Abstract
Mutations in ATP13A2 (PARK9), encoding a lysosomal P-type ATPase, are associated with both Kufor–Rakeb syndrome (KRS) and neuronal ceroid lipofuscinosis (NCL). KRS has recently been classified as a rare genetic form of Parkinson's disease (PD), whereas NCL is a lysosomal storage disorder. Although the transport activity of ATP13A2 has not been defined, in vitro studies show that its loss compromises lysosomal function, which in turn is thought to cause neuronal degeneration. To understand the role of ATP13A2 dysfunction in disease, we disrupted its gene in mice. Atp13a2−/− and Atp13a2+/+ mice were tested behaviorally to assess sensorimotor and cognitive function at multiple ages. In the brain, lipofuscin accumulation, α-synuclein aggregation and dopaminergic pathology were measured. Behaviorally, Atp13a2−/− mice displayed late-onset sensorimotor deficits. Accelerated deposition of autofluorescent storage material (lipofuscin) was observed in the cerebellum and in neurons of the hippocampus and the cortex of Atp13a2−/− mice. Immunoblot analysis showed increased insoluble α-synuclein in the hippocampus, but not in the cortex or cerebellum. There was no change in the number of dopaminergic neurons in the substantia nigra or in striatal dopamine levels in aged Atp13a2−/− mice. These results show that the loss of Atp13a2 causes sensorimotor impairments, α-synuclein accumulation as occurs in PD and related synucleinopathies, and accumulation of lipofuscin deposits characteristic of NCL, thus providing the first direct demonstration that null mutations in Atp13a2 can cause pathological features of both diseases in the same organism.
INTRODUCTION
ATP13A2 (PARK9) encodes a lysosomal P5-ATPase of undetermined substrate specificity (1,2). Mutations in ATP13A2 are associated with Kufor–Rakeb syndrome (KRS), a juvenile onset, autosomal recessive Parkinsonism in humans (3), and are also associated with both adult onset neuronal ceroid lipofuscinosis (NCL) in dogs (4,5) and juvenile onset NCL in humans (6). KRS patients display motor deficits, including rigidity and bradykinesia, and by brain imaging show pallido-pyramidal degeneration followed later by widespread cerebral atrophy (7–9). NCLs are a group of neurodegenerative diseases characterized by the accumulation of autofluorescent storage material (ceroid lipofuscin) within lysosomes of neurons and other cell types. Common clinical features include motor deterioration (cerebellar ataxia), dementia, seizures and visual impairment (4,5,10). Of particular interest is a recent study showing that a juvenile onset form of NCL was caused by an ATP13A2 missense mutation (6). Affected individuals developed typical NCL pathology (11) but also presented with clinical features commonly seen in KRS (12), suggesting that KRS and NCL are related disorders in which lysosomal dysfunction plays a central role (6).
Insights into the function of ATP13A2 have come from studies in yeast, Caenorhabditis elegans and mammalian cell lines. Ypk9p, the yeast homologue of human ATP13A2, protects yeast cells from Mn2+ toxicity (13,14). Similarly, ATP13A2 protects mammalian cell lines and rat primary neuronal cultures from Mn2+-induced cell death, thus establishing a link between ATP13A2 activity and an environmental risk factor (manganese exposure) for Parkinsonism (15). Studies also demonstrate a potential protective function of ATP13A2 against α-synuclein toxicity in C. elegans and rodent primary neuronal cell cultures (14,16). Aggregation of α-synuclein is associated with both inherited and sporadic forms of Parkinson's disease (PD) (17,18), and intratstriatal inoculation of synthetic misfolded fibrilar α-synuclein leads to nigrostriatal cell death in vivo (19), suggesting a direct role for α-synuclein in the pathophysiology of PD. Studies in fibroblasts from KRS patients and dopaminergic cell lines show that the loss of ATP13A2 leads to the accumulation of α-synuclein and neurotoxicity in vitro, impaired lysosomal acidification and degradation capacity, decreased proteolytic processing of lysosomal enzymes, accumulation and enlargement of lysosomes and decreased clearance of autophagosomes (16,20).
Other investigators (6) have noted that although NCL histopathology has been identified in patients with ATP13A2 mutations, there are currently no histopathological studies from KRS patients. Thus, it is unclear whether KRS patients develop Lewy body and nigrostriatal pathology similar to that found in PD. To assess the function of ATP13A2 and its potential role in KRS and NCL, we generated mice with a targeted disruption of the Atp13a2 gene. These mice were evaluated with a battery of sensorimotor and cognitive tests, and various histological and biochemical analyses were performed to determine whether the loss of Atp13a2 function affected behavior, α-synuclein aggregation, lipofuscin accumulation and nigrostriatal dopaminergic integrity. We show that the loss of Atp13a2 in mice results in sensorimotor deficits in old age, widespread neuronal accumulation of lipofuscin and aggregation of α-synuclein in hippocampus.
RESULTS
Atp13a2 gene targeting
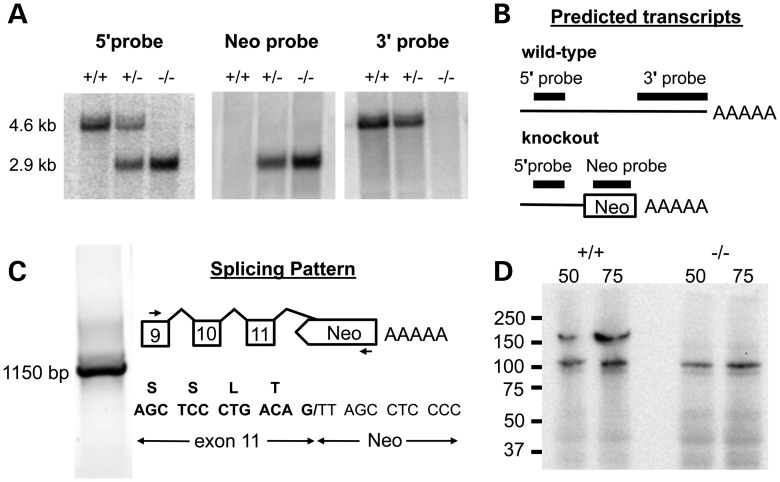
The gene targeting strategy led to a global knockout of Atp13a2 (Fig. 1A). The deleted exons included sequences encoding the catalytic phosphorylation site (Asp 559), which is essential for enzyme activity. After germline transmission of the targeted allele, heterozygous mice were bred to obtain offspring of all three genotypes. Southern blot (Fig. 1B) and PCR (Fig. 1C) analyses of tail DNA from offspring of a heterozygous mating showed that the Atp13a2 gene was correctly targeted.
Figure 1.
Atp13a2 gene targeting and genotype analysis. (A) Targeting strategy. Top, region of the wild-type gene encompassing exons 2–23. Middle, targeting construct. Exons 12–15 and a portion of exon 16 were replaced by the Neomycin resistance gene (Neo). The herpes simplex virus thymidine kinase gene (tk) was included for negative selection. Dashed lines represent vector sequences. Bottom, predicted structure of the targeted allele. 5′ and 3′ diagnostic probes (solid bars below the wild-type allele) used in the Southern blot analysis and Bgl II restriction fragments unique to the wild-type (7.9 and 6.2 kb) and targeted (11.1 kb) alleles are indicated. (B) Southern blot analysis of tail DNA from representative offspring of a heterozygous mating. DNA was digested with Bgl II and hybridized with the 5′ and 3′ outside probes. (C) PCR genotyping of tail biopsy DNAs from mice of all three genotypes.
Northern blot analysis (Fig. 2A) revealed a transcript of the expected size in brains of wild-type mice, a truncated transcript in null mutants and wild-type and truncated transcripts at similar expression levels in heterozygous mice. The 2.9 kb mutant transcript hybridized with the Neo and 5′ probes but not with the 3′ probe, indicating that it terminates at an SV40 poly A signal sequence (Fig. 2B), which was included immediately beyond the promoter of the Neo gene to ensure the termination of the transcript. RT–PCR and sequence analysis (Fig. 2C) showed that the donor splice site of exon 11 was spliced to a cryptic acceptor site within the Neo gene, thereby eliminating the last 828 codons of the 1169 codon mRNA, but adding an alternative 168 codons before a stop codon was encountered.
Figure 2.
Northern blot and immunoblot analysis of Atp13a2 transcripts and protein. (A) Northern blot analysis of Atp13a2 transcripts in brain poly(A) RNA of all three genotypes using probes indicated in the schematic diagram (B) of wild-type and mutant transcripts. The 2.9 kb mutant transcript hybridizes with the Neo and 5′ probes but not the 3′ probe indicating that it terminates at an SV40 poly(A) signal sequence that precedes sequences from exon 16. (C) RT–PCR amplification of mutant first-strand cDNA using primers (small arrows) specific for Atp13a2 exon 9 and the Neo gene yielded an 1150 bp product. Sequence analysis showed that the donor splice site of exon 11 was spliced to a cryptic acceptor site in the non-coding strand of the Neo gene (top). The region containing the cryptic splice is shown (bottom), with the Atp13a2 nucleotide sequence and its deduced amino acid sequence in bold. (D) Western blot analysis of Atp13a2 protein using brain extracts (50 and 75 μg) from Atp13a2+/+ and Atp13a2−/− mice. An antibody directed against amino acids 195–213 of human ATP13A2 detected the wild-type protein (∼160 kDa) in Atp13a2+/+ but not Atp13a2−/− mice extracts and a non-specific band at 100 kDa in both genotypes, but did not detect a truncated mutant protein in Atp13a2−/− brains.
The protein encoded by the mutant mRNA is predicted to consist of the first 341 amino acids of Atp13a2 and 168 amino acids derived from the antisense strand of the Neo gene. Immunoblot analysis using an antibody that would identify both wild-type and mutant proteins detected a 160 kDa band in Atp13a2+/+ brain extracts, which was absent in extracts from Atp13a2−/− brains (Fig. 2D). The protein detected in wild-type extracts has an apparent molecular weight that is larger than predicted (127 kDa) on the basis of amino acid sequence, possibly due to post-translational modifications. A non-specific band of 110 kDa was identified in both genotypes; however, a 50–60 kDa protein was not observed in extracts of mutant brains, suggesting that the predicted mutant protein was not stable.
Expression of other P5-ATPases is unaffected by Atp13a2 ablation
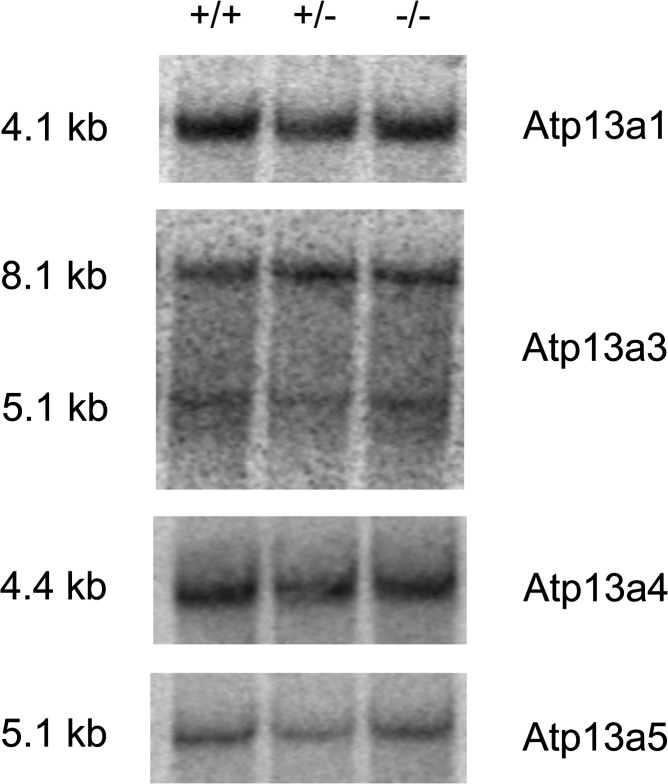
Because all the five P5-ATPases (Atp13a1-Atp13a5) are expressed in the mouse brain, it was important to determine whether compensatory upregulation of Atp13a1, Atp13a3, Atp13a4 or Atp13a5 might occur in the brains of Atp13a2−/− mice. However, northern blot analysis of brain RNA revealed no differences in the expression of any of these genes (Fig. 3), suggesting that compensation by another isoform does not occur.
Figure 3.
mRNA expression of Atp13a1, Atp13a3, Atp13a4 and Atp13a5 in the brain of wild-type (+/+), heterozygous (+/−) and homozygous (−/−) mutant mice. Northern blot analysis was performed using cDNA probes corresponding to mRNAs for each of the four other Atp13a isoforms. Each lane contains 5 µg of poly(A) brain RNA from the mice of the indicated Atp13a2 genotype. The size of each mRNA, in kilobases, is indicated on the left and the isoform is indicated on the right.
Gross phenotype
Offspring from heterozygous matings were born at the expected 1:2:1 Mendelian ratios (73 wild-type, 136 heterozygous, 70 homozygous), indicating that the null mutation does not cause embryonic or fetal lethality. Both male and female homozygous mutant mice were fertile. Atp13a2−/− mice grew normally and were indistinguishable from Atp13a2+/+ mice in outward appearance, size and behavior until old age. Behavioral problems first became apparent at ∼2 years-of-age, when several mutant animals exhibited an unsteady gait. Consequently, we subjected mice of varying ages to a battery of sensorimotor and cognitive tests including challenging beam, spontaneous activity, gait analysis, nest building, Morris water maze and novel object recognition to determine whether they display behavioral phenotypes characteristic of KRS or NCL.
Challenging beam traversal
At 5–12 months of age, there were no significant differences in time to traverse, number of steps taken while traversing, or the number of errors made on the beam between Atp13a2+/+ and Atp13a2−/− mice (Supplementary Material, Fig. S1). However, at 20–29 months of age several differences were noted. Analysis of the mean of five trials showed that Atp13a2−/− mice took more steps and made more errors per step while traversing the beam compared with age-matched Atp13a2+/+ mice (P < 0.05; Fig. 4A–C). Further analysis in the older cohort by trial was also performed on traversal time, steps and errors per step and showed differences in all three parameters (Fig. 4D–F).
Figure 4.
Motor performance and coordination in Atp13a2+/+ and Atp13a2−/− mice at 20–29 months of age. Motor performance and coordination were assessed in Atp13a2+/+ (n = 13) and Atp13a2−/− (n = 18) mice using a challenging beam test and measured mean time to traverse, number of steps and errors per step (A–C). Student's t-test showed a significant increase in number of steps and errors per step in Atp13a2−/− compared with Atp13a2+/+ mice (P < 0.05). Analysis of performance by trial was also measured (D–F). A 2×5 mixed design ANOVA for traversal time showed a main effect of trial F(4,116) = 3.83, P < 0.01 and a significant genotype × trial interaction F(4,116) = 3.98, P < 0.01. Post hoc analysis showed that time to traverse between Atp13a2+/+ and Atp13a2−/− mice was comparable during the first two trials; however, Atp13a2+/+ mice were significantly faster than Atp13a2−/− mice during trials 3–5 (T3–T5, P < 0.05–0.01). Trial analysis of steps revealed main effects of both genotype F(1,29) = 12.65, P < 0.01 and trial F(4,116) = 2.75, P < 0.05 and post hoc analysis showed that Atp13a2−/− mice made significantly more steps compared with Atp13a2+/+ mice at each trial (T1–T5 P < 0.01). For errors per step, there was a main effect of the genotype F(1,29) = 4.61, P < 0.05 with post hoc analysis showing that Atp13a2+/+ and Atp13a2−/− mice made a similar number of errors per step on the first trial but that Atp13a2+/+ mice made significantly fewer errors per step compared with mutants in subsequent trials (for T2–T5 P < 0.05–0.01). *, **P < 0.05, 0.01 compared with Atp13a2+/+ mice, Fisher's LSD post hoc. The values shown are means ± SEM.
Spontaneous activity
At 5–12 months of age, there were no significant differences in forelimb steps, hindlimb steps, rearing or grooming between Atp13a2+/+ and Atp13a2−/− mice (Supplementary Material, Fig. S2). At 20–29 months of age, there were no differences between genotypes in forelimb stepping, rearing or grooming. However, hindlimb stepping was significantly decreased in Atp13a2−/− mice compared with Atp13a2+/+ mice (P < 0.05; Fig. 5A–D).
Figure 5.
Spontaneous activity and Gait in Atp13a2+/+ and Atp13a2−/− mice at 20–29 months of age. Spontaneous activity was assessed in the cylinder and measured forelimb and hindlimb stepping, rears and time spent grooming in Atp13a2 +/+ (n = 12) and Atp13a2−/− (n = 18) mice (A–D). Atp13a2−/− mice showed a significant reduction in hindlimb stepping compared with Atp13a2+/+ mice (B). Gait analyses assessed stride length and stride variability in Atp13a2 +/+ (n = 15) and Atp13a2−/− (n = 20) mice (E and F). Atp13a2−/− mice had a significantly shorter stride length than Atp13a2+/+ mice (E). No difference in stride variability was observed when females and males were analyzed together, but when analyzed separately, stride length was significantly more variable in Atp13a2−/− (n = 5) than Atp13a2+/+ (n = 3) males (F). *, **P < 0.05, 0.01 compared with Atp13a2+/+mice. Student's t-test. The values shown are means ± SEM.
Gait analysis
Stride length and stride difference (the variability in stride lengths) were measured in both Atp13a2+/+ and Atp13a2−/− mice. Analysis at 5–12 months of age showed no difference in stride length or stride variability between genotypes (Supplementary Material, Fig. S2). Student's t-test at 20–29 months comparing genotypes showed that Atp13a2−/− mice made significantly shorter stride lengths compared with wild-type controls. Stride variability scores with females and males combined did not differ between genotypes at 20–29 months, however, analysis of only males (Atp13a2+/+ = 3, Atp13a2−/− = 5) showed that Atp13a2−/− males had significantly more variable stride lengths, which is indicative of ataxic movements (Fig. 5E–F).
Nest building
The amount of cotton used to build nests was measured in Atp13a2+/+ and Atp13a2−/− mice at 20–29 months of age. Mann–Whitney U analysis showed that Atp13a2−/− mice used significantly less cotton compared with WT mice (P < 0.05; Fig. 6).
Figure 6.
Nestbuilding in Atp13a2+/+ and Atp13a2−/− mice at 20–29 months of age. Complex orofacial and fine motor skills were assessed in Atp13a2 +/+ (n = 12) and Atp13a2−/− (n = 17) mice using a nestbuilding test. The amount of nestbuilding material was measured over a 24 h period. Mann–Whitney U analysis showed that Atp13a2−/− mice used significantly less cotton compared with Atp13a2+/+ mice (P < 0.05). The values shown are means ± SEM.
Cognitive testing
In addition to sensorimotor deficits, cognitive and neuropsychiatric impairments have been shown to develop in KRS, NCL and PD. Therefore, cognitive function and emotional reactivity were compared between 13–17-month-old Atp13a2−/− and Atp13a2+/+ mice in several tests.
In the novel object and spatial recognition task, measures of the time spent exploring a novel object or an object in a novel spatial location minus the time spent investigating the older, matching object did not reveal an effect of genotype (Supplementary Material, Fig. S3A). However, when the total amount of time spent exploring both objects during the familiarization and recognition phase of the test was compared, Atp13a2−/− mice spent significantly more time investigating the objects than the Atp13a2+/+ mice (P < 0.05; Supplementary Material, Fig. S3B). To further analyze this phenotype, mice were tested in a latency to explore task that assesses how quickly a mouse will move toward the perimeter of a tabletop when placed in the center. While there was no difference between the genotypes in their initiation of movement (Supplementary Material, Fig. S3C) or the percentage of time spent moving while inside of a square (Supplementary Material, Fig. S3D), Atp13a2−/− mice took significantly less time to walk outside of the square in comparison with the Atp13a2+/+ mice (P < 0.05; Supplementary Material, Fig. S3E). Locomotor activity in an open-field was also measured in Atp13a2−/− and Atp13a2+/+ mice. There were no group differences in overall activity or time spent in the center of the cage. Finally, in a light–dark test of anxiety conducted over 3 days, Atp13a2−/− and Atp13a2+/+ mice displayed similar patterns of behavior.
In hidden and visible versions of a water-maze task, a two-way analysis of variance comparing the effects of genotype and test day did not reveal a significant genotype effect on path length (Supplementary Material, Fig. S4A). A comparison of escape latency over test days revealed a significant effect of the genotype (P < 0.05; Supplementary Material, Fig. S4B). Atp13a2−/− mice found the platform faster on Days 2 and 3 of testing when compared with the Atp13a2+/+ mice. There was a trend toward a genotype effect on swim speed (P = 0.14, Supplementary Material, Fig. S4C). The mean swim speed in the Atp13a2−/− mice was greater on Days 2 and 3 of testing in comparison with the swim speed recorded in the Atp13a2+/+ mice. No genotype difference was observed in any of the measures taken during the probe trial or visible platform testing.
Stereology and analysis of striatal dopamine content
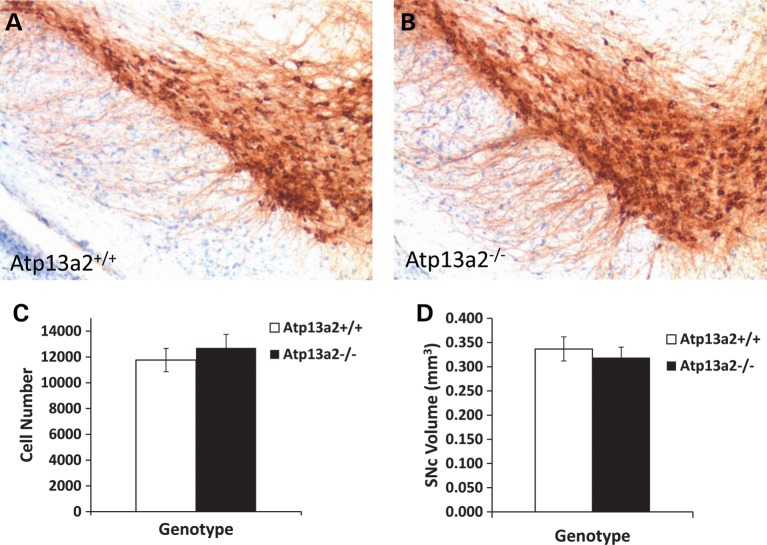
To determine the effect of the loss of Atp13a2 function on the nigrostriatal dopamine (DA) system, immunohistochemical staining and stereological counting of tyrosine hydroxylase-positive (TH+) neurons in the substantia nigra pars compacta (SNc) was carried out in aged animals (20–29 months). Staining of TH was similar in both genotypes (Fig. 7A and B) and they had a comparable number of TH+ neurons (Fig. 7C; Atp13a2+/+: 11 760 ± 905; Atp13a2−/−: 12 704 ± 1054) indicating that the loss of Atp13a2 does not lead to a significant loss of TH-immunoreactive neurons. The total volume of the SNc in Atp13a2+/+ and Atp13a2−/− brains was also similar (Fig. 7D; Atp13a2+/+: 0.337 ± 0.025 mm3; Atp13a2−/−: 0.319 ± 0.022 mm3). Because changes in DA neurotransmission can occur without loss of dopaminergic neurons per se, we also examined striatal DA levels by HPLC. No differences in striatal DA levels or other biogenic amines were observed (Table 1).
Figure 7.
Tyrosine hydroxylase immunostaining and stereological analysis. TH immunostaining of representative mid-brain sections showing dopaminergic neurons in the SNc of Atp13a2+/+ (A) and Atp13a2−/− (B) mice at 27 months of age; sections were counterstained with cresyl violet; ×100 magnification. (C) Average number of tyrosine hydroxylase-positive neurons in the SNc of Atp13a2+/+ and Atp13a2−/− mice at 22–27 months of age and (D) volume of SNc in the same mice, as determined by stereological analysis. n = 6 mice of each genotype for all analyses. The values shown are means ± SEM.
Table 1.
Striatal biogenic amine concentrations
| Norepinephrine | DOPAC | DA | 5-HIAA | HVA | 5-HT | |
|---|---|---|---|---|---|---|
| Atp13a2+/+ | 1.34 ± 0.36 | 8.50 ± 0.41 | 112.13 ± 7.40 | 2.81 ± 0.16 | 9.35 ± 0.56 | 6.73 ± 0.57 |
| Atp13a2−/− | 1.64 ± 0.45 | 8.17 ± 0.69 | 112.83 ± 10.55 | 3.31 ± 0.33 | 9.72 ± 0.82 | 7.28 ± 0.98 |
Values are given in ng/mg protein and represent the average ± SEM. n = 10 for each genotype; mice were 20–29 months of age. DOPAC, 3,4-dihydroxyphenylacetic acid; DA, dopamine; 5-HIAA, 5-hydroxyindole acetic acid; HVA, homovanillic acid; 5-HT, 5-hydroxytryptamine (serotonin).
Accelerated accumulation of intraneuronal storage material in Atp13a2−/− mice
The intraneuronal storage material (lipofuscin) that accumulates in all types of NCL is autofluorescent and stains positively with luxol fast blue (LFB) (21). Compared with Atp13a2+/+ mice, Atp13a2−/− mice show an accelerated deposition of autofluorescent storage material in the cerebellum (Fig. 8A and B) and in the hippocampus at 18 months of age (Fig. 8C and D). Though not as pronounced as in the cerebellum or hippocampus, enhanced autofluorescence was also observed in the cortex (data not shown). The autofluorescent signal was quenched with Sudan black staining (data not shown) and the storage material was also visualized by Luxol fast blue-periodic acid Schiff-hematoxylin (LFB-PAS) staining (Fig. 8E–H), both of which are characteristic of lipofuscin.
Figure 8.
Accelerated accumulation of intraneuronal storage material (lipofuscin) in the cerebellum, hippocampus and cortex of Atp13a2−/− mice. Autofluorescent storage material in the cerebellum (A and B), and in neurons of the CA-3 region of the hippocampus (C and D) of an 18-month-old Atp13a2−/− mouse and age-matched control; representative fluorescence images of 7 μm paraffin sections are shown. Particularly large autofluorescent inclusions (arrows) were seen in the cerebellum of Atp13a2−/− mice (E–H). LFB-PAS stained paraffin sections. Lipofuscin staining (dark blue) is markedly increased in Atp13a2−/− brain neurons relative to controls. Sections from the hippocampus (E and F) and the cortex (G and H) are shown.
Accumulation of insoluble α-synuclein in hippocampus of Atp13a2−/− mice
Accumulation of aggregated, insoluble α-synuclein within the central nervous system is a characteristic feature of PD. To determine whether the loss of Atp13a2 leads to changes in the levels of soluble and insoluble α-synuclein species, homogenates prepared from the hippocampus (Fig. 9A–C), cortex (Fig. 9D–F) and cerebellum (Fig. 9G–I) were subjected to sequential extraction, first in Triton X-100 buffer and then in 2% SDS buffer, and analyzed by western blot analysis. Increased levels of Triton X-100 insoluble α-synuclein were observed in the hippocampus of Atp13a2−/− mice (Fig. 9A and B). However, no change in α-synuclein protein solubility was observed in either the cortex (Fig. 9D and E) or cerebellum (Fig. 9G and H). The levels and solubility of tau and huntingtin, proteins prone to aggregation in other neurodegenerative disorders, were not significantly changed in any of the brain regions analyzed. These results suggest that the loss of Atp13a2 results in the preferential accumulation of α-synuclein in the hippocampus.
Figure 9.
α-Syn accumulation in hippocampus of Atp13a2−/− mice. Western blot analysis of T-sol (A, D and G) and SDS-sol fractions (B, E and H) from hippocampal (A and B), cortical (D and E) and cerebellar (G and H) lysates. Endogenous α-synuclein, tau and huntingtin levels were compared in lysates prepared from Atp13a2+/+ (n = 5), Atp13a2−/− (n = 4) and Atp13a2+/− (n = 1) mice at 18–20 months of age. Relative protein levels in the hippocampus (C), cortex (F) and cerebellum (I) are shown in the panels to the right of the blots (*P < 0.03). Neuron-specific enolase (NSE) was used as a loading control. The values shown are means ± SEM.
Glucocerebrosidase enzyme activity is not significantly down-regulated in Atp13a2−/− mouse brains
It has been suggested that glucocerebrosidase (GCase) enzyme activity and protein levels are reduced in brain samples from patients with PD and Parkinson mouse models (22–25). We measured GCase activity in brain lysates from five Atp13a2+/+ and four Atp13a2−/− mice at 20–23 months of age and three Atp13a2+/+ and three Atp13a2−/− mice at 16–18 months of age. Within the different age groups, there was no significant difference in GCase enzyme activity detected between genotypes (Supplementary Material, Fig. S5A). The activity of acid alpha-glucosidase (GAA), the enzyme involved in the breakdown of lysosomal glycogen (26), was measured as a control for lysosomal activity and no significant differences were observed (Supplementary Material, Fig. S5B). Furthermore, when measuring the GCase protein content in the different mouse brains with a previously described GCase-specific fluorescent probe (27), no significant difference between genotypes was detected among animals of the same age (Supplementary Material, Fig. S6).
Quantitative RT–PCR array analyses
To determine whether the loss of Atp13a2 leads to changes in the expression of genes implicated in PD and autophagy, gene expression levels in the striatum and SN of Atp13a2+/+ and Atp13a2−/− mice were compared using autophagy and Parkinson's RT2 Profiler PCR arrays (SABiosciences). The expression of Nr4a2 (Nurr1) and brain-derived neurotrophic factor (Bdnf) was down-regulated 6.14 and 3.16-fold, respectively, in the striatum of Atp13a2−/− mice but unchanged in the SN. Slc6a3 (dopamine transporter, DAT) and tyrosine hydroxylase (Th) expression did not change in either brain region, whereas Snca (α-synuclein) was significantly down-regulated in the striatum, but only by 1.15-fold (Table 2). The expression of cathespsin S (Ctss), tumor necrosis factor α (Tnf) and Irgm1 (immunity-related GTPase family, M1) was up-regulated 1.75, 1.60 and 1.56-fold, respectively, in the SN of Atp13a2−/− mice (Table 3). Similar expression results were obtained for these genes in the striatum but the results were based on only two animals of each genotype and are not reported in the table.
Table 2.
Parkinson's PCR array
| Gene | ΔCT (n = 6) |
Expression relative to +/+ | Fold regulation relative to +/+ | |
|---|---|---|---|---|
| Atp13a2+/+ | Atp13a2−/− | |||
| Striatum | ||||
| Nr4a2 | 6.29 ± 0.36 | 8.91 ± 0.57 | 0.163* | −6.14 |
| Bdnf | 9.01 ± 0.26 | 10.68 ± 0.45 | 0.316* | −3.16 |
| Slc6a3 | 11.39 ± 0.14 | 11.71 ± 0.24 | 0.801 | −1.25 |
| Snca | 0.79 ± 0.03 | 0.99 ± 0.02 | 0.871** | −1.15 |
| Th | 8.64 ± 0.14 | 8.74 ± 0.08 | 0.936 | −1.07 |
| ΔCT (n = 4) | Expression relative to +/+ | Fold regulation relative to +/+ | ||
| Atp13a2+/+ | Atp13a2−/− | |||
| Substantia nigra | ||||
| Nr4a2 | 6.10 ± 0.27 | 6.06 ± 0.08 | 1.03 | 1.03 |
| Bdnf | 7.89 ± 0.27 | 8.04 ± 0.38 | 0.901 | −1.11 |
| Slc6a3 | 2.45 ± 0.21 | 2.33 ± 0.20 | 1.09 | 1.09 |
| Snca | 3.13 ± 0.22 | 3.38 ± 0.15 | 0.844 | −1.19 |
| Th | 4.84 ± 0.29 | 4.71 ± 0.25 | 1.10 | 1.10 |
The Nr4a2 and Bdnf genes are down-regulated in the striatum but not the substantia nigra of Atp13a2−/− mice. Values obtained from Parkinson's RT2 Profiler PCR array analysis of tissues from wild-type and Atp13a2−/− animals. ΔCT represents the difference in cycle threshold between the mRNA of interest and the average of a set of housekeeping mRNAs whose expression was found not to vary as a function of the Atp13a2 genotype. Values are shown as mean ± SEM and the number of replicates is indicated. Expression relative to +/+ is the mean normalized gene expression (2−ΔCT) in the −/− replicates divided by the mean normalized expression in the +/+ replicates. Differences in expression that were found to be significant are marked with one (P < 0.05) or two (P < 0.005) asterisks. Fold regulation is equal to relative expression for values ≥1 and to the negative reciprocal of relative expression for values <1.
Table 3.
Autophagy PCR array
| Gene | ΔCT (n = 4) |
Expression relative to +/+ | Fold regulation relative to +/+ | |
|---|---|---|---|---|
| Atp13a2+/+ | Atp13a2−/− | |||
| Substantia nigra | ||||
| Ctss | 3.09 ± 0.05 | 2.28 ± 0.12 | 1.75** | 1.75 |
| Tnf | 13.86 ± 0.09 | 13.18 ± 0.25 | 1.60* | 1.60 |
| Irgm1 | 6.92 ± 0.12 | 6.28 ± 0.15 | 1.56* | 1.56 |
The Ctss, Tnf and Irgm1 genes are up-regulated in the substantia nigra of Atp13a2−/− mice. Values obtained from Autophagy RT2 Profiler PCR array analysis of tissue from wild-type and Atp13a2−/− animals. ΔCT represents the difference in cycle threshold between the mRNA of interest and the average of a set of housekeeping mRNAs whose expression was found not to vary as a function of the Atp13a2 genotype. Values are shown as mean ± SEM and the number of replicates is indicated. Expression relative to +/+ is the mean normalized gene expression (2−ΔCT) in the −/− replicates divided by the mean normalized expression in the +/+ replicates. Differences in expression that were found to be significant are marked with one (P < 0.05) or two (P < 0.005) asterisks. Fold regulation is equal to relative expression for values ≥1 and to the negative reciprocal of relative expression for values <1.
DISCUSSION
The Atp13a2-null mouse model was developed using a global targeting strategy that eliminated much of the coding sequence from the mutant mRNA, including exons encoding the catalytic phosphorylation site, the ATP binding site and most of the transmembrane domains. The mutant and wild-type mRNAs were expressed at similar levels in heterozygous mice, indicating that non-sense-mediated decay does not contribute to degradation of the mutant mRNA, as reported for an ATP13A2 mutation in humans (28). For some ATP13A2 mutations, there is evidence that the expression or accumulation of the mutant protein and resulting neurodegeneration from endoplasmic reticulum stress might contribute to the disease phenotype (3,29). Despite the stability of the mRNA, however, a mutant protein was not detected, suggesting that the protein is rapidly degraded or that the mutant mRNA is inefficiently translated. Thus, the phenotype of the mouse model is likely due to lysosomal dysfunction resulting from the loss of Atp13a2, with little, if any, contribution from secondary effects of a mutant protein.
Homozygous or compound heterozygous mutations in ATP13A2 are now known to cause both Kufor–Rakeb syndrome (3) and NCL (6). The clinical presentation in KRS varies among patients but can include bradykinesia, rigidity, ataxia, dementia, spasticity and supranuclear gaze palsy (8,30–32). To date only one family with NCL has been linked to mutations in ATP13A2 (6). In an earlier study, members of this Belgian family variably presented with rigidity, akinesia, resting tremor, uncoordinated movements and impaired intellect (11), a spectrum of symptoms that overlap with those of individuals affected by KRS. The Tibetan terrier model of NCL has also been associated with mutations in ATP13A2, and develops behavioral problems such as mild ataxia, confusion and occasional aggressiveness (33,34); however, Parkinsonian features, such as bradykinesia, rigidity and tremor, have not been reported in affected dogs (4,5). Therefore, wild-type and mutant mice were subjected to a variety of sensorimotor and cognitive tests at different ages.
Sensorimotor function was measured using tests sensitive to varying types of motor dysfunction and included challenging beam traversal, spontaneous activity, gait and nest building (35–37). Sensorimotor function did not differ between genotypes at 5–12 months of age; however, at 20–29 months of age Atp13a2−/− mice displayed significant impairments in all of the motor tests. On the beam mutant mice made more errors per step compared with wild-type mice and analysis of trial-by-trial performance revealed potential motor learning deficits as well. In the trial-by-trial analysis, both genotypes made a similar number of errors on the first trial, but while wild-type mice improved performance on subsequent trials, Atp13a2−/− mice did not. A similar pattern was also observed in time to traverse, suggesting the aged mutant mice have impaired aspects of motor learning. In addition, Atp13a2−/− mice showed reduced hindlimb stepping in the cylinder, and exhibited impairments in fine motor skills and orofacial movements involved in nestbuilding. In analyses of gait, aged mutant mice exhibited a shortened stride length and while there was no statistical difference in the variability of stride lengths between genotypes, there was a slight trend for mutants to have more variable stride lengths. However, when males and females were analyzed separately we found male Atp13a2−/− mice had significantly more variable stride lengths compared with male Atp13a2+/+ mice suggesting ataxia-like movements in aged males. The sensorimotor deficits observed in aged mutant mice are similar to deficits seen in genetic mouse models of PD and ataxia (36–38) and resemble aspects of motor dysfunction observed in KRS, NCL and PD.
Cognitive function and emotional reactivity were also compared in Atp13a2−/− and Atp13a2+/+ mice. In the water-maze task, mutant mice found the platform faster, which would typically indicate improved spatial learning capacity and hippocampal function. However, the improved escape latency was likely due to mean increases in swim speed displayed by the mutant mice. Moreover, the distance traveled before finding the platform did not distinguish mutant and wild-type mice. In the object recognition and latency to explore tests, mutant mice demonstrated greater exploratory/investigatory activities that were not accompanied by changes in general locomotor activity or anxiety, as assessed in open-field and light-dark tests, respectively. It is possible that such changes in exploratory behavior are linked to altered hippocampal function. Hippocampal neurons fire during spatial exploration (39,40), and it is possible that lipofuscin and α-synuclein accumulation in or near these neurons disrupts their spatial processing function. It is noteworthy that enhanced exploration has also been observed in other mouse models of PD (41), suggesting that different cellular pathways implicated in PD can lead to a similar behavioral phenotype.
Although there is currently no reported postmortem analysis of brain histopathology in KRS, imaging studies show that some patients develop diffuse cerebral and cerebellar atrophy and decreased DAT activity in the striatum, but there is no evidence of abnormalities in the SN (42,43). In the one known family with ATP13A2-associated NCL in humans, brain histopathology has not been reported, but in a peripheral nerve biopsy these patients exhibited autofluorescent inclusions (11). In canine NCL, diffuse atrophy in the brain and accelerated accumulation of autofluorescent storage material, or lipofuscin, has been shown in both the cortex and the cerebellum (4). Lipofuscin develops in the lysosomes of cells over the course of aging and accumulates preferentially in the cytosol of postmitotic cells. Its formation is negatively associated with cell survival (44–46) and it is known from lysosomal storage disorders that substantial deposition of lipofuscin is associated with cellular dysfunction (47). Therefore, we measured autofluorescent material in multiple brain regions of Atp13a2−/− mice. We found enhanced autofluorescence and luxol blue-positive staining indicative of lipofuscin in the cortex, cerebellum and the CA3 region of the hippocampus in Atp13a2−/− mice compared with age-matched controls. Thus, the results of the current study, and previous studies of ATP13A2-deficiency in dogs and humans, clearly demonstrate that the loss of this P-type ATPase causes lipofuscinosis in all three species.
A characteristic feature of PD is the accumulation of aggregated forms of α-synuclein (48,49). Although there is no confirmation of α-synuclein pathology in KRS or ATP13A2-associated NCL, dysfunction of the lysosomal degradation pathway is associated with both PD and α-synuclein aggregation. For example, mutations in GCase cause Gaucher's disease, a rare inherited lysosomal storage disorder, and are the most common genetic risk factor for PD (50,51). The lysosomal degradation pathway is particularly important for clearance of aggregated α-synuclein (52), and studies in yeast (14) and in mammalian cell culture, including mouse primary cortical neurons and rat primary mesencephalic dopaminergic neurons, show that the loss of Atp13a2 function results in α-synuclein aggregation and cellular toxicity (16,20). Consistent with those studies, we show for the first time that depletion of Atp13a2 in vivo leads to the accumulation of α-synuclein in the mouse brain. Loss of Atp13a2 did not cause changes in levels of soluble α-synuclein in the hippocampus, cortex or cerebellum, but did result in ∼3-fold increase in Triton-insoluble α-synuclein in the hippocampus. It is important to note that lipofuscin accumulation in Atp13a2−/− brains was most pronounced in the hippocampus, where α-synuclein accumulation was also observed. This suggests that there may be a relationship, most likely lysosomal dysfunction, between lipofuscin accumulation and the formation of insoluble α-synuclein. Tau and huntingtin proteins were also analyzed and showed no changes in protein solubility, indicating that the loss of Atp13a2 causes specific aggregation of α-synuclein. These results highlight the functional importance of Atp13a2 in lysosomal degradation of α-synuclein in a mammalian brain in vivo. Recent studies have also shown that GCase enzyme activity is reduced in PD patients (22–27). Therefore, we examined GCase enzyme activity and protein levels in whole brain extracts from Atp13a2−/− and Atp13a+/+ mice. However, no significant differences were observed across genotypes. It will be worthwhile in future studies to measure GCase enzyme activity and protein levels in specific brain regions of the knockout mice.
We also compared the expression of genes implicated in PD and autophagy in the striatum and SN of Atp13a2+/+ and Atp13a2−/− mice at 8–15 months of age using pathway-specific real-time PCR arrays. The observed down-regulation of Nurr1 and Bdnf in the striatum of Atp13a2−/− mice is particularly noteworthy as these genes play an important role in the development and maintenance of dopaminergic neurons (53,54). Moreover, previous studies have shown that Nurr1 expression is decreased in PD and may play an important role in the pathogenesis of the disease (53). Though it is unclear why these genes are down-regulated in the striatum and not the SN, it suggests that the loss of Atp13a2 leads to perturbations in the basal ganglia region of the brain. Indeed, imaging studies suggest iron accumulation in caudate and putamen in KRS (42,43). It is also noteworthy that the absence of changes in Nurr1 expression in the SN is consistent with the analysis of nigrostriatal integrity showing no differences in nigrostriatal dopaminergic neuron numbers or in the striatal DA content in the mutant mice (53).
Because the behavioral phenotype was not detectable until old age, it seemed possible that it might be modulated to some extent by compensatory mechanisms associated with other members of the P5-ATPase family. We examined mRNA levels for the other members of the P5-ATPase family (Atp13a1 and Atp13a3-5) and found no compensatory up-regulation. This was not surprising, as there is no evidence that the other isoforms are expressed in lysosomes and, in fact, the yeast homologue of ATP13A1 is expressed in the endoplasmic reticulum (55). Furthermore, there is suggestive evidence that substrate specificities may differ between isoforms. As noted previously (1), P-type pumps that share the same substrate specificity and stoichiometry, such as isoforms of the sarco(endo)plasmic reticulum Ca2+-ATPase, plasma membrane Ca2+-ATPase or Na+,K+-ATPase, have a high degree of amino acid sequence identity in their transmembrane domains. In contrast, for the P5-ATPases, including those whose genes are tandemly linked on the same chromosome, the transmembrane domains exhibit a high degree of sequence divergence (1). This suggests that ATP13A2 and the other P5-ATPases, rather than transporting a specific inorganic cation, may transport substrates belonging to a general class of molecules, with each isoform having different substrate specificities. Other investigators have also speculated that ATP13A2 may transport substrates other than inorganic cations, such as a cofactor for lysosomal enzymes (56), or that its transport activity might affect lysosomal pathways for ceramide metabolism (57). This is a reasonable hypothesis, as some P4-ATPases are known to mediate phospholipid translocation (58) and the yeast homolog of ATP13A1 affects regulation of the HMG-CoA reductase, a critical enzyme of the mevalonate pathway (59), raising the possibility that it handles lipid substrates. Further studies are needed to address this important issue.
In summary, our data show that the loss of Atp13a2 in mice leads to lipofuscinosis that is a critical feature of NCL and supports recent in vitro studies showing that ATP13A2 deficiency compromises lysosomal function and results in both lipofuscinosis and abnormal accumulation of α-synuclein (14,16,20). Additionally, these mice will serve as a useful model for clarifying the relationship between NCL and KRS and furthering our understanding of the pathogenic mechanisms underlying these disease states, and others, such as PD.
MATERIALS AND METHODS
Generation of mutant mice
A replacement-type vector was used to exchange exons 12–15 and the 5′ half of exon 16 with a neomycin resistance (Neo) gene, which was in the opposite orientation relative to the Atp13a2 transcription unit. Homologous arms flanking the Neo gene were amplified by PCR from mouse genomic DNA. The 3.2 kb 5′ arm was amplified using forward (5′-GGTGCAGACTGAGGCTGTTGTTCAG-3′) and reverse (5′-TCCTGCATTATGCAGAGTCTCAGCC-3′) primers complementary to sequences in exon 4 and intron 11, respectively, whereas the forward (5′-GCTTGGACGTAATGGGCGTGGTGC-3′) and reverse primers (5′-GATGACAAGGAATGGGGTCACTCTG-3′) used to amplify the 2.2 kb 3′ arm correspond to sequences in exon 16 and intron 21. Gene targeting and blastocyst-mediated transgenesis were performed using standard protocols, and chimeras were bred with Black Swiss mice to obtain ES cell-derived offspring. The mice are available to interested researchers directly through the corresponding author.
Southern blot analysis and PCR genotyping
Southern blot analysis of ES cell and mouse tail DNA was carried out as described previously (60). The 419 base pair (bp) 5′ probe began at nucleotide (nt) 879 of intron 3 and ended at nt 22 of exon 4, and the 304 bp 3′ probe began at nt 98 of intron 21 and ended at nt 80 of exon 23. Primers used to amplify the probes were as follows: 5′ probe, (5′-AGGATGCTTGCTGCATAGGTG-3′ and 5′-TGAAGAGCTGTCTGGAGCTAC-3′); 3′ probe, (5′-CTGAGCTGAGTTTCCTGACTG-3′ and 5′-TGAAGCCTCTGCAGTTCGCAC-3′). PCR genotyping was performed using DNA from tail biopsies. Forward (5′-AACACTCCGTGCTTCAGTTTCC-3′) and reverse (5′-AAGGACACGGGACGCAAGCAG-3′) primers corresponding to nucleotides 133–154 and 277–297 of intron 11, respectively, amplified a 165 bp product from the wild-type gene. A reverse primer (5′-CCAATACGCCCGCGTTTCTTCC-3′) complementary to sequences within the Neo gene, and the forward primer described above amplified a 284 bp product from the targeted allele.
RNA isolation and northern blot analysis
Total RNA and Poly(A) RNA was isolated using Tri-Reagent and Oligo (dT)-Cellulose Columns (Molecular Research Center, Inc.). Poly(A) RNA (5 μg) was denatured with glyoxal and dimethyl sulfoxide, fractionated in 1% agarose and transferred to a nylon membrane. Hybridization with 32P-labeled probes was performed as described previously (60). cDNA probes for wild-type and mutant Atp13a2 transcripts included 652 bp (5′ probe) and 1775 bp (3′ probe) fragments spanning codons 60–276 and 568–1158, respectively, and a 760 bp probe corresponding to codons 1–240 of the Neo gene. cDNA probes for the other P5-ATPases (Atp13a1, Atp13a3, Atp13a4 and Atp13a5) have been described previously (60).
RT–PCR and sequence analysis of the homozygous mutant transcript
Oligo(dT) primed cDNA was generated from Atp13a2−/− brain poly(A) RNA (∼250 ng) using the Invitrogen SuperScript First-Strand Synthesis System for RT–PCR. PCR amplification was carried out using AccuPrime Taq DNA polymerase (Invitrogen) and primers specific for Atp13a2 exon 9 (5′-ACTACTGGTATGCTCTCTGC-3′) and the Neo gene (5′-GCATATTAAGGTGACGCGTGTG-3′). The resultant PCR product was purified using a QIAquick Gel Extraction Kit (Qiagen) and sequenced (Retrogen, Inc.).
Western blot analysis
For the detection of Atp13a2 protein, brains were sonicated in 2% SDS, 50 mm Tris–HCl (pH 6.8) and the protein content was determined by the BCA assay. Lysates (50 and 75 µg) were mixed with 4× Laemmli buffer, boiled for 10 min, resolved on a 4–20% acrylamide gel, transferred to nitrocellulose and blocked for 1 h in 5% skim milk in Tris-buffered saline (TBS). The membrane was incubated overnight at 4°C with a 1:500 dilution of rabbit anti-human ATP13A2 antibody (Sigma A9607, which identifies amino acids 195–213), washed and incubated in anti-rabbit horseradish peroxidase labeled secondary antibody (1:5000 dilution) for 1 h at room temperature (RT). After washes in TBS, the membranes were treated with ECL plus reagent for 1 min, and signals were detected with the Protein Simple Imager.
Breeding and behavioral testing
Heterozygous male and female mice were bred together to produce knockout and wild-type mice used in all experiments. Separate cohorts of Atp13a2+/+ and Atp13a2−/− mice were tested at 5–12 and 20–29 months of age for motor performance and coordination, spontaneous activity and gait.
Challenging beam traversal
Motor performance and coordination were measured with the challenging beam traversal test (36,61–65). Briefly, the beam consists of four sections (25 cm each, 1 m total length), each section having a different width. The beam starts at a width of 3.5 cm and gradually narrows by 1 cm increments a final width of 0.5 cm. Animals were trained to traverse the length of the beam starting at the widest section and ending at the narrowest section. Animals received 2 days of training prior to testing; on the day of the test, a mesh grid (1 cm2) of corresponding width was placed over the beam surface leaving ∼1 cm space between the grid and the beam surface. Animals were then videotaped, while traversing the grid-surfaced beam and videotapes were rated in slow motion by an experimenter blind to genotype. Errors, steps and time to traverse the beam were measured across five trials, and the means of those trials were included in the analysis.
Spontaneous activity
Spontaneous movements of the mice were measured in a small, transparent cylinder 15.5 cm high and 12.7 cm in diameter (36,61–65). The cylinder was placed on a piece of glass with a mirror positioned at an angle beneath the cylinder to allow a clear view of movements along the ground and walls of the cylinder. Videotapes were viewed and rated in slow motion by an experimenter blind to mouse genotype. The number of rears, forelimb and hindlimb steps, and time spent grooming over a 3-min period were measured for each mouse.
Gait analysis
To measure gait, animals were trained to walk through a narrow alley leading into their home cage. Once trained, paper was placed along the alley floor and each animal's hindlimbs were brushed with non-toxic paint. Animals were then placed at the beginning of the alley. As they walked into their home cage they left their paw prints on the paper underneath (35,36,61,66–68). The stride length was determined by measuring the distance between hindlimb prints. Only strides made while continuously walking (no stopping) were included in the analysis. Stride lengths at the beginning and end of the alley were not counted since animals tend to make irregular steps at the beginning and typically stop and make smaller steps just before entering the cage. In addition, the range of stride lengths for each animal was measured by subtracting the shortest stride length from the longest stride length (69).
Nest building
Nest building, a natural mouse behavior was measured in Atp13a2+/+ and Atp13a2−/− mice at 20–29 months of age. The behavior requires the use of orofacial and complex forelimb movements, where animals pull the nesting material apart with their forelimbs and teeth and then break down the material in their mouths and incorporate it into their bedding (36,70–73). Pre-weighed cotton was placed into the feeder of the cage in individually housed mice. Pulling the cotton from the feeder requires rearing up in addition to complex fine motor skills. Cotton use was monitored at 24 h by weighing the remaining cotton in the feeder.
Cognitive testing
Water-maze learning and memory, novel object and spatial recognition and locomotor/exploratory activity tests are described in detail in the Supplementary Material, Methods.
Quantitative RT–PCR array analyses
Total RNA was extracted from freshly dissected mouse brain tissues from 8- to 15-month-old mice using TRI Reagent® (Molecular Research Center). Prior to use in cDNA synthesis reactions, RNAs were further purified to remove DNA and other contaminants by using an RNeasy® Mini kit (Qiagen). RNA concentration and purity were assessed spectrophotometrically using a NanoDrop ND-1000 (Thermo Scientific NanoDrop Products). RNA samples were reverse-transcribed using an RT2 First-Strand cDNA kit (SABiosciences) and gene expression levels in Atp13a2+/+ and Atp13a2−/− tissues were compared using Autophagy (PAMM-084) and Parkinson's (PAMM-124) RT2 Profiler PCR arrays (SABiosciences) according to the manufacturer's procedures. Complete listings of the genes in each array are available at http://www.sabiosciences.com/ArrayList.php?pline=PCRArray. Thermocycling and data collection were carried out with a model 7300 Real-Time PCR system (Applied Biosystems) and data were analyzed by the ΔΔCT method (74) using the Web-based RT2 Profiler PCR Array Data Analysis tool (version 3.5, SABiosciences). For analyses of gene expression in the mouse striatum, the Gusb, Hsp90ab1, Gapdh and Actb transcripts were used as internal housekeeping controls. For analyses of expression in the SN, the Hsp90ab1, Gapdh and Actb transcripts were used.
Soluble and insoluble α-synuclein accumulation
To detect Triton and SDS soluble forms of α-synuclein, tau and huntingtin proteins, lysates from the frontal cortex, hippocampus and cerebellum of mutant and wild-type mice at 18–20 months of age were sequentially extracted as described previously (75). Frozen brains were homogenized in Triton X-100 solution with a homogenizer (Glas-Col), and subjected to four freeze-thaw cycles. Homogenates were then centrifuged at 100 000g for 30 min. After taking supernatants, the pellets were homogenized in Triton X-100 solution and centrifuged again. The second supernatants were mixed with the first one and used as Triton-soluble (T-sol) fractions. The pellets were resuspended with 2% SDS buffer, sonicated and boiled for 10 min. After centrifugation at 16 000g for 30 min, the supernatants were removed and used as SDS-soluble (SDS-sol) fractions. Thirty micrograms of protein was loaded on 4–12% Tris–glycine gels or 3–8% Tris–acetate gels. After separation, samples were transferred to PVDF membranes (Millipore), which were fixed in 0.4% PFA/TBS for 10 min, and then blocked in 5% milk/TBS with 0.1% Tween20 (TBS-T) for 1 h at RT. Membranes were incubated with α-synuclein antibody (202; Covance), tau antibody (A0024; Dako) or huntingtin antibody (5656; Cell Signaling) in TBS-T with 5% BSA at 4°C overnight. After washing 3 times with TBS-T, the membranes were incubated with secondary antibodies for 1 h at RT. The membranes were visualized using a LI-COR Odyssey infrared fluorescence scanner.
GCase enzyme activity assay
The mouse brain tissue was diluted 1:10 (w/v) in citrate-phosphate extraction buffer [150 mm citrate-phosphate buffer pH 5.4, 0.25% Triton X-100, protease inhibitor mix (Roche Diagnostics, Indianapolis, IN, USA)] and homogenized with a mechanical tissue homogenizer (Omni International, Kennesaw, GA, USA). The homogenate was sonicated for 2 × 10 s at 50% amplitude followed by centrifugation at 10 000g for 15 min at 4°C. The GCase enzyme activity assay was based on a previously described method (76). GCase or GAA activity was measured in assay buffer which consisted of citrate-phosphate buffer (pH 5.4) with 10 mm 4-methylumbelliferyl-β-d-glucopyranoside (4MU-β-glc) or 10 mm 4-methylumbelliferyl-α-d-glucopyranoside (4MU-α-glc), respectively. NaTC was not added to the buffer since this chemical is known to over-activate the GCase enzyme (77), which could skew read-out of the assay. Briefly, 5 μl of brain extract was added to wells of black 384-well plates (Greiner Bio-One, Inc, Monroe, NC, USA). On each plate, each sample was read in triplicate. For the GCase assay, conduritol-B-epoxide (CBE) triplicates were included for correction of GBA2 activity; 5 µl of 100 µm CBE was added followed by brief centrifugation and incubation at 37°C for 15 min at 600 rpm on a plate shaker. Assay buffer was added to each well to a total volume of 30 μl and incubated at 37°C for 1 h at 600 rpm. Thirty milliliters of stop solution (1 m glycine, pH 12.5) was added to each well and the fluorescence was measured using a Victor plate reader (Perkin-Elmer, Waltham, MA, USA), at 365 nm excitation and 440 nm emission. Enzyme activity was normalized to total protein amount.
Measurement of GCase protein levels in mouse brain extracts
A GCase-specific chemical probe (MDW933) was synthesized at the Imaging Probe Development Center (IPDC, NHLBI, NIH) as previously described (27). Total protein amount in each sample was determined by the Bradford assay according to the manufacturer's guidelines (Bio-Rad, Hercules, CA, USA). Ten micrograms of mouse brain tissue homogenate was incubated with 100 nm green fluorescent MDW933 probe in NaAc buffer (pH 5.0) for 90 min at 37°C followed by analysis on SDS–PAGE. As a control, we incubated 60 nm Cerezyme (Genzyme, Cambridge, MA, USA) with 100 nm MDW933 probe. Fluorescent signal in the gel was detected with a Typhoon Variable Mode Imager (Amersham Biosciences, Piscataway, NJ, USA) with λex at 488 nm and λem at 520 nm. ImageJ was used for densitometry analysis (version 1.44o; NIH, Bethesda, MD, USA).
Measurement of striatal dopamine content
Brains were removed from 20 to 29-month-old Atp13a2+/+ and Atp13a2−/− mice (n = 10 for each genotype), and one hemisphere was flash frozen in liquid nitrogen for striatal DA measurements and the other was fixed in 4% paraformaldehyde for 20 h and then processed for stereology. Striatal DA levels were analyzed by the Neurochemistry Core Laboratory in the Center for Molecular Neuroscience at Vanderbilt University. Striatal punches were homogenized in 100–750 μl of 0.1 m trichloracetic acid (TCA), 10−2 m sodium acetate, 10−4 m EDTA, 5 ng/ml isoproterenol (as an internal standard) and 10.5% methanol (pH 3.8). Samples were centrifuged at 10000g for 20 min, and the pellet and supernatant were stored separately at −80°C. Supernatant was thawed and spun for 20 min and then analyzed for biogenic amines using an HPLC assay utilizing an Antec Decade II (oxidation: 0.5) electrochemical detector at 33°C. Twenty microlitre samples were injected using a Water 717+ autosampler onto a Phenomenex Nucleosil (5 μm, 100 Å) C18 HPLC column (150 × 4.60 mm). Biogenic amines were eluted with a mobile phase consisting of 89.5% 0.1 m TCA, 10−2 m sodium acetate, 10−4 m EDTA and 10.5% methanol (pH 3.8). Solvent was delivered at 0.6 ml/min using a Waters 515 HPLC pump. Using this HPLC solvent the following biogenic amines were eluted in the following order: noradrenaline, 3-methoxy-4-hydroxyphenylglycol, adrenaline, 3,4-dihydroxyphenylacetic acid (DOPAC), DA, 5-hydroxyindole acetic acid (5-HIAA), homovanillic acid (HVA), 5-hydroxytryptamine (serotonin, 5-HT) and 3-methoxytyramine. HPLC control and data acquisition were managed by the Millennium 32 software. Total protein concentration of the brain extracts was determined by the standard BCA Protein Assay (Pierce Chemical, Rockford, IL, USA).
Tyrosine hydroxylase immunostaining and stereology
After fixation (see above) brain hemispheres from 6 Atp13a2+/+ mice and 6 Atp13a2−/− mice, ages 20–29 months were processed for stereology. TH immunostaining was performed by FD Neurotechnologies (Ellicott City, MD, USA). Serial cryostat sections (40 μm) were cut coronally through the cerebrum and the brain stem containing the striatum, the SN and the locus coeruleus, ∼1.94 to −5.88 mm from Bregma (78). TH immunohistochemistry was performed on sections from the SN (∼−2.30 to −4.16 mm from Bregma). After inactivating the endogenous peroxidase activity with hydrogen peroxidase, sections were incubated separately with avidin and biotin solutions (Vector Lab, Burlingame, CA, USA) to block non-specific binding of endogenous biotin, biotin-binding protein and lectins. Sections were then incubated free-floating in PBS-containing normal blocking serum, Triton X-100 and specific antibody for 3 days at 4°C. Subsequently, the immunoreaction product was visualized by the avidin–biotin complex method using a Vectastin elite ABC kit (Vector Lab., Burlingame, CA, USA) and 3′,3′-diaminobenzidine as a chromogen. After washing, all sections were mounted on gelatin-coated slides, counterstained with thionin, dehydrated in ethanol, cleared in xylene and coverslipped in Permount® (Fisher Scientific, Fair Lawn, NJ, USA).
The total number of TH-immunoreactive neurons in the SN and the total volume of SN were quantified using the optical fractionator method (79) and the Cavalieri-point counting method (80), as previously reported (81–83). These studies were carried out with assistance from a commercially available computerized stereology system (Stereologer) by trained personnel blind to group and genotype at a contract research organization (Stereology Resource Center, Chester, MD, USA).
Histology
Brains were isolated from 15- to 18-month-old mutant and wild-type mice. LFB-PAS staining was performed on paraffin sections as previously described (84). For autofluorescence, deparafinized and rehydrated sections were coverslipped with fluorescent mounting medium and viewed under a fluorescent microscope.
Statistical analysis
Challenging beam, spontaneous activity and gait data collected from Atp13a2+/+ and Atp13a2−/− mice at 5–12 months of age were analyzed using Student's t-test or the Mann–Whitney U test depending on whether the data maintained homogeneity of variance. Similarly, beam, activity, gait and nestbuilding data collected in a separate cohort of mice at 20–29 months of age were analyzed using Student's t-test or the Mann–Whitney U test. Beam data at 20–29 months were also analyzed by trial. Here, a 2 × 5 mixed design ANOVA was used to compare scores between genotypes across each of the five trials. Post hoc analysis was done using Fisher's LSD. All analyses were conducted with the GB-STAT software (Dynamic Microsystems, Inc., Silver Spring, MD, 2000) for Macintosh. The level of significance was set at P < 0.05.
SUPPLEMENTARY MATERIAL
Funding
This work was supported by the National Institutes of Health (NS070268 to P.J.S., GM103436 to P.J.S. and M.E.B., DK050594 and HL061974 to G.E.S., NS077022 to S.M.F., MH094955 to M.E.B., NS076054 to D.K.); the UF CTRND and Department of Neuroscience; the UC Gardner Family Center for Parkinson's Disease and Movement Disorders and the Intramural Research program of the National Human Genome Research Institute at the National Institutes of Health, Bethesda, MD, USA.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Gene Targeted Mouse Service Core of the University of Cincinnati College of Medicine for design and construction of the targeting vector and generation of mice heterozygous for Atp13a2. Additional thanks to Abbey Bailes, Christina Basinger and Kelsey Carnahan of the Schultheis lab for their help with maintenance of the Atp13a2 mouse colony. We also thank Dr Gary Griffiths and Dr Zhen-Dan Shi (IPDC, NHLBI, NIH) for synthesis of the MDW933 probe.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Schultheis P.J., Hagen T.T., O'Toole K.K., Tachibana A., Burke C.R., McGill D.L., Okunade G.W., Shull G.E. Characterization of the P5 subfamily of P-type transport ATPases in mice. Biochem. Biophys. Res. Comm. 2004;323:731–738. doi: 10.1016/j.bbrc.2004.08.156. [DOI] [PubMed] [Google Scholar]
- 2.Sørensen D.M., Buch-Pedersen M.J., Palmgren M.G. Structural divergence between the two subgroups of P5 ATPases. Biochim. Biophys. Acta. 2010;1797:846–855. doi: 10.1016/j.bbabio.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Ramirez A., Heimbach A., Grundemann J., Stiller B., Hampshire D., Cid L.P., Goebel I., Mubaidin A.F., Wriekat A.-L., Roeper J., et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nature Genet. 2006;38:1184–1191. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 4.Farias F.H.G., Zeng R., Johnson G.S., Wininger F.A., Taylor J.F., Schnabel R.D., McKay S.D., Sanders D.N., Lohi H., Seppala E.H., et al. A truncating mutation in ATP13A2 is responsible for adult-onset neuronal ceroid lipofuscinosis in Tibetan terriers. Neurobiol. Dis. 2011;42:468–474. doi: 10.1016/j.nbd.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Wohlke A., Philipp U., Bock P., Beineke A., Lichtner P., Meitinger T., Distl O. A one base pair deletion in the canine ATP13A2 gene causes exon skipping and Late-onset Neuronal Ceroid Lipofusccinosis in the Tibetan Terrier. PLoS Genet. 2001;7:e1002304. doi: 10.1371/journal.pgen.1002304. doi:10.1371/journal.pgen1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bras J., Verloes A., Schneider S.A., Mole S.E., Guerreiro R.J. Mutation of the parkinsonism gene ATP13A2 causes neuronal ceroid-lipfuscinosis. Hum. Mol. Gen. 2012;21:2646–2650. doi: 10.1093/hmg/dds089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Najim Al-Din A.S., Wriekat A., Mubaidin A., Dasouki M., Hiari M. Pallido-pyramidal degeneration, supranuclear upgaze paresis and dementia: Kufor-Rakeb syndrome. Acta Neurol. Scand. 1994;89:347–352. doi: 10.1111/j.1600-0404.1994.tb02645.x. [DOI] [PubMed] [Google Scholar]
- 8.Williams D.R., Hadeed A., Al-Din A.S., Wreikat A.L., Lees A.J. Kufor Rakeb disease: autosomal recessive, levodopa-responsive parkinsonism with pyramidal degeneration, supranuclear gaze palsy, and dementia. Mov. Disord. 2005;20:1264–1271. doi: 10.1002/mds.20511. [DOI] [PubMed] [Google Scholar]
- 9.Lees A.J., Singleton A.B. Clinical heterogeneity of ATP13A2 linked disease (Kufor-Rakeb) justifies a PARK designation. Neurology. 2007;68:1553–1554. doi: 10.1212/01.wnl.0000265228.66664.f4. [DOI] [PubMed] [Google Scholar]
- 10.Jalanko A., Braulke T. Neuronal ceroid lipfuscinoses. Biochem. Biophys. Acta. 2009;1793:697–709. doi: 10.1016/j.bbamcr.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Tome F.M., Brunet P., Fardeau M., Hentati F., Reix J. Familial disorder of the central and peripheral nervous systems with particular cytoplasmic lamellated inclusions in peripheral nerves, muscle satellite cells, and blood capillaries. Acta Neuropathol. 1985;68:209–217. doi: 10.1007/BF00690197. [DOI] [PubMed] [Google Scholar]
- 12.De Volder A.G., Cirelli S., de Barsy T., Brucher J.M., Bol A., Michel C., Goffinet A.M. Neuronal ceroid-lipofuscinosis: preferential metabolic alterations in thalamus and posterior association cortex demonstrated by PET. J. Neurol. Neurosurg. Psychiatry. 1990;53:1063–1067. doi: 10.1136/jnnp.53.12.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt K., Wolfe D.M., Stiller B., Pearce D.A. Cd2+, Mn2+, Ni2+, and Se2+ toxicity to Saccharomyces cerevisiae lacking YPK9p the orthologue of human ATP13A2. Biochem. Biophys. Res. Comm. 2009;383:198–202. doi: 10.1016/j.bbrc.2009.03.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gitler A.D., Chesi A., Geddie M.L., Strathearn K.E., Hamamichi S., Hill K.J., Caldwell K.A., Caldwell G.A., Cooper A.A., Rochet J.C., et al. Alpha-synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nat. Genet. 2009;41:308–315. doi: 10.1038/ng.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burton N.C., Guilarte T.R. Manganese neurotoxicity: lessons learned from longitudinal studies in nonhuman primates. Environ. Health. Perspect. 2009;117:325–332. doi: 10.1289/ehp.0800035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Usenovic M., Tresse E., Mazzulli J.R., Taylor J.P., Krainc D. Deficiency of ATP13A2 leads to lysosomal dysfunction, α-synuclein accumulation, and neurotoxicity. J. Neurosci. 2012;32:4240–4246. doi: 10.1523/JNEUROSCI.5575-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polymeropoulos M.H., Lavedan C., Leroy E., Ide S.E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., et al. Mutation in the α-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 18.Spillantini M.G., Crowther R.A., Jakes R., Hasegawa M., Goedert M. α-Synuclein in filamentous inclusions of Lewy bodies from Parkinson's disease and dementia with Lewy bodies. Proc. Natl Acad. Sci. USA. 1998;95:6469–6473. doi: 10.1073/pnas.95.11.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luk K.C., Kehm V., Carroll J., Zhang B., O'Brien P., Trojanowski J.Q., Lee V.M. Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012;338:949–953. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dehay B., Ramirez A., Martinez-Vicente M., Perier C., Canron M.H., Doudnikoff E., Vital A., Vila M., Klein C., Bezard E. Loss of P-type ATPase ATP13A2/PARK9 function induces general lysosome deficiency and leads to Parkinson disease neurodegeneration. Proc. Natl Acad. Sci. USA. 2012;109:9611–9616. doi: 10.1073/pnas.1112368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haltia M. The neuronal ceroid-liposuscinoses: from past to present. Biochim. Biophys. Acta. 2006;1762:850–856. doi: 10.1016/j.bbadis.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Balducci C., Pierguidi L., Persichetti E., Parnetti L., Sbaragli M., Tassi C., Orlacchio A., Calabresi P., Beccari T., Rossi A. Lysosomal hydrolases in cerebrospinal fluid from subjects with Parkinson's disease. Mov. Disord. 2007;22:1481–1484. doi: 10.1002/mds.21399. [DOI] [PubMed] [Google Scholar]
- 23.Gegg M.E., Burke D., Heales S.J., Cooper J.M., Hardy J., Wood N.W., Schapira A.H. Glucocerebrosidase deficiency in substantia nigra of Parkinson disease brains. Ann. Neurol. 2012;72:455–463. doi: 10.1002/ana.23614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parnetti L., Balducci C., Pierguidi L., De Carlo C., Peducci M., D'Amore C., Padiglioni C., Mastrocola S., Persichetti E., Paciotti S., et al. Cerebrospinal fluid beta-glucocerebrosidase activity is reduced in Dementia with Lewy Bodies. Neurobiol. Dis. 2009;34:484–486. doi: 10.1016/j.nbd.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Sardi S.P., Clarke J., Viel C., Chan M., Tamsett T.J., Treleaven C.M., Bu J., Sweet L., Passini M.A., Dodge J.C., et al. Augmenting CNS glucocerebrosidase activity as a therapeutic strategy for parkinsonism and other Gaucher-related synucleinopathies. Proc. Natl Acad. Sci. USA. 2013 doi: 10.1073/pnas.1220464110. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raben N., Wong A., Ralston E., Myerowitz R. Autophagy and mitochondria in Pompe disease: nothing is so new as what has long been forgotten. Am. J. Med. Genet. C Semin. Med. Genet. 2012;160:13–21. doi: 10.1002/ajmg.c.31317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Witte M.D., Kallemeijn W.W., Aten J., Li K.Y., Strijland A., Donker-Koopman W.E., van den Nieuwendijk A.M., Bleijlevens B., Kramer G., Florea B.I., et al. Ultrasensitive in situ visualization of active glucocerebrosidase molecules. Nat. Chem. Biol. 2010;6:907–913. doi: 10.1038/nchembio.466. [DOI] [PubMed] [Google Scholar]
- 28.Park J.S., Mehta P., Cooper A.A., Veivers D., Heimbach A., Stiller B., Kubisch C., Fung V.S., Krainc D., Mackay-Sim A., et al. Pathogenic effects of novel mutations in the P-type ATPase ATP13A2 (PARK9) causing Kufor-Rakeb syndrome, a form of early-onset parkinsonism. Hum. Mutat. 2011;32:956–64. doi: 10.1002/humu.21527. [DOI] [PubMed] [Google Scholar]
- 29.Ugolino J., Fang S., Kubisch C., Monteiro M.J. Mutant Atp13a2 proteins involved in parkinsonism are degraded by ER-associated degradation and sensitize cells to ER-stress induced cell death. Hum. Mol. Genet. 2011;20:3565–3577. doi: 10.1093/hmg/ddr274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eiberg H., Hansen L., Korbo L., Nielsen I.M., Svenstrup K., Bech S., Pinborg L.H., Friberg L., Hjermind L.E., Olsen O.R., et al. Novel mutation in ATP13A2 widens the spectrum of Kufor-Rakeb syndrome (PARK9) Clin. Genet. 2012;82:256–263. doi: 10.1111/j.1399-0004.2011.01745.x. [DOI] [PubMed] [Google Scholar]
- 31.Hampshire D.J., Roberts E., Crow Y., Bond J., Mubaidin A., Wriekat A.L., Al-Din A., Woods C.G. Kufor-Rakeb syndrome, pallido-pyramidal degeneration with supranuclear upgaze paresis and dementia, maps to 1p36. J. Med. Genet. 2001;38:680–682. doi: 10.1136/jmg.38.10.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Behrens M.I., Brüggemann N., Chana P., Venegas P., Kägi M., Parrao T., Orellana P., Garrido C., Rojas C.V., Hauke J., et al. Clinical spectrum of Kufor-Rakeb syndrome in the Chilean kindred with ATP13A2 mutations. Mov. Disord. 2010;25:1929–1937. doi: 10.1002/mds.22996. [DOI] [PubMed] [Google Scholar]
- 33.Riis R.C., Cummings J.F., Loew E.R., de Lahunta A. Tibetan terrier model of canine ceroid lipofuscinosis. Am. J. Med. Genet. 1992;42:615–621. doi: 10.1002/ajmg.1320420437. [DOI] [PubMed] [Google Scholar]
- 34.Katz M.L., Shibuya H., Johnson G.S. Animal models for the ceroid lipofuscinoses. Adv. Genet. 2001;45:183–203. doi: 10.1016/s0065-2660(01)45011-5. [DOI] [PubMed] [Google Scholar]
- 35.Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J.N., Ried T., Tagle D., et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 36.Fleming S.M., Salcedo J., Fernagut P.O., Rockenstein E., Masliah E., Levine M.S., Chesselet M.F. Early and progressive sensorimotor anomalies in mice overexpressing wild-type human alpha-synuclein. J. Neurosci. 2004;24:9434–9440. doi: 10.1523/JNEUROSCI.3080-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glajch K.E., Fleming S.M., Surmeier D.J., Osten P. Sensorimotor assessment of the unilateral 6-hydroxydopamine mouse model of Parkinson's disease. Behav. Brain Res. 2012;230:309–316. doi: 10.1016/j.bbr.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reliene R., Fleming S.M., Chesselet M.F., Schiestl R.H. Effects of antioxidants oncancer prevention and neuromotor performance in Atm deficient mice. Food Chem. Toxicol. 2008;46:1371–1377. doi: 10.1016/j.fct.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henriksen E.J., Colgin L.L., Barnes C.A., Witter M.P., Moser M.B., Moser E.I. Spatial representation along the proximodistal axis of CA1. Neuron. 2010;68:127–137. doi: 10.1016/j.neuron.2010.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartley M., Taylor N., Taylor J. Subfield variations in hippocampal processing-components of a spatial navigation system. Neural. Netw. 2005;18:611–619. doi: 10.1016/j.neunet.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 41.Navarro P., Guerrero R., Gallego E., Avila J., Luquin R., Garcia Ruiz P.J., Sanchez M.P. Motor alterations are reduced in mice lacking the PARK2 gene in the presence of a human FTDP-17 mutant form of four-repeat tau. J. Neurol. Sci. 2008;275:139–144. doi: 10.1016/j.jns.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 42.Schneider S.A., Paisan-Ruiz C., Quinn N.P., Lees A.J., Houlden H., Hardy J., Bhatia K.P. ATP13A2 mutations (PARK9) cause neurodegeneration with brain iron accumulation. Mov. Disord. 2010;25:979–984. doi: 10.1002/mds.22947. [DOI] [PubMed] [Google Scholar]
- 43.Brüggemann N., Hagenah J., Reetz K., Schmidt A., Kasten M., Buchmann I., Eckerle S., Bähre M., Münchau A., Djarmati A., et al. Recessively inherited parkinsonism: effect of ATP13A2 mutations on the clinical and neuroimaging phenotype. Arch. Neurol. 2010;67:1357–1363. doi: 10.1001/archneurol.2010.281. [DOI] [PubMed] [Google Scholar]
- 44.Munnel J.F., Getty R. Rate of accumulation of cardiac lipofuscin in the aging canine. J. Gerontol. 1968;23:154–158. doi: 10.1093/geronj/23.2.154. [DOI] [PubMed] [Google Scholar]
- 45.Nakano M., Gotoh S. Accumulation of cardiac lipofuscin depends on metabolic rate of mammals. J. Gerontol. 1992;47:B126–B129. doi: 10.1093/geronj/47.4.b126. [DOI] [PubMed] [Google Scholar]
- 46.Sheehy M.R., Greenwood J.G., Fielder D.R. Lipofuscin as a record of rate of living in an aquatic poikilotherm. J. Gerontol. 1995;50:B327–B336. doi: 10.1093/gerona/50a.6.b327. [DOI] [PubMed] [Google Scholar]
- 47.Neufeld E.F. Lysosomal storage diseases. Annu. Rev. Biochem. 1991;60:257–280. doi: 10.1146/annurev.bi.60.070191.001353. [DOI] [PubMed] [Google Scholar]
- 48.Cookson M.R. α-Synuclein and neuronal cell death. Mol. Degeneration. 2009;4:9. doi: 10.1186/1750-1326-4-9. doi:10.1186/1750–1326–4–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Auluck P.K., Caraveo G., Lindquist S. α synuclein: membrane interactions and toxicity in Parkinson's disease. Annu. Rev. Cell Dev. Biol. 2010;26:211–233. doi: 10.1146/annurev.cellbio.042308.113313. [DOI] [PubMed] [Google Scholar]
- 50.Mazzulli J.R., Xu Y.H., Sun Y., Knight A.L., McLean P.J., Caldwell G.A., Sidransky E., Grabowski G.A., Krainc D. Gaucher disease glucocerebrosidase and α-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell. 2011;146:37–52. doi: 10.1016/j.cell.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sidransky E., Nalls M.A., Aasly J.O., Aharon-Peretz J., Annesi G., Barbosa E.R., Bar-Shira A., Berg D., Bras J., Brice A., et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N.Engl. J. Med. 2009;361:1651–1661. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cuervo A.M., Stefanis L., Fredenburg R., Lansbury P.T., Sulzer D. Impaired degradation of mutant α-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 53.Jankovic J., Chen S., Le W.D. The role of Nurr1 in the development of dopaminergic neurons and Parkinson's disease. Prog. Neurobiol. 2005;77:128–138. doi: 10.1016/j.pneurobio.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 54.Howells D.W., Porritt M.J., Wong J.Y., Batchelor P.E., Kalnins R., Hughes A.J., Donnan G.A. Reduced BDNF mRNA expression in the Parkinson's disease substantia nigra. Exp. Neurol. 2000;166:127–135. doi: 10.1006/exnr.2000.7483. [DOI] [PubMed] [Google Scholar]
- 55.Cronin S.R., Rao R., Hampton R.Y. Cod1p/Spf1p is a P-type ATPase involved in ER function and Ca2+ homeostasis. J. Cell Biol. 2002;157:1017–1028. doi: 10.1083/jcb.200203052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Covy J.P., Waxman E.A., Giasson B.I. Characterization of cellular protective effects of ATP13A2/PARK9 expression and alterations resulting from pathogenic mutants. J. Neurosci. Res. 2012;90:2306–2316. doi: 10.1002/jnr.23112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bras J., Singleton A., Cookson M.R., Hardy J. Emerging pathways in genetic Parkinson's disease: potential role of ceramide metabolism in Lewy body disease. FEBS J. 2008;275:5767–5773. doi: 10.1111/j.1742-4658.2008.06709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Folmer D.E., Elferink R.P., Paulusma C.C. P4 ATPases—lipid flippases and their role in disease. Biochim. Biophys. Acta. 2009;1791:628–35. doi: 10.1016/j.bbalip.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 59.Cronin S.R., Khoury A., Ferry D.K., Hampton R.Y. Regulation of HMG-CoA reductase degradation requires the P-type ATPase Cod1p/Spflp. J. Cell Biol. 2000;148:915–924. doi: 10.1083/jcb.148.5.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schultheis P.J., Clarke L.L., Meneton P., Harline M., Boivin G.P., Stemmermann G., Duffy J.J., Doetschman T., Miller M.L., Shull G.E. Targeted disruption of the murine Na+/H+ exchanger isoform 2 gene causes reduced viability of gastric parietal cells and loss of net acid secretion. J. Clin. Invest. 1998;101:1243–1253. doi: 10.1172/JCI1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fleming S.M., Salcedo J., Hutson C.B., Rockenstein E., Masliah E., Levine M.S., Chesselet M.F. Behavioral effects of dopaminergic agonists in transgenic mice overexpressing human wildtype alpha-synuclein. Neuroscience. 2006;142:1245–1253. doi: 10.1016/j.neuroscience.2006.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fleming S.M., Mulligan C.K., Richter F., Mortazavi F., Lemesre V., Frias C., Zhu C., Stewart A., Gozes I., Morimoto B., et al. A pilot trial of the microtubule-interacting peptide (NAP) in mice overexpressing alpha-synuclein shows improvement in motor function and reduction of alpha-synuclein inclusions. Mol. Cell Neurosci. 2011;46:597–606. doi: 10.1016/j.mcn.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goldberg M.S., Fleming S.M., Palacino J.J., Cepeda C., Lam H.A., Bhatnagar A., Meloni E.G., Wu N., Ackerson L.C., Klapstein G.J., et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J. Biol. Chem. 2003;278:43628–43635. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- 64.Hwang D.Y., Fleming S.M., Ardayfio P., Moran-Gates T., Kim H., Tarazi F.I., Chesselet M.F., Kim K.S. 3,4-dihydroxyphenylalanine reverses the motor deficits in Pitx3-deficient aphakia mice: behavioral characterization of a novel genetic model of Parkinson's disease. J. Neurosci. 2005;25:2132–2137. doi: 10.1523/JNEUROSCI.3718-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu X.H., Fleming S.M., Meurers B., Ackerson L.C., Mortazavi F., Lo V., Hernandez D., Sulzer D., Jackson G.R., Maidment N.T., et al. Bacterial artificial chromosome transgenic mice expressing a truncated mutant parkin exhibit age-dependent hypokinetic motor deficits, dopaminergic neuron degeneration, and accumulation of proteinase K-resistant alpha-synuclein. J. Neurosci. 2009;29:1962–1976. doi: 10.1523/JNEUROSCI.5351-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schallert T., Whishaw I.Q., Ramirez V.D., Teitelbaum P. Compulsive, abnormal walking caused by anticholinergics in akinetic, 6-hydroxydopamine-treated rats. Science. 1978;199:1461–1463. doi: 10.1126/science.564552. [DOI] [PubMed] [Google Scholar]
- 67.Fernagut P.O., Diguet E., Labattu B., Tison F. A simple method to measure stride length as an index of nigrostriatal dysfunction in mice. J. Neurosci. Methods. 2002;113:123–130. doi: 10.1016/s0165-0270(01)00485-x. [DOI] [PubMed] [Google Scholar]
- 68.Tillerson J.L., Caudle W.M., Reverón M.E., Miller G.W. Detection of behavioral impairments correlated to neurochemical deficits in mice treated with moderate doses of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Exp. Neurol. 2002;178:80–90. doi: 10.1006/exnr.2002.8021. [DOI] [PubMed] [Google Scholar]
- 69.Dunnett S.B. Assessment of motor impairments in transgenic mice. In: Crawley J.N., editor. Mouse Behavioral Phenotyping. Washington, DC: Society for Neuroscience; 2003. pp. 1–12. [Google Scholar]
- 70.Upchurch M., Schallert T. A behavior analysis of the offspring of “haloperidol-sensitive” and “haloperidol-resistant” gerbils. Behav. Neural. Biol. 1983;39:221–228. doi: 10.1016/s0163-1047(83)90876-2. [DOI] [PubMed] [Google Scholar]
- 71.Sedelis M., Hofele K., Auburger G.W., Morgan S., Huston J.P., Schwarting R.K. MPTP susceptibility in the mouse: behavioral, neurochemical, and histological analysis of gender and strain differences. Behav. Genet. 2000;30:171–182. doi: 10.1023/a:1001958023096. [DOI] [PubMed] [Google Scholar]
- 72.Hofele K., Sedelis M., Auburger G.W., Morgan S., Huston J.P., Schwarting R.K. Evidence for a dissociation between MPTP toxicity and tyrosinase activity based on congenic mouse strain susceptibility. Exp. Neurol. 2001;168:116–122. doi: 10.1006/exnr.2000.7588. [DOI] [PubMed] [Google Scholar]
- 73.Szczypka M.S., Kwok K., Brot M.D., Marck B.T., Matsumoto A.M., Donahue B.A., Palmiter R.D. Dopamine production in the caudate putamen restores feeding in dopamine-deficient mice. Neuron. 2001;30:819–828. doi: 10.1016/s0896-6273(01)00319-1. [DOI] [PubMed] [Google Scholar]
- 74.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 75.Mazzulli J.R., Mishizen A.J., Giasson B.I., Lynch D.R., Thomas S.A., Nakashima A., Nagatsu T., Ota A., Ischiropoulos H. Cytosolic catechols inhibit alpha-synuclein aggregation and facilitate the formation of intracellular soluble oligomeric intermediates. J. Neurosci. 2006;26:10068–10078. doi: 10.1523/JNEUROSCI.0896-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marshall J., McEachern K.A., Kyros J.A., Nietupski J.B., Budzinski T., Ziegler R.J., Yew N.S., Sullivan J., Scaria A., van Rooijen N., et al. Demonstration of feasibility of in vivo gene therapy for Gaucher disease using a chemically induced mouse model. Mol. Ther. 2002;6:179–189. doi: 10.1006/mthe.2002.0650. [DOI] [PubMed] [Google Scholar]
- 77.Goldin E., Zheng W., Motabar O., Southall N., Choi J.H., Marugan J., Austin C.P., Sidransky E. High throughput screening for small molecule therapy for Gaucher disease using patient tissue as the source of mutant glucocerebrosidase. PLoS One. 2012;7:e29861. doi: 10.1371/journal.pone.0029861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Paxinos G., Franklin K.B.J. The Mouse Brain in Stereotaxic Coordinates. 2nd edn. San Diego, CA: Elsevier Academic Press; 2004. [Google Scholar]
- 79.West M.J., Slomianka L., Gundersen H.J. Unbiased stereological estimation of the total number of neurons in the subdivisions of the rat hippocampus using the optical fractionator. Anat. Rec. 1991;231:482–497. doi: 10.1002/ar.1092310411. [DOI] [PubMed] [Google Scholar]
- 80.Gundersen H.J.G., Jensen E.B.V., Kieu K., Nielsen J. The efficiency of systematic sampling in stereology–reconsidered. J. Microsc. 1999;193:199–211. doi: 10.1046/j.1365-2818.1999.00457.x. [DOI] [PubMed] [Google Scholar]
- 81.Manaye K.F., Mouton P.R., Xu G., Drew A., Lei D.L., Sharma Y., Rebeck G.W., Turner S. Age-related loss of noradrenergic neurons in the brains of triple transgenic mice. Age. 2013;35:139–147. doi: 10.1007/s11357-011-9343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mouton P.R., Kelley-Bell B., Tweedie D., Spangler E.L., Perez E., Carlson O.D., Short R.G., Decabo R., Chang J., Ingram D.K., et al. Effects of age and LPS-mediated inflammation on central catecholaminergic neuron number in B6 mice. Neurobiol. Aging. 2012;423:27–36. doi: 10.1016/j.neurobiolaging.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mouton P.R. Unbiased Stereology: A Concise Guide. Baltimore, MD: The Johns Hopkins University Press; 2011. [Google Scholar]
- 84.Ahmed Z., Sheng H., Xu Y.F., Lin W.L., Innes A.E., Gass J., Yu X., Wuertzer C.A., Hou H., Chiba S., et al. Accelerated lipofuscinosis and ubiquitination in granulin knockout mice suggest a role for progranulin in successful aging. Am. J. Pathol. 2010;177:311–324. doi: 10.2353/ajpath.2010.090915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.