Abstract
Autism spectrum disorder (ASD) is a phenotypically and genetically heterogeneous condition characterized by the presence of repetitive/restrictive behaviors and variable deficits in language and social behavior. Many genes predisposing an individual to ASD have been identified, and understanding the causal disease mechanism(s) is critical to be able to develop treatments. Neurobiological, genetic, and imaging data provide strong evidence for the CNTNAP2 gene as a risk factor for ASD and related neurodevelopmental disorders. This review discusses the clinical genetics and current understanding of the biology of CNTNAP2 as related to ASD and illustrates how the integration of multiple research approaches, from human studies to animal models, converge to inform functional biology focused on novel treatment development.
Autism Spectrum Disorder (ASD)
Autism is a neurodevelopmental behavioral disorder with deficits in three main core domains: social behavior, language development, and repetitive behaviors and restrictive interests. The variability and heterogeneity observed in the phenotypes across these domains has led to the term Autism Spectrum Disorder (ASD), which includes autism, Asperger syndrome and pervasive developmental disorder not otherwise specified (PDD-NOS). These disorders share deficits in social communication and show variability in the language and repetitive behavior domains [1]. In addition to the core domains, a number of other behavioral and neurological abnormalities are frequently associated with ASD, including epilepsy, sensory abnormalities, hyperactivity, motor abnormalities, sleep disturbances, and gastrointestinal symptoms [1].
Autism was first described in 1943 as a disorder of affective contact [2]. However, it was not until many decades later that family and twin studies [3–5] revealed its high heritability and, therefore, genetic basis. Due to the clinically and genetically heterogeneous nature of the disorder, the search for genes involved in the development of ASD has been challenging. So far, no major gene has been identified as responsible for the disorder. Rather, many genes with either common variants that have small effect sizes or rare variants with larger effects have been identified, further emphasizing ASD's etiological heterogeneity [6]. Nevertheless, recent genetic studies have identified one of many causal genetic factors in up to 20% of patients [7–9], opening up a new perspective on the disorder [6]. Understanding the biological and molecular pathways affected by these genes could provide a unifying theme for autism pathophysiology [10,11], facilitating the development of targeted treatments.
The CNTNAP2 gene and Autism Spectrum Disorder
Given the growing list of genes associated with autism, one might ask the very reasonable question: how do we prioritize or select genes for further in depth mechanistic study? There obviously is not a simple answer for this question, but a reasonable criterion would be to focus on genes for which the evidence is the most diverse and strong. Several years ago, this basic rubric pointed us to the gene contactin associated protein-like 2 (CNTNAP2; also known asCASPR2) because it was expressed in human brain regions related to the disorder [12,13] and was among the first genes with evidence for both rare and common variation contributing to ASD [13–19]. We review some of this evidence below.
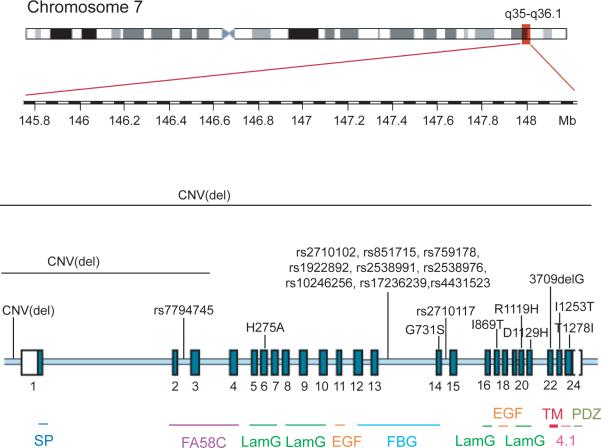
In 2006, a recessive mutation in CNTNAP2 was identified in an Amish family with what is clearly a syndromic form of ASD called cortical dysplasia-focal epilepsy (CDFE) syndrome [14]. CDFE is a rare neuronal migration disorder resulting in epileptic seizures, language regression, intellectual disability, hyperactivity and, in nearly two-thirds of the patients, ASD (Box 1). Subsequently, several laboratories provided converging evidence that common and rare variation in CNTNAP2 confer risk to ASD or ASD-related endophenotypes, such as language delay or developmental language disorders (Table 1). However, in contrast to the CDFE mutation, where the disorder is clearly transmitted in an autosomal recessive manner, the pathogenicity of these variants is uncertain, and their effect on protein function or gene expression remains to be elucidated. Most of the common variants associated with language endophenotypes in ASD and specific language impairment (SLI) cluster in a narrow intronic region between exons 13–14 (Figure 1) and are likely to be in linkage disequilibrium with the causal variant, rather than causal themselves. The rare variants identified are heterozygous and inherited from an apparently unaffected parent. So, even in the case of non-synonymous coding changes that are predicted to be deleterious, the variant must be viewed as a risk factor, rather than causal. Thus, variation on the other CNTNAP2 allele, or other transgenetic, epigenetic or environmental factors must also be invoked in those with ASD.
Table 1.
Summary of neurodevelopmental disorder-related genetic variation in CNTNAP2
| Genetic change | Location | Phenotype | Study/association | Sample | Refs. | |
|---|---|---|---|---|---|---|
| 3709delG | Coding region Exon 22 |
ASD Epilepsy Hyperactivity Language regression |
Autozygosity scanning plus sequencing (premature stop codon) | 13 patients 105 controls |
14 | |
| G731S I869T* R1119H D1129H I1253T T1278I |
Coding region Exon 14 Exon 17 Exon 20 Exon 21 Exon 23 Exon 24 |
ASD | Exome sequencing (changes predicted deleterious) | 635 patients 942 controls |
17 | |
| H275R** | Coding region Exon 6 |
ASD | Exome sequencing (change predicted deleterious) | 20 families | 19 | |
| CNV (deletion) | Promoter | ASD | Array CGH (reduced transcript) | 41 cases 367 controls |
18 | |
| rs7794745 | Major allele T | Intron 2 | ASD | QTDT, p<0.006 | 145 families | 15 |
| rs2710102 | Major allele C | Intron 13 | Age at first word in ASD | Quantitative regression p=0.028 | 304 families | 13 |
| Non-word repetition in SLI | QTDT p=0.002, p=0.0005 | 184, 181 families | 22,23 | |||
| Expressive language abilities in SLI | QTDT, p=0.02 | 181 families | 23 | |||
| Receptive language abilities in SLI | QTDT, p=0.03 | |||||
| Minor allele T | Intron 13 | Non-word repetition in dyslexia | QTDT, p=0.017 | 188 families | 20 | |
| Selective mutism | FBAT, p=0.018 | 99 families | 21 | |||
| Social anxiety | Quantitative regression p=0.015 | 1028 cases | ||||
| rs851715 | Major allele A | Intron 13 | Non-word repetition in SLI | QTDT, p=0.002 QTDT, p=0.002 QTDT, p=0.002 QTDT, p=0.002 QTDT, p=0.002 |
184 families | 22 |
| rs759178 | Major allele G | |||||
| rs1922892 | Major allele T | |||||
| rs2538991 | Major allele C | |||||
| rs2538976 | Major allele G | |||||
| rs10246256 | Major allele T | Intron 13 | Non-word repetition in SLI | QTDT p=0.001, p=0.0008 | 184,181 families | 22,23 |
| Expressive language abilities in SLI | QTDT, p=0.006 QTDT, p=0.003 |
181 families | 23 | |||
| Receptive language abilities in SLI | ||||||
| rs2710117 | Major allele A | Intron 14 | Non-word repetition in SLI | QTDT p=0.0004, p=0.001 | 181, 184 families | 22,23 |
| Expressive language abilities in SLI | QTDT, p=0.018 | 181 families | 23 | |||
| Receptive language abilities in SLI | QTDT, p=0.005 | |||||
| rs17236239 | Minor allele G | Intron 13 | Non-word repetition in SLI | QTDT p=0.00005, p=8×10(−5) | 181, 184 families | 22,23 |
| Expressive language abilities in SLI | QTDT p=0.008, p=0.007 | |||||
| Receptive language abilities in SLI | QTDT, p=0.033 | 181 families | 23 | |||
| rs4431523 | Minor allele G | Intron 13 | Receptive language abilities in SLI | QTDT, p=0.003 | 184 families | 22 |
| Complex chromosomal rearrangement | 7q32.1–7q35 | Language delay ASD |
Karyotyping plus FISH | Case report | 24 | |
| CNV (deletion) | 7q34–7q36.2 Several genes |
Language delay Epilespy Intellectual disability |
Array CGH | Case report | 25 | |
| CNV (deletion) | 7q33–q35 Several genes Exons 1–3 CNTNAP2 |
Stuttering | Array CGH | Case report | 26 | |
Variant significantly associated with ASD (p = 0.014)
FOXP1 is also mutated in proband
CGH: Comparative Genomic Hybridization
QTDT: Quantitative Transmission Disequilibrium Test
FBAT: Family Based Association Test
Figure 1. Schematic representation of the location of the mutations/variants found within CNTNAP2 associated to neurodevelopmental disorders.
The CNTNAP2 gene spans 2.3 Mb on chromosome 7. Exons are presented as numbered dark blue boxes, introns as a light blue line. Variants from references listed in Table 1 are indicated on top. The protein structural domains encoded by the specific exons are presented at the bottom. SP, signal peptide; FA58C, coagulation factor 5/8 C-terminal domain; LamG, laminin G; EGF, epidermal growth factor; FBG, fibrinogen-like domain; TM, transmembrane domain; 4.1, protein 4.1B binding domain; PDZ, PSD95/DlgA/ZO-1 homology protein-protein interaction domain.
CNTNAP2 and language
The heterogeneity of the autistic phenotype has led to the study of ASD endophenotypes to investigate genetic risks and pathophysiology of the disorder, together with the fact that the individual components of the disorder (language, social cognition, and repetitive-restricted behavior) are independently inherited in the general population [27] and that subtle ASD-like discrete symptoms are present in siblings and parents of affected patients [28]. This approach assumes that distinct factors control normal variation in each ASD-related behavioral domain and that ASD emerges as a convergence of abnormalities in all of them [29]. Indeed, endophenotype analysis has led to the identification of CNTNAP2 as having a major role in language development in ASD and other language related disorders (Table 1) [13,20–23].
Alarcon and colleagues were the first to show that a variant in CNTNAP2 (rs2710102) was associated with the language endophenotype age at first word in ASD [13]. Interestingly, this same variant has been linked to the endophenotype nonsense-word repetition, a task that requires reproducing a pronounceable but meaningless word in response to a spoken model, in developmental dyslexia, although in this case the association was driven by the opposite allele [20]. In addition, several other variants very nearby rs2710102 have been associated with the same endophenotype, nonsense-word repetition, in subjects with specific language impairment [22,23]. Interestingly, some of these variants have recently been associated with early language development in a normal population [30]. This finding is consistent with the notion that genetic risk variants are likely to affect normal variation in cognition and behavior and supports the idea that ASD emerges as the extreme of a continuum of normal behavioral variation [29]. Additional complex chromosomal rearrangements and large deletions affecting CNTNAP2 (in addition to other genes) have been reported in other language related deficits such as language delay [24,25] and stuttering [26].
Further support for CNTNAP2 in language development is provided by the fact that disruption of the transcription factor forkhead box P2 (FOXP2) has been shown to cause developmental speech and language disorder [31,32]. FOXP2 has been shown to directly bind intron 1 of the CNTNAP2 gene and regulate its expression [22]. This provides an intriguing connection between two genes related to human language function, suggesting a molecular pathway related to this circuitry critical for human higher cognition.
Although there are no animal parallels to human speech and language, several species (e.g. some marine mammals and birds) present learned vocal communication through auditory guided, vocal imitation, which itself is a prerequisite for language development. The zebra finch (Taeniopygia guttata) is a species with well-characterized, learned vocal communication [33]. Interestingly Panaitof and colleagues [34] showed punctuated expression of Cntnap2 in key song control nuclei relative to adjacent brain tissue. Importantly, this punctuated expression was observed in males, but not females, in accordance with the sexual dimorphism of neural circuitry and vocal learning in this species [35]. In addition, Foxp2 expression in zebra finch brain parallels its expression in developing human brain [36], which includes the striatal region dedicated to song learning in the finch (Area X). Foxp2 expression levels have been reported to be dynamically regulated in song nuclei based on the amount of vocal learning required, being highly expressed in learning juvenile zebra finches and decreasing as a result of singing activity in both juvenile and adults when learning is not required and songs become more stereotyped [37,38]. The most direct evidence that Foxp2 is necessary for song learning is that knockdown of Foxp2 in Area X during song learning leads to inaccurate and incomplete reproduction of the tutor's song [39]. Because FOXP2 is a transcription factor, its role in shaping vocal learning circuits likely depends on its ability to regulate expression of its targets (e.g. Cntnap2 among other Foxp2 targets). Although Cntnap2 regulation by Foxp2 has not been confirmed in the zebrafinch, their punctuate expression in song brain areas suggests this is a strong possibility worth further investigation. If conservation of Cntnap2-Foxp2 molecular circuitry is confirmed, additional work on this species will provide insight into the molecular contributions of Cntnap2-Foxp2 in auditory-guided vocal motor learning [40].
As has been presented above for the language-related circuitry, the genetic contribution to ASD should be viewed as an influence on the development and function of frontal-striatal circuits mediating social cognition and language. Unless the circuit is restricted to one function, a certain genetic mutation will give rise to a range of distinct, but overlapping neurobehavioral phenotypes that rely on the intact circuit for optimal functioning. From this perspective, it should not be surprising that the association of CNTNAP2 variants with the language endophenotype is not specific. Recent studies have suggested that these same variants are also associated with social behavior endophenotypes, such as sociability and social inhibition in normal and autistic populations [16] and selective mutism (e.g. failure to speak in one or more social settings despite speaking normally in other settings) in social anxiety disorder [21]. Such a pleiotropic effect might be expected for a gene whose loss of function leads to the abnormal development of frontal-striatal brain circuits that are involved in many behavioural and cognitive processes (see below) [14,41]. The availability of new brain imaging techniques is helping to provide insight into the genetic contribution to the function of brain circuits affected in the disorder.
Neuroimaging: Unraveling the functionality of CNTNAP2 variants
Non-invasive human brain imaging allows assessment of the brain in vivo and is currently one of the few ways to investigate functional genotype-phenotype in humans. Using structural magnetic resonance imaging (MRI), despite its limitations (often small sample sizes and difficulty to have a completely matched control sample) has provided solid evidence of alterations in brain structures and functions associated with many psychiatric disorders. Tan and colleagues [42] investigated variation in white and grey matter morphology in normal individuals carrying a single nucleotide polymorphism (SNP) in the CNTNAP2 gene (rs7794745) that had been previously reported to confer risk to ASD [15]. The authors found that despite the absence of behavioral abnormalities homozygotes for the risk allele (T) showed significant cerebral morphological variation, including reductions in grey and white matter volume in several regions that have already been implicated in ASD (cerebellum, fusiform gyrus, and occipital and frontal cortices), suggesting that this polymorphism could disrupt fronto-occipital connections.
Using functional neuroimaging, Scott-Van Zeeland et al. [43] demonstrated that the variant in CNTNAP2 (rs2710102) that was shown to increase risk for the language endophenotype age at first word in ASD [13] leads to abnormal frontal lobe functional brain connectivity in human subjects. The authors found that the medial prefrontal cortex (mPFC) showed differential activity as a function of genotype during implicit learning tasks known to engage fronto-striatal circuits, a region previously shown to have enriched CNTNAP2 expression [12]. In addition, functional connectivity analysis between mPFC and more posterior cortical regions showed that non-risk individuals had greater long-range anterior-posterior connectivity, while risk allele (C) carriers had increased local connectivity [43]. It has been recently shown that weakening of local and strengthening of long-range connectivity are natural steps in brain maturation [44], which could indicate more immature connectivity patterns in CNTNAP2 risk allele carriers.
Brain connectivity has been proposed as a potential unifying abnormality in ASD pathophysiology [45]. The heterogeneity in the genetics and neurobiology of ASD (e.g. genes involved in different neurological processes converge in the behavioural phenotype) has led to the proposal that ASD is a “developmental disconnection” syndrome [46], where different molecular mechanisms, including neuronal development and migration, dendritic maturation, axon pathfinding and synapse formation, could lead to the disconnection of brain areas important for higher order cognitive function (Box 2). Because disconnectivity related to CNTNAP2 genotype is independent of diagnosis [43], disconnectivity is likely a risk factor for ASD, but is not itself sufficient for the disorder.
CNTNAP2 function: Linking genetic variation to molecular mechanisms
Given the association of rare and common genetic variation with ASD and a number of allied neurodevelopmental disorders, and the effects of common variants on brain function, it is important to understand the molecular role of CNTNAP2. Most of our current knowledge on the function of the gene is based on postnatal studies of mouse brain. The human CNTNAP2 gene is thought to be the largest gene in the genome, spanning approximately 2.3 Mb at chromosomal region 7q35–q36.1 [49]. The CNTNAP2 protein (CASPR2) was identified in 1999 in rodents as a new member of the neurexin superfamily (Box 3) and specifically as the mammalian homolog of the fruit fly (Drosophila melanogaster) neurexin IV, involved in neuron-glia interactions in myelinated axons [50].
The fact that the gene is expressed embryonically [12,13,50], and that myelination takes place postnatally, together with the increasing number of reports that link the gene to ASD, suggests an additional role for CNTNAP2 in early brain development. Its developmental role is further supported by the imaging and pathology data in patients with CDFE syndrome [14]. CNTNAP2 is expressed predominantly in brain and spinal cord. In situ hybridization in human fetal brain reveals that CNTNAP2 is highly expressed in a cortico-striato-thalamic circuit that is involved in diverse higher order cognitive functions. Within the cortex it is particularly enriched in frontal and prefrontal cortical areas during development, a pattern that appears to be preserved in adulthood [12,13]. In addition, genome-wide microarray studies of regional gene expression in human fetal brain demonstrated CNTNAP2 to be enriched in the perisylvian language-related association cortex [12], consistent with its role in language development. In the mouse, the expression of the Cntnap2 gene shows a lack of frontal enrichment and much broader cortical expression, although its expression parallels what is observed in humans in the striatum, thalamus and amygdala [13]. In addition, in situ hybridization in mouse embryos shows expression in the ventricular proliferative zones of the developing cortex and ganglionic eminence (where excitatory neurons and inhibitory interneurons arise, respectively), as well as in areas of migrating neurons and post-migratory cells [41], suggesting a possible role in neuronal development and/or migration and, therefore, in the formation of neuronal circuits.
Insights from the Cntnap2 mouse model of ASD
The difficulty in studying gene function in human subjects with neuropsychiatric disorders makes animal models an essential component of research. As mammals, mice (Mus musculus) may be sufficiently close to humans to present similar neurobiological characteristics and, remarkably, the behavioral deficits associated with ASD. Social interaction, vocal communication, restricted interests, and repetitive behavior can be paralleled, to some extent, in mice [58]. The study of mouse models that recapitulate the human behavioral symptoms can help us understand the effects of these genes in ASD development. A mouse knockout for the Cntnap2 gene (Caspr2 null mouse) was generated by the Peles laboratory in 2003, long before the initial associations of the gene with ASD, as part of work on the structural organization of myelinated axons [55]. When a recessive mutation (suggesting loss of function) was reported on this gene as causing CDFE syndrome with a high penetrance of ASD [14], we backcrossed the original mouse (kept in a mixed background) to the C57/B6 background to make them more amenable to behavioral evaluation [59]. Remarkably, the Cntnap2 knockout mouse in the C57/B6 background demonstrates striking parallels not only to the core behavioral features of ASD but also to the major neuropathological features observed in CDFE patients [14]. Cntnap2 mutant mice show deficits in the three core domains of ASD: reduced vocal communication, repetitive and restrictive behaviors, and abnormal social interactions. In addition, they show hyperactivity and epileptic seizures [41], both features described in CDFE patients.
Neuropathologically, Cntnap2 mutant mice show defects in the migration of cortical projection neurons [41], confirming the observation of migration abnormalities in human patients with CDFE syndrome [14]. Neuronal migration plays an essential role during brain development, and the extraordinary degree of organization of the mammalian brain reflects the precise manner in which those migration movements should occur. Defects in neuronal migration during development have been reported to cause epilepsy, intellectual disability and other neurodevelopmetal disorders [60]. In addition to neuronal migration abnormalities, the Cntnap2 mouse model shows a reduction in the number of GABAergic interneurons [41]. This interneuron phenotype adds a new perspective to CNTNAP2 function because to date the study of the functional role of the gene has been restricted to excitatory pyramidal cells and no such deficit was reported in CDFE patients. In addition, these new data open up the possibility that the behavioral phenotype observed in this animal model is a consequence of an excitatory/inhibitory imbalance, which has been proposed to be a primary event in ASD pathophysiology [61]. Supporting this notion, it has recently been reported that altering the neocortical excitation/inhibition balance leads to deficits in social behavior and information processing [62]. Further, one of the neurophysiological alterations accompanying the abnormal brain structure in the Cntnap2 mouse model is a highly asynchronous cortical neuronal activity [41]. There is increasing evidence of abnormal neural synchrony as a pathophysiological mechanism in ASD [63] and, interestingly, one prominent candidate mechanism for abnormalities in neural synchrony is a dysfunction of GABAergic interneurons [64], which are deficient in this mouse model. Taken together, these data suggest that a disorganization of neuronal circuits is the cause of alterations in information processing. The specific contributions of the distinct neuropathologies observed in this mouse model to the phenotype remain to be elucidated. Further studies on the structure of neuronal networks and interneuron function in Cntnap2 KO mice may have important implications both for the understanding and the treatment of ASD.
It is also likely that the Cntnap2 mouse will be a useful model for pharmacological research because treatment of the mice with risperidone, one of the only two drugs approved by the FDA for autism treatment, rescues their increased repetitive behavior but not the social deficits, a dissociation similar to what is seen in human patients [41]. In autism, no drug has yet been proven to consistently improve communication and social behaviors. Current pharmacotherapy is used to mostly reduce stereotypic behaviors and non-core associated phenotypes. The dissociation observed in the mouse model between social and repetitive behavior with regards to treatment response suggests i) that this model has the potential of dissecting and studying the distinct circuitries involved in the core components of autistic related abnormal behavior, and ii) it may be a good model for testing new pharmacological treatments. Furthermore, the discovery of an interneuron phenotype in the mouse model, if confirmed in humans, would open up a new direction in pharmacotherapy targeting the GABAergic system.
Concluding Remarks and Future Perspectives
Because the understanding of CNTNAP2 function was previously focused primarily on postnatal development, the recent human genetic and mouse model findings set a new direction for investigating the role of CNTNAP2 during development and in the formation and function of neuronal circuits (Figure 2). This is a critical point, as we view genes as influencing the development and function of brain circuits; it is dysfunction in these circuits, or the inability to effectively compensate that may variably lead to clinical phenotypes. Although ASD is a complex neurodevelopmental disorder of which only 10–15% is due to known major gene mutations, the studies described throughout this review support the integrative use of animal models and human studies of `single gene' causes of ASD to understand its pathophysiology and develop targeted treatments. CNTNAP2 loss of function is responsible not only for the behavioral deficits associated with ASD but also leads to hyperactivity and epilepsy, two neuropsychiatric disorders that show high comorbidity with ASD. Additional electrophysiological, brain imaging, and cognitive studies in both animal models and humans carrying risk variants (with and without behavioral symptoms) will be important to identify the specific circuits and the underlying pathophysiology that underlie the different behaviors.
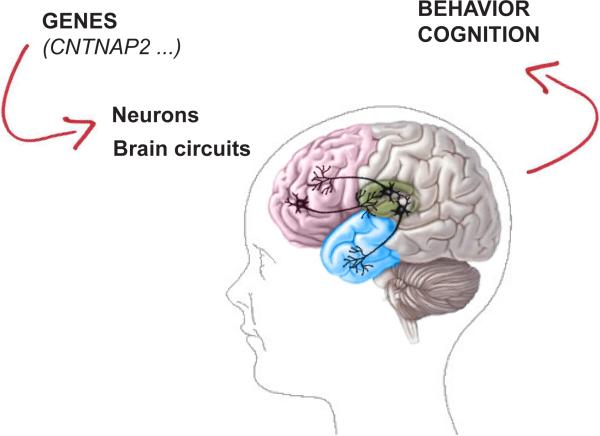
Figure 2. Genetic contribution of CNTNAP2 to Autism Spectrum Disorders.
CNTNAP2 is expressed in brain regions important for social cognition, language and implicit learning such as the frontal cortex (red), the anterior temporal cortex (blue) and the basal ganglia (green). Its contribution to the clinical phenotype is seen as an influence on the development and function of these brain circuits.
Outstanding Questions
What is the contribution of the neuropathology observed in humans [14] and mice [41] with loss of function of CNTNAP2 to each distinct behavioral domain?
What brain circuits are responsible for each of the ASD related behaviors?
How does CNTNAP2 affect the functioning of those circuits?
How do common CNTNAP2 risk variants act; where are the causal variants? What is their role in gene expression or protein function?
Can we pharmacologically or genetically rescue the social and communication phenotypes in the adult similar to the repetitive behavior?
Box 1. CNTNAP2 and CDFE syndrome.
Patients with CDFE showed a single base deletion (3709delG) in exon 22 of CNTNAP2. The frameshift mutation results in a premature stop codon and predicts a nonfunctional protein. All patients were homozygous for the mutation and inherited it from heterozygous unaffected parents. The recessive nature of the syndrome, with a Mendelian inheritance pattern, strongly indicates loss of function of the CNTNAP2 protein as causative. Clinically, patients were reported to develop normally until intractable focal seizures began in early childhood (1–9 years old), after which language regression, hyperactivity, impulsive and aggressive behavior, and mental retardation developed in all children. In addition, nearly two-thirds of the patients fulfilled the diagnostic criteria for pervasive developmental delay (PDD), the most generic ASD diagnosis. This is one of the most highly penetrant forms of `single gene' or syndromic autism. Neuropathological analysis of brain tissue from patients who underwent surgery for epilepsy (all patients had a recurrence of seizures 6–15 months after surgery) showed abnormalities in neuronal migration, as had been presumed from MRI analysis in which 43% of patients had evidence of cortical dysplasia.
Box 2. Brain disconnection and ASD.
Clinical, neuropathological and neuroimaging studies support the hypothesis that ASD is a disorder of neuronal-cortical organization with abnormalities at different levels, from neuronal migration to synaptic deficits, which would result in alteration of information processing [47]. The cerebral cortex has been attributed to play a key role in cognitive and emotional processes, such as attention, social behavior and language. The formation of the cerebral cortex is roughly divided into three main steps: cell proliferation, cell migration and cortical organization. The neocortex is the largest region of the cortex, where neurons are precisely organized in six layers. During migration and after migrating into their proper layers the neurons extend processes to establish synaptic connections with other neurons and create functional neuronal circuits that will allow the processing of cognitive and emotional information. Approximately 80% of neurons in the neocortex are glutamatergic projection neurons (excitatory) that extend their axons long distance to their targets. The rest are mostly GABAergic interneurons (inhibitory), which generally establish synapses on nearby neurons [48]. The neurons in each layer establish distinct connections within the cortex and with subcortical structures, some of which have also been implicated in ASD, such as the basal ganglia and amygdala. Therefore, the precise development, spatial location, synaptic connections and functioning (e.g. synaptic activity) of these neurons is required for the proper functioning of the circuit and disruption of any of these steps could lead to the abnormal behavioral outcome seen in ASD. Understanding the specific cortical-cortical and cortical-subcortical circuits involved in the distinct ASD behaviors, their function and how they can be modulated (e.g. identifying molecular drug targets) is critical for the development of pharmacological treatments.
Box 3. CNTNAP2 (CASPR2) and the neurexin superfamily.
Neurexins are presynaptic cell adhesion molecules implicated in synapse development and function by binding their postsynaptic partners, neuroligins [51,52], although all neurexins are unlikely to share the same function [53]. At a molecular level, they are single pass transmembrane proteins with a long extracellular and a short cytoplasmic region characterized by different protein-protein interacting domains. Contactin associated proteins (CNTNAPs) are also transmembrane cell adhesion molecules that resemble neurexins, but their function is believed to be restricted to outside the synapse, particularly to neuron-glia interactions in myelinated axons. In humans, there are five CNTNAP genes (CNTNAP1-CNTNAP5). Contactin associated protein 1 (CNTNAP1) receives its name from its interaction with contactin, which is essential for the generation of the axo-glial junction, while the rest of the family members do not interact with contactin but are named contactin associated protein-like after their resemblance to CNTNAP1, and have different roles in the neuron-glia junction [54]. In particular, CNTNAP2 function at this junction resides in clustering K+ channels at the juxtaparanodal region of the nodes of Ranvier [50,55]. The specialization of subcellular domains (i.e. nodes, paranodes and juxtaparanodes) within this structure, to which different sets of ion channels are localized, permits a rapid and efficient propagation of action potentials in myelinated axons. At the juxtaparanodes CNTNAP2 forms a neuron-glia cell adhesion complex with contactin 2 (CNTN2), which seems to be necessary for the proper localization of K+ channels in this structure [55]. In addition to the nodes of Ranvier, CNTNAP2 has also been shown to associate with K+ channels in the distal region of the axon initial segment of pyramidal cells, which is a critical region for the generation of action potentials and for the control of pyramidal cell activity [56], although CNTNAP2 is not necessary for K+ channel clustering in this region of the cell [57], as it is in myelinated axons.
Acknowledgments
Research on CNTNAP2 in the Geschwind laboratory is supported by grants NIH/NIMH R01 MH081754-02R and National Institutes of Health ACE Center 1P50-HD055784-01 (Project II) and the ACE Network grant 5R01-MH081754-04.
Glossary
- Asperger Syndrome
Neurodevelopmental disorder characterized by poor social skills and presence of restrictive/repetitive behaviors, but normal language development and IQ.
- Copy number variation (CNV)
The insertion or deletion of a region greater than 1kb.
- Dyslexia
Communication disorder that affects comprehension and formulation of written language.
- Endophenotype
Measurable features associated with an underlying psychiatric disorder, which are heritable and observed in non-affected family members at a higher rate than in the general population.
- Mendelian inheritance
Transmission of genetic traits from parents to offspring as described by Gregor Mendel. They include dominant inheritance (when the inheritance of only one copy of the specific genetic variant is enough to express the phenotype) and recessive inheritance (when two copies, one from each parent, are necessary to express the phenotype).
- Penetrance
The frequency with which individuals that carry an allele of a given gene will show the manifestations associated with the variant. If the penetrance of a disease allele is 100% then all individuals carrying that allele will express the associated disorder.
- Pervasive Developmental Delay (PDD)
Group of neurodevelopmental disorders characterized by a greater or lesser delay in the development of socialization and communication skills.
- Pleiotropy
A single gene influencing multiple phenotypes
- Single Nucleotide Polymorphism
A one base pair change in DNA sequence. The term common polymorphism refers to a variant that is present in at least 1% of the population.
- Social Anxiety Disorder
Anxiety (intense nervousness) disorder characterized by an excessive and unreasonable fear of social situations.
- Specific Language Impairment
Communication disorder that affects comprehension and formulation of spoken language with otherwise normal intellectual functioning.
- Stuttering
Communication disorder that affects the fluency of spoken language.
- Syndromic ASD
An ASD case that is observed in the context of a recognized syndrome (for example, fragile × syndrome).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Geschwind DH. Advances in autism. Annu. Rev. Med. 2009;60:367–380. doi: 10.1146/annurev.med.60.053107.121225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanner L. Autistic disturbances of affective contact. Nerv. Child. 1943;2:217–250. [PubMed] [Google Scholar]
- 3.Folstein S, Rutter M. Infantile autism: a genetic study of 21 twin pairs. J. Child Psychol. Psychiatry. 1977;18:297–321. doi: 10.1111/j.1469-7610.1977.tb00443.x. [DOI] [PubMed] [Google Scholar]
- 4.Ritvo ER, et al. Concordance for the syndrome of autism in 40 pairs of afflicted twins. Am. J. Psychiatry. 1985;142:74–77. doi: 10.1176/ajp.142.1.74. [DOI] [PubMed] [Google Scholar]
- 5.Smalley SL, et al. Autism and genetics. A decade of research. Arch. Gen. Psychiatry. 1988;45(10):953–961. doi: 10.1001/archpsyc.1988.01800340081013. [DOI] [PubMed] [Google Scholar]
- 6.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat. Rev. Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy D, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70(5):886–97. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Sanders SJ, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70(5):863–85. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2009;466(7304):368–72. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bill BR, Geschwind DH. Genetic Advances in Autism: Heterogeneity and Convergence on Shared Pathways. Curr. Opin. Genet. Dev. 2009;19:271–278. doi: 10.1016/j.gde.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474(7351):380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abrahams BS, et al. Genome-wide analyses of human perisylvian cerebral cortical patterning. Proc. Natl. Acad. Sci. 2007;104(45):17849–17854. doi: 10.1073/pnas.0706128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alarcón M, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am. J. Hum. Genet. 2008;82(1):150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strauss KA, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N. Engl. J. Med. 2006;354(13):1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 15.Arking DE, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am. J. Hum. Genet. 2008;82(1):160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steer CD, et al. Traits contributing to the autistic spectrum. PLoS One. 2010;5(9):e12633. doi: 10.1371/journal.pone.0012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakkaloglu B, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am. J. Hum. Genet. 2008;82(1):165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nord AS, et al. Reduced transcript expression of genes affected by inherited and de novo CNVs in autism. Eur. J. Hum. Genet. 2011;19(6):727–731. doi: 10.1038/ejhg.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 2011;46(3):585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peter B, et al. Replication of CNTNAP2 association with nonword repetition and support for FOXP2 association with timed reading and motor activities in a dyslexia family sample. J. Neurodev. Disord. 2011;3(1):39–49. doi: 10.1007/s11689-010-9065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein MB, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 is associated with increased risk for selective mutism and social anxiety-related traits. Biol. Psychiatry. 2011;69(9):825–831. doi: 10.1016/j.biopsych.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vernes SC, et al. A functional genetic link between distinct developmental language disorders. N. Engl. J. Med. 2008;359(22):2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newbury D, et al. Investigation of dyslexia and SLI risk variants in reading- and language-impaired subjects. Behav. Genet. 2011;41(1):90–104. doi: 10.1007/s10519-010-9424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poot M, et al. Disruption of CNTNAP2 and additional structural genome changes in a boy with speech delay and autism spectrum disorder. Neurogenetics. 2010;11(1):81–89. doi: 10.1007/s10048-009-0205-1. [DOI] [PubMed] [Google Scholar]
- 25.Sehested LT, et al. Deletion of 7q34–q36.2 in two siblings with mental retardation, language delay, primary amenorrhea, and dysmorphic features. Am. J. Med. Genet. 2010;152A(12):3115–9. doi: 10.1002/ajmg.a.33476. [DOI] [PubMed] [Google Scholar]
- 26.Petrin AL, et al. Identification of a microdeletion at the 7q33–q35 disrupting the CNTNAP2 gene in a Brazilian stuttering case. Am. J. Med. Genet. 2010;152A(12):3164–72. doi: 10.1002/ajmg.a.33749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ronald A, et al. Genetic heterogeneity between the three components of the autism spectrum: a twin study. J. Am. Acad. Child Adolesc. Psychiatry. 2006;45:691–699. doi: 10.1097/01.chi.0000215325.13058.9d. [DOI] [PubMed] [Google Scholar]
- 28.Bishop DV, et al. Using self-report to identify the broad phenotype in parents of children with autistic spectrum disorders: a study using the Autism-Spectrum Quotient. J. Child Psychol. Psychiatry. 2004;45(8):1431–6. doi: 10.1111/j.1469-7610.2004.00849.x. [DOI] [PubMed] [Google Scholar]
- 29.Geschwind DH. Autism: many genes, common pathways? Cell. 2008;135:391–395. doi: 10.1016/j.cell.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitehouse AJ, et al. CNTNAP2 variants affect early language development in the general population. Genes Brain Behav. 2011;10:451–456. doi: 10.1111/j.1601-183X.2011.00684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai CS, et al. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 32.MacDermot KD, et al. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am. J. Hum. Genet. 2005;76:1074–1080. doi: 10.1086/430841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams H. Birdsong and singing behavior. Ann N Y Acad Sci. 2004;1016:1–30. doi: 10.1196/annals.1298.029. [DOI] [PubMed] [Google Scholar]
- 34.Panaitof SC, et al. Language-related Cntnap2 gene is differentially expressed in sexually dimorphic song nuclei essential for vocal learning in songbirds. J. Comp. Neurol. 2010;518(11):1995–2018. doi: 10.1002/cne.22318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nottebohm F, Arnold AP. Sexual dimorphism in vocal control areas of the songbird brain. Science. 1976;194:211–213. doi: 10.1126/science.959852. [DOI] [PubMed] [Google Scholar]
- 36.Teramitsu I, et al. Parallel FoxP1 and FoxP2 expression in songbird and human brain predicts functional interaction. J. Neurosci. 2004;24(13):3152–63. doi: 10.1523/JNEUROSCI.5589-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teramitsu I, White SA. FoxP2 regulation during undirected singing in adult songbirds. J. Neurosci. 2006;26(28):7390–4. doi: 10.1523/JNEUROSCI.1662-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teramitsu I, et al. Striatal FoxP2 is actively regulated during songbird sensorimotor learning. PLoS One. 2010;5(1):e8548. doi: 10.1371/journal.pone.0008548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haesler S, et al. Incomplete and inaccurate vocal imitation after knockdown of FoxP2 in songbird basal ganglia nucleus Area X. PLoS Biol. 2007;5(12):e321. doi: 10.1371/journal.pbio.0050321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolhuis JJ, et al. Twitter evolution: converging mechanisms in birdsong and human speech. Nat. Rev. Neurosci. 2010;11(11):747–759. doi: 10.1038/nrn2931. [DOI] [PubMed] [Google Scholar]
- 41.Peñagarikano O, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities and core autism-related deficits. Cell. 2011;147(1):235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan GC, et al. Normal variation in fronto-occipital circuitry and cerebellar structure with an autism-associated polymorphism of CNTNAP2. Neuroimage. 2010;53(3):1030–1042. doi: 10.1016/j.neuroimage.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott-Van Zeeland AA, et al. Altered functional connectivity in frontal lobe circuits is associated with variation in the autism risk gene CNTNAP2. Sci. Transl. Med. 2010;2(56):56–80. doi: 10.1126/scitranslmed.3001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dosenbach NU, et al. Prediction of individual brain maturity using fMRI. Science. 2010;329:1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belmonte MK, et al. Autism and abnormal development of brain connectivity. J. Neurosci. 2004;24(42):9228–9241. doi: 10.1523/JNEUROSCI.3340-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geschwind DH, Levitt P. Autism spectrum disorders: Developmental disconnection syndromes. Curr. Opin. Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 47.Pardo CA, Eberhart CG. The neurobiology of autism. Brain Pathol. 2007;17(4):434–447. doi: 10.1111/j.1750-3639.2007.00102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubenstein JL. Development of the cerebral cortex: implications for neurodevelopmental disorders. J Child Psychol Psychiatry. 2011;52(4):339–55. doi: 10.1111/j.1469-7610.2010.02307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakabayashi K, Scherer SW. The human contactin-associated protein-like 2 gene (CNTNAP2) spans over 2 Mb of DNA at chromosome 7q35. Genomics. 2001;73(1):108–112. doi: 10.1006/geno.2001.6517. [DOI] [PubMed] [Google Scholar]
- 50.Poliak S, et al. Caspr2, a new member of the neurexin superfamily, is localized at the juxtaparanodes of myelinated axons and associates with K+ channels. Neuron. 1999;24(4):1037–1047. doi: 10.1016/s0896-6273(00)81049-1. [DOI] [PubMed] [Google Scholar]
- 51.Ushkaryov YA, et al. Neurexins: synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science. 1992;257(5066):50–56. doi: 10.1126/science.1621094. [DOI] [PubMed] [Google Scholar]
- 52.Ichtchenko K, et al. Neuroligin 1: a splice site-specific ligand for beta-neurexins. Cell. 1995;81(3):435–443. doi: 10.1016/0092-8674(95)90396-8. [DOI] [PubMed] [Google Scholar]
- 53.Konopka G, et al. Modeling the functional genomics of autism using human neurons. Mol. Psychiatry. 2011 doi: 10.1038/mp.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poliak S, Peles E. The local differentiation of myelinated axons at nodes of Ranvier. Nat. Rev. Neurosci. 2003;4(12):968–980. doi: 10.1038/nrn1253. [DOI] [PubMed] [Google Scholar]
- 55.Poliak S, et al. Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J. Cell. Biol. 2003;162(6):1149–1160. doi: 10.1083/jcb.200305018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Inda MC, et al. Voltage-gated ion channels in the axon initial segment of human cortical pyramidal cells and their relationship with chandelier cells. Natl. Acad. Sci. 2006;103(8):2920–2925. doi: 10.1073/pnas.0511197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ogawa Y, et al. Postsynaptic density-93 clusters Kv1 channels at axon initial segments independently of Caspr2. J. Neurosci. 2008;28(22):5731–5739. doi: 10.1523/JNEUROSCI.4431-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crawley JN. Behavioral phenotyping strategies for mutant mice. Neuron. 2008;57(6):809–818. doi: 10.1016/j.neuron.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 59.Crawley JN, et al. Behavioral phenotypes of inbred mouse strains: implications and recommendations for molecular studies. Psychopharmacology. 1997;132(2):107–124. doi: 10.1007/s002130050327. [DOI] [PubMed] [Google Scholar]
- 60.Guerrini R, Barba C. Malformations of cortical development and aberrant cortical networks: epileptogenesis and functional organization. J. Clin. Neurophysiol. 2010;27(6):372–9. doi: 10.1097/WNP.0b013e3181fe0585. [DOI] [PubMed] [Google Scholar]
- 61.Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/ inhibition in key neural systems. Genes Brain Behav. 2003;2(5):255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yizhar O, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477(7363):171–8. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uhlhaas PJ, Singer W. Neural synchrony in brain disorders: Relevance for cognitive dysfunctions and pathophysiology. Neuron. 2006;52:155–168. doi: 10.1016/j.neuron.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 64.Sohal VS, et al. GABAergic hub neurons orchestrate synchrony in developing hippocampal networks. Nature. 2009;459(7247):698–702. doi: 10.1126/science.1175509. [DOI] [PubMed] [Google Scholar]