes were filled with aCSF solution. Slices were continuously perfused with aCSF with a flow rate of 1–2 ml/min. and bubbled with 95% O2/5% CO2. Neurons were identified by bright-field Nikon Eclipse e600FN microscopy with a 40× water-immersion len and infrared illumination. Recorded cells were located in the barrel hollows of the somatosensory cortex. The action potential firing rate was recorded in gap-free voltage-clamp mode. Current was injected into cells to adjust the basal firing rate to within a range of 1–2 Hz. The mean firing frequency (2 min periods) was obtained before and after 1 min bath application of 10 or 50 μm DHK.
Data analysis
In vitro astrocyte Ca2+ imaging was analyzed with Leica TCS SP2 software (Leica Microsystems). Fractional fluorescence (F/Fo) was determined by dividing the fluorescence intensity (F) within a region of interest (ROI) by a baseline fluorescence value (Fo) determined from 20 images showing no activity. The number of peaks over a given time was automatically detected from oscillations crossing a set threshold value (>1.30 F/F0). Ca2+ imaging on slices was analyzed with the MATLAB software (MathWorks, Natick, MA, USA). Ca2+ average peak intensity and fractional fluorescence (F/Fo) was determined by dividing the fluorescence intensity (F) within an ROI (10 × 10 pixels or 2.5 × 2.5 μm) by a baseline fluorescence value (Fo) determined from 300 images showing no activity. Electrophysiological recording was analyzed by pClamp version 9.2 and Clampfit 9.2 (Molecular Devices, Sunnyvale, CA, USA). All the resulting raw data were graphed and plotted in GraphPad 5 program (GraphPed software, La Jolla, CA, USA). All values are presented as mean ± SEM, with n indicating number of replicates. Corresponding statistical analysis was described in each figure legend.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by NIH (MH 099554 to Y.Y.) and by a joint post doc fellowship from Autism Science Foundation and FRAXA Research Foundation (12-1003 to H.H.).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Joshua Ainsley's technical help on isolating translating mRNA from BAC ALDH1L1 TRAP mice; Dr Leon Reijmers for providing CaMKIIα TRAP mice samples; Dr Laura Liscum's laboratory for the scintillation counter in the glutamate uptake assay; Jerry Hsu's help in analyzing the GLT1 promoter activity in fmr1−/− cortex; Dr Jeffrey Rothstein for providing BAC GLT1 eGFP mice and GLT1/GLAST/EAAC1 antibodies; GENSAT project for the BAC ALDH1L1 TRAP mice; Dr Mark Bear for the mGluR5−/− mice; Dr Elizabeth Berry-Kravis (Rush University Medical Center, Chicago, IL, USA) for providing fmr1 cDNA construct; Drs Rob Jackson and Kathleen Dunlap for reading the manuscripts; Tufts Center for Neuroscience Research (NIH P30 NS047243; PI, Rob Jackson) for providing valuable core facilities; new faculty recruitment grant (NIH P30 5P30NS069254-02; PI, Phil Haydon) in Tufts Neuroscience Department.
Conflicts of Interest statement: None declared.
REFERENCES
- 1.Verkerk A.J., Pieretti M., Sutcliffe J.S., Fu Y.H., Kuhl D.P., Pizzuti A., Reiner O., Richards S., Victoria M.F., Zhang F.P., et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 2.Bakker C.E., Verheij C., Willemsen R., Vanderhelm R., Oerlemans F., Vermey M., Bygrave A., Hoogeveen A.T., Oostra B.A., Reyniers E., et al. Fmr1 knockout mice—a model to study Fragile-X mental-retardation. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- 3.Chen L., Toth M. Fragile X mice develop sensory hyperreactivity to auditory stimuli. Neuroscience. 2001;103:1043–1050. doi: 10.1016/s0306-4522(01)00036-7. [DOI] [PubMed] [Google Scholar]
- 4.Comery T.A., Harris J.B., Willems P.J., Oostra B.A., Irwin S.A., Weiler I.J., Greenough W.T. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc. Natl Acad. Sci. USA. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhakar A.L., Dolen G., Bear M.F. The pathophysiology of Fragile X (and what it teaches us about synapses) Annu. Rev. Neurosci. 2012;35:417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassell G.J., Warren S.T. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng Y., Absher D., Eberhart D.E., Brown V., Malter H.E., Warren S.T. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 8.Till S.M. The developmental roles of FMRP. Biochem. Soc. Trans. 2010;38:507–510. doi: 10.1042/BST0380507. [DOI] [PubMed] [Google Scholar]
- 9.Huber K.M., Gallagher S.M., Warren S.T., Bear M.F. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc. Natl Acad. Sci. USA. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronesi J.A., Huber K.M. Metabotropic glutamate receptors and fragile X mental retardation protein: partners in translational regulation at the synapse. Sci. Signal. 2008;1:pe6. doi: 10.1126/stke.15pe6. [DOI] [PubMed] [Google Scholar]
- 11.Bear M.F., Huber K.M., Warren S.T. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Wang H., Ku L., Osterhout D.J., Li W., Ahmadian A., Liang Z., Feng Y. Developmentally-programmed FMRP expression in oligodendrocytes: a potential role of FMRP in regulating translation in oligodendroglia progenitors. Hum. Mol. Genet. 2004;13:79–89. doi: 10.1093/hmg/ddh009. [DOI] [PubMed] [Google Scholar]
- 13.Pacey L.K., Doering L.C. Developmental expression of FMRP in the astrocyte lineage: implications for fragile X syndrome. Glia. 2007;55:1601–1609. doi: 10.1002/glia.20573. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs S., Nathwani M., Doering L.C. Fragile X astrocytes induce developmental delays in dendrite maturation and synaptic protein expression. BMC Neurosci. 2010;11:132. doi: 10.1186/1471-2202-11-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobs S., Doering L.C. Astrocytes prevent abnormal neuronal development in the fragile x mouse. J. Neurosci. 2010;30:4508–4514. doi: 10.1523/JNEUROSCI.5027-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danbolt N.C. Glutamate uptake. Prog. Neurobiol. 2001;65:1–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 17.Rothstein J.D., Martin L., Levey A.I., Dykes-Hoberg M., Jin L., Wu D., Nash N., Kuncl R.W. Localization of neuronal and glial glutamate transporters. Neuron. 1994;13:713–725. doi: 10.1016/0896-6273(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka K., Watase K., Manabe T., Yamada K., Watanabe M., Takahashi K., Iwama H., Nishikawa T., Ichihara N., Kikuchi T., et al. Epilepsy and exacerbation of brain injury in mice lacking glutamate transporter GLT-1 (Vol 276, pg 1699, 1997) Science. 1997;276:1699–1702. doi: 10.1126/science.276.5319.1699. [DOI] [PubMed] [Google Scholar]
- 19.Huang Y.H., Bergles D.E. Glutamate transporters bring competition to the synapse. Curr. Opin. Neurobiol. 2004;14:346–352. doi: 10.1016/j.conb.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Tzingounis A.V., Wadiche J.I. Glutamate transporters: confining runaway excitation by shaping synaptic transmission. Nat. Rev. Neurosci. 2007;8:935–947. doi: 10.1038/nrn2274. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y.H., Sinha S.R., Tanaka K., Rothstein J.D., Bergles D.E. Astrocyte glutamate transporters regulate metabotropic glutamate receptor-mediated excitation of hippocampal interneurons. J. Neurosci. 2004;24:4551–4559. doi: 10.1523/JNEUROSCI.5217-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Omrani A., Melone M., Bellesi M., Safiulina V., Aida T., Tanaka K., Cherubini E., Conti F. Up-regulation of GLT-1 severely impairs LTD at mossy fibre–CA3 synapses. J. Physiol. 2009;587:4575–4588. doi: 10.1113/jphysiol.2009.177881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takasaki C., Okada R., Mitani A., Fukaya M., Yamasaki M., Fujihara Y., Shirakawa T., Tanaka K., Watanabe M. Glutamate transporters regulate lesion-induced plasticity in the developing somatosensory cortex. J. Neurosci. 2008;28:4995–5006. doi: 10.1523/JNEUROSCI.0861-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christie S.B., Akins M.R., Schwob J.E., Fallon J.R. The FXG: a presynaptic fragile X granule expressed in a subset of developing brain circuits. J. Neurosci. 2009;29:1514–1524. doi: 10.1523/JNEUROSCI.3937-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doyle J.P., Dougherty J.D., Heiman M., Schmidt E.F., Stevens T.R., Ma G., Bupp S., Shrestha P., Shah R.D., Doughty M.L., et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y., Vidensky S., Jin L., Jie C., Lorenzini I., Frankl M., Rothstein J.D. Molecular comparison of GLT1+ and ALDH1L1+ astrocytes in vivo in astroglial reporter mice. Glia. 2011;59:200–207. doi: 10.1002/glia.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Regan M.R., Huang Y.H., Kim Y.S., Dykes-Hoberg M.I., Jin L., Watkins A.M., Bergles D.E., Rothstein J.D. Variations in promoter activity reveal a differential expression and physiology of glutamate transporters by glia in the developing and mature CNS. J. Neurosci. 2007;27:6607–6619. doi: 10.1523/JNEUROSCI.0790-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furuta A., Rothstein J.D., Martin L.J. Glutamate transporter protein subtypes are expressed differentially during rat CNS development. J. Neurosci. 1997;17:8363–8375. doi: 10.1523/JNEUROSCI.17-21-08363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutherland M.L., Delaney T.A., Noebels J.L. Glutamate transporter mRNA expression in proliferative zones of the developing and adult murine CNS. J. Neurosci. 1996;16:2191–2207. doi: 10.1523/JNEUROSCI.16-07-02191.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reiss A.L., Aylward E., Freund L.S., Joshi P.K., Bryan R.N. Neuroanatomy of fragile X syndrome: the posterior fossa. Ann. Neurol. 1991;29:26–32. doi: 10.1002/ana.410290107. [DOI] [PubMed] [Google Scholar]
- 31.Robinson M.B., Hunter-Ensor M., Sinor J. Pharmacologically distinct sodium-dependent L-[3H]glutamate transport processes in rat brain. Brain Res. 1991;544:196–202. doi: 10.1016/0006-8993(91)90054-y. [DOI] [PubMed] [Google Scholar]
- 32.Dowd L.A., Coyle A.J., Rothstein J.D., Pritchett D.B., Robinson M.B. Comparison of Na+-dependent glutamate transport activity in synaptosomes, C6 glioma, and Xenopus oocytes expressing excitatory amino acid carrier 1 (EAAC1) Mol. Pharmacol. 1996;49:465–473. [PubMed] [Google Scholar]
- 33.Yang Y., Gozen O., Watkins A., Lorenzini I., Lepore A., Gao Y., Vidensky S., Brennan J., Poulsen D., Won Park J., et al. Presynaptic regulation of astroglial excitatory neurotransmitter transporter GLT1. Neuron. 2009;61:880–894. doi: 10.1016/j.neuron.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlag B.D., Vondrasek J.R., Munir M., Kalandadze A., Zelenaia O.A., Rothstein J.D., Robinson M.B. Regulation of the glial Na+-dependent glutamate transporters by cyclic AMP analogs and neurons. Mol. Pharmacol. 1998;53:355–369. doi: 10.1124/mol.53.3.355. [DOI] [PubMed] [Google Scholar]
- 35.Swanson R.A., Liu J., Miller J.W., Rothstein J.D., Farrell K., Stein B.A., Longuemare M.C. Neuronal regulation of glutamate transporter subtype expression in astrocytes. J. Neurosci. 1997;17:932–940. doi: 10.1523/JNEUROSCI.17-03-00932.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen W., Aoki C., Mahadomrongkul V., Gruber C.E., Wang G.J., Blitzblau R., Irwin N., Rosenberg P.A. Expression of a variant form of the glutamate transporter GLT1 in neuronal cultures and in neurons and astrocytes in the rat brain. J. Neurosci. 2002;22:2142–2152. doi: 10.1523/JNEUROSCI.22-06-02142.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agulhon C., Petravicz J., McMullen A.B., Sweger E.J., Minton S.K., Taves S.R., Casper K.B., Fiacco T.A., McCarthy K.D. What is the role of astrocyte calcium in neurophysiology. Neuron. 2008;59:932–946. doi: 10.1016/j.neuron.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palma-Cerda F., Auger C., Crawford D.J., Hodgson A.C., Reynolds S.J., Cowell J.K., Swift K.A., Cais O., Vyklicky L., Corrie J.E., et al. New caged neurotransmitter analogs selective for glutamate receptor sub-types based on methoxynitroindoline and nitrophenylethoxycarbonyl caging groups. Neuropharmacology. 2012;63:624–634. doi: 10.1016/j.neuropharm.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Todd P.K., Mack K.J., Malter J.S. The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc. Natl Acad. Sci. USA. 2003;100:14374–14378. doi: 10.1073/pnas.2336265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muddashetty R.S., Kelic S., Gross C., Xu M., Bassell G.J. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J. Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berry-Kravis E., Ciurlionis R. Overexpression of fragile X gene (FMR-1) transcripts increases cAMP production in neural cells. J. Neurosci. Res. 1998;51:41–48. doi: 10.1002/(SICI)1097-4547(19980101)51:1<41::AID-JNR4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 42.Bushong E.A., Martone M.E., Jones Y.Z., Ellisman M.H. Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. J. Neurosci. 2002;22:183–192. doi: 10.1523/JNEUROSCI.22-01-00183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A., et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia A.D., Doan N.B., Imura T., Bush T.G., Sofroniew M.V. GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nat. Neurosci. 2004;7:1233–1241. doi: 10.1038/nn1340. [DOI] [PubMed] [Google Scholar]
- 45.Darnell J.C., Van Driesche S.J., Zhang C., Hung K.Y., Mele A., Fraser C.E., Stone E.F., Chen C., Fak J.J., Chi S.W., et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen W., Mahadomrongkul V., Berger U.V., Bassan M., DeSilva T., Tanaka K., Irwin N., Aoki C., Rosenberg P.A. The glutamate transporter GLT1a is expressed in excitatory axon terminals of mature hippocampal neurons. J. Neurosci. 2004;24:1136–1148. doi: 10.1523/JNEUROSCI.1586-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michalon A., Sidorov M., Ballard T.M., Ozmen L., Spooren W., Wettstein J.G., Jaeschke G., Bear M.F., Lindemann L. Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron. 2012;74:49–56. doi: 10.1016/j.neuron.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dolen G., Osterweil E., Rao B.S., Smith G.B., Auerbach B.D., Chattarji S., Bear M.F. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hays S.A., Huber K.M., Gibson J.R. Altered neocortical rhythmic activity states in Fmr1 KO mice are due to enhanced mGluR5 signaling and involve changes in excitatory circuitry. J. Neurosci. 2011;31:14223–14234. doi: 10.1523/JNEUROSCI.3157-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gibson J.R., Bartley A.F., Hays S.A., Huber K.M. Imbalance of neocortical excitation and inhibition and altered UP states reflect network hyperexcitability in the mouse model of fragile X syndrome. J. Neurophysiol. 2008;100:2615–2626. doi: 10.1152/jn.90752.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cleveland D.W., Rothstein J.D. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat. Rev. Neurosci. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 52.Sherman S.L., Jacobs P.A., Morton N.E., Froster-Iskenius U., Howard-Peebles P.N., Nielsen K.B., Partington M.W., Sutherland G.R., Turner G., Watson M. Further segregation analysis of the fragile X syndrome with special reference to transmitting males. Hum. Genet. 1985;69:289–299. doi: 10.1007/BF00291644. [DOI] [PubMed] [Google Scholar]
- 53.Ghosh M., Yang Y., Rothstein J.D., Robinson M.B. Nuclear factor-{kappa}B contributes to neuron-dependent induction of glutamate transporter-1 expression in astrocytes. J. Neurosci. 2011;31:9159–9169. doi: 10.1523/JNEUROSCI.0302-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X., Spandidos A., Wang H., Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic. Acids Res. 2012;40:D1144–D1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perkins K.L. Cell-attached voltage-clamp and current-clamp recording and stimulation techniques in brain slices. J. Neurosci. Methods. 2006;154:1–18. doi: 10.1016/j.jneumeth.2006.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.