Abstract
Human tumors often display startling intratumor heterogeneity in various features including histology, gene expression, genotype, and metastatic and proliferative potential. This phenotypic and genetic heterogeneity plays an important role in neoplasia, cancer progression, and therapeutic resistance. In this issue of the journal (beginning on page XXX), Merlo et al. report their use of molecular data from 239 patients with Barrett’s esophagus to evaluate the propensity of major diversity indices for predicting progression to esophageal adenocarcinoma. This work helps elucidate the implications of molecular heterogeneity for the evolution of neoplasia.
Neoplasia originates from normal cells that accumulate genetic and epigenetic alterations. Although the types and numbers of alterations necessary for transformation differ between tumor types, most types share a common feature: A noteworthy variability among the cancer cells within a single neoplastic lesion (1–3). These cells can be distinguished from each other by characteristics such as size, morphology, and antigen expression, as well as by behaviors like cell turnover, cell-cell interaction, invasive and metastatic ability, and sensitivity to pharmacological interventions (4, 5). This intratumor heterogeneity impedes the investigation and treatment of cancer since individual tumor-tissue samples may not be representative of the whole tumor, and predictions about its evolution frequently are inaccurate.
The origins of intratumor heterogeneity have been the subject of much discussion (6, 7). Both the cancer stem cell hypothesis and the clonal evolution model have been proposed as descriptions for the establishment and maintenance of intratumor heterogeneity (7). The cancer stem cell hypothesis suggests that a subset of cells with stem-cell properties drive tumor initiation and progression. These “cancer stem cells” are the only cells within the tumor that possess indefinite self-renewal abilities (5, 8–11). Their differentiation, which leads to the production of all cell types in the tumor, generates intratumor heterogeneity. In contrast, the clonal evolution model states that a premalignant or malignant cell population accumulates various hereditary changes over time that may confer advantages or disadvantages to the cell, which is hence subjected to natural selection. Carcinogenesis is initiated by the accumulation of several mutations in a single cell and is driven by the emergence of further genetic and epigenetic alterations that confer more aggressive, invasive, and drug-resistant phenotypes. In the context of this model, the emergence of new hereditary traits in premalignant and tumor cells gives rise to heterogeneity.
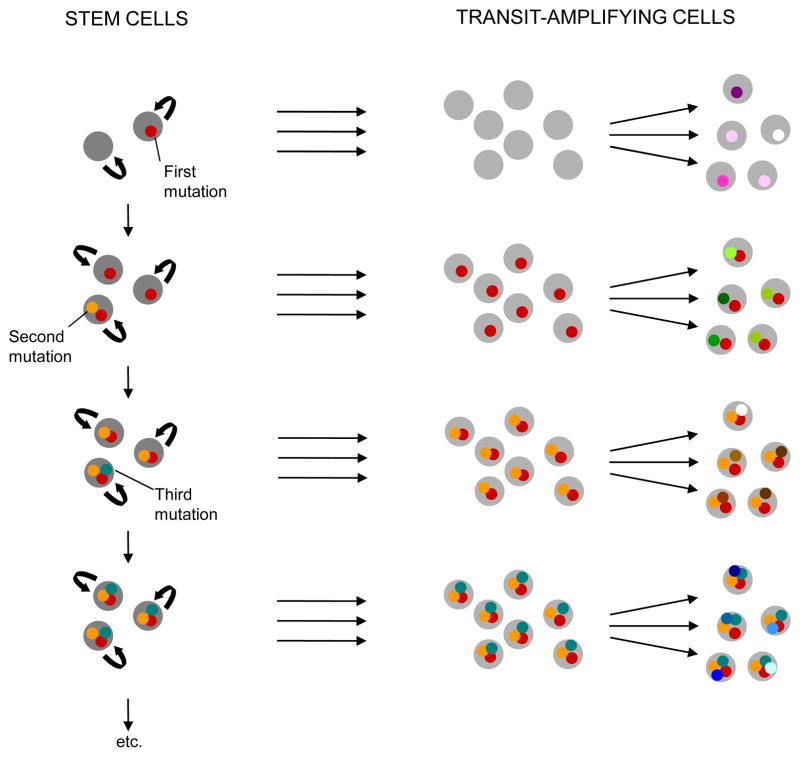
Although these two hypotheses have been presented as mutually exclusive explanations of tumor heterogeneity, they are easily reconciled and are both an integral part of cancer evolution and the generation of diversity (Fig. 1). Only cells that have self-renewal capabilities are able to persist over time and accumulate the genetic and epigenetic changes necessary for cancer initiation and progression. Such cancer stem cells give rise to distinct types of transit-amplifying cells and more differentiated cancer cells. Transit-amplifying cells may also accumulate genetic changes, but unless a mutation conferring self-renewal capabilities emerges, these changes are unable to persist in the cell population and will be “washed out” of the system. Nevertheless, they can be responsible for a fraction of the variation detected in a snapshot analysis of a tumor (Fig. 1).
Fig 1.
Emergence and maintenance of tumor diversity. Cancer cell diversity arises both from the differentiation of cancer stem cells (parallel horizontal arrows) and the accumulation of mutations in cancer stem cells (vertical arrows) and transit-amplifying cells (diverging arrows). Unless a mutation conferring self-renewal capabilities arises in a transit-amplifying cell, genetic changes emerging in those cells are unable to persist in the population and will disappear.
An alternative model for the ability of only a subset of cells to propagate the tumor cell population and give rise to intratumor heterogeneity is tumor cell plasticity. According to this model, all or most tumor cells have varying degrees of stem cell–like characteristics, which may depend on microenvironmental conditions and/or cell-intrinsic stochasticity (12). This idea is supported by recent evidence indicating that signaling within the perivascular niche of glioma cells acted as a driving force for tumor cells to acquire stem cell characteristics (13). In this study, nitric oxide was demonstrated to activate Notch signaling in a subset of the glioma cells. This signaling resulted in the acquisition of the side population phenotype and led to increased neurosphere and tumor formation. These alterations occurred within as few as two hours after nitric oxide stimulation and had long-term effects on the stem-like character of cells. Such plasticity of tumor stem cells may also apply to liquid tumors since leukemia-initiating cells in acute myeloid leukemia patients harboring mutations in nucleophosmin can reside in the CD34+ as well as CD34− fraction (14). Furthermore, the model of a rigid differentiation hierarchy is not supported by experimental evidence even in normal tissues. Certain extracellular signals, for instance, can induce oligodendrocyte precursor cells to dedifferentiate into multipotential neural stem cells (15). Similarly, a single extracellular factor was sufficient to induce differentiated cells of the central nervous system to regress into a stem cell–like stage in a study of mature astrocytes exposed to transforming growth factor α (16).
In contrast to its origins, the implications and clinical importance of intratumor diversity have been widely accepted. Since tumor cell populations are highly heterogeneous and continuously evolve towards more aggressive phenotypes, the identification of effective treatment modalities poses a major challenge; similarly difficult is the stratification of a patient population with regard to risk of progression from a premalignant lesion such as Barrett’s esophagus to cancer such as esophageal adenocarcinoma. Tumors consisting of multiple distinct clones display different sensitivities to therapeutic interventions as compared with monoclonal tumors, and the pre-existence of resistance mutations may render certain treatment options ineffective for a subset of patients. For instance, patients with chronic myeloid leukemia sometimes harbor mutations that confer resistance to one or more tyrosine kinase inhibitors. The specific therapeutic strategy of choice for such patients depends on the identity and composition of the resistant cell populations. Knowledge of the composition of tumors or premalignant lesions at diagnosis therefore may determine both the prognosis and therapeutic response of cancer patients. Intratumor diversity also complicates the molecular classification of tumors into clinically relevant subtypes predicting prognosis since diagnostic biopsies sample only small regions of the premalignant lesion or tumor. These small regions may not be representative of the whole lesion, and the treatment selection derived from a single-biopsy–based diagnosis might not elicit responses in all areas of the tumor.
Because of its clinical importance, intratumor diversity has been at the center of attention of investigators from diverse fields. Several computational models have been devised to study aspects of intratumor diversity, from the impact of differentiation hierarchies (17, 18) and tissue architecture (19–22) on heterogeneity to the consequences of somatic evolution (23–25), mutator phenotypes, the evolution of resistance (26–28), and interactions of cancer cells with the microenvironment (29). The application of computational and statistical models to data from human cancer samples, however, has only recently been initiated. In 2006, Maley and colleagues demonstrated for the first time a direct relationship between clonal diversity in premalignant lesions and progression to cancer (23). They studied the extent of genetic heterogeneity in patient samples of Barrett’s esophagus using ecological measures of diversity. Diversity was measured by three different indices: The number of clones in a neoplasm; the Shannon diversity index, which represents the information content of a message in computer science (30); and genetic divergence as measured by the number of loci showing differences divided by the number of informative loci. These measures incorporated information about loss of heterozygosity, microsatellite shifts, and sequence mutations across the genome. High degrees of clonal diversity were found to correlate with increased risk of progression to malignancy, thereby suggesting that diversity measures can be used to identify high-risk patients.
To investigate the relative importance of the clonal evolution model versus the stem cell model and to quantify diversity in human tumor samples, we recently performed combined analyses of markers associated with cellular differentiation and genotypic alterations in human breast carcinomas (31). Similar to earlier studies (23), several different measures of diversity were utilized to analyze the samples. These studies uncovered a high degree of genetic heterogeneity both within and between distinct tumor cell populations that were defined based on markers of cellular phenotypes including stem cell–like characteristics. In several tumors, stem cell–like and more differentiated cancer cell populations were genetically distinct; this observation questioned the validity of a simple differentiation hierarchy–based cancer stem cell model. The degree of diversity correlated with clinically relevant breast-tumor subtypes and in some tumors was markedly different between the in situ and invasive cell populations. We also found that diversity measures were associated with clinical variables. Therefore, the hypothesis that the degree of intratumor heterogeneity in in situ and invasive breast tumors predicts the risk of progression and therapeutic resistance would be worth investigating in larger cohorts, as also suggested by Tlsty et al. based on their studies of DCIS (3).
In this issue of the journal, Maley and colleagues present a new analysis of clonal diversity in Barrett’s esophagus (32). The authors utilized molecular data from patient samples to systematically test all major diversity measurement methods for their accuracy in predicting progression to esophageal adenocarcinoma. The molecular data included microsatellite shifts, loss of heterozygosity, tetraploidy and aneuploidy, and methylation and sequence mutations. Since diversity indices are based on the number and relative abundance of distinct cellular clones within a sample, the definition of clones may influence the magnitude and predictive power of individual indices. The authors systematically investigated this issue by considering several distinct definitions of clones, ranging from cellular groups distinguished by all available molecular data to those that differed by alterations in neutral loci only. The choice of diversity measurement method may also influence the ability of intratumor heterogeneity to predict tumor progression. To address this issue, the authors considered a generalized expression for diversity indices incorporating the total number and frequency of distinct clones in the sample. By adjusting the order (i.e., a parameter q) of the diversity index by scaling the parameter q, the relative importance of frequent clones in the sample is altered. This generalized expression allows an organic way of investigating a large range of diversity indices, including the frequently used Simpson and Shannon indices (30). The authors discovered that all measures of diversity were predictive of future progression, regardless of which definition of a clone was used. This result is important from both the basic science and clinical perspective. Moreover, since progression to cancer in different sites is driven by evolutionary mechanisms that share the same basic principles, these findings may also be applicable to cancers of other organs.
Clinically, the assessment of clonal diversity of human tumors may represent a unified method for identifying patients at a high risk of progression to cancer, to more advanced stages of cancer, and of relapse due to the existence of drug-resistant cells in the tumor cell population. Solidifying the utility of diversity measures for diagnosis, prognosis, and treatment choices will require the elucidation of the extent of diversity in a large number of tumor samples as well as in additional tumor types and premalignant lesions. The magnitude of intratumor heterogeneity must then be correlated with clinical variables such a survival, proliferative indices, treatment sensitivity, and the risk of acquired resistance. Furthermore, studies of single cell-based heterogeneity in different situations during the evolution of a neoplasm, such as before and after treatment and in premalignancy, primary cancer, and metastasis, have not been performed to date and are needed in order to obtain an accurate picture of the dynamics of diversity.
A comprehensive analysis of intratumor heterogeneity depends on more than just the availability of appropriate analysis tools such as those described by Merlo et al. (32). Cost-efficient ways to profile single cells from neoplasms at multiple stages of their evolution are also necessary. Although not yet available, these methodologies, in combination with appropriate analysis tools, would significantly improve the clinical management of patients with premalignant lesions or cancer.
Acknowledgments
Grant Support
This work was supported by the Breast Cancer Research Foundation (K.P.) and the NCI initiative to found Physical Science-Oncology Centers (grant U54CA143798; F.M.).
Footnotes
Disclosure of Potential Conflicts of Interest
K. Polyak receives research support from Novartis Oncology, holds stocks in Aveo Pharmaceuticals Inc., is a consultant to Novartis Oncology, and serves on the Scientific Advisory Boards of Metamark Genetics Inc. and Theracrine Inc.
References
- 1.Fidler IJ. Tumor heterogeneity and the biology of cancer invasion and metastasis. Cancer Res. 1978;38:2651–2660. [PubMed] [Google Scholar]
- 2.Heppner GH. Tumor heterogeneity. Cancer Res. 1984;44:2259–2265. [PubMed] [Google Scholar]
- 3.Berman HK, Gauthier ML, Tlsty TD. Premalignant breast neoplasia: a paradigm of interlesional and intralesional molecular heterogeneity and its biological and clinical ramifications. Cancer Prev Res. 2010;3:579–587. doi: 10.1158/1940-6207.CAPR-10-0073. [DOI] [PubMed] [Google Scholar]
- 4.Axelson H, Fredlund E, Ovenberger M, Landberg G, Pahlman S. Hypoxia-induced dedifferentiation of tumor cells a mechanism behind heterogeneity and aggressiveness of solid tumors. Semin Cell Dev Biol. 2005;16:554–563. doi: 10.1016/j.semcdb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Ichim CV, Wells RA. First among equals: The cancer cell hierarchy. Leuk Lymphoma. 2006;47:2017–2027. doi: 10.1080/10428190600733325. [DOI] [PubMed] [Google Scholar]
- 6.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 7.Campbell LL, Polyak K. Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle. 2007;6:2332–2338. doi: 10.4161/cc.6.19.4914. [DOI] [PubMed] [Google Scholar]
- 8.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 9.Al-Hajj M, Clarke MF. Self-renewal and solid tumor stem cells. Oncogene. 2004;23:7274–7282. doi: 10.1038/sj.onc.1207947. [DOI] [PubMed] [Google Scholar]
- 10.Bjerkvig R, Tysnes BB, Aboody KS, Najbauer J, Terzis AJ. The origin of the cancer stem cell: current controversies and new insights. Nat Rev Cancer. 2005;5:899–904. doi: 10.1038/nrc1740. [DOI] [PubMed] [Google Scholar]
- 11.Bapat S. A. Evolution of cancer stem cells Semin Cancer Biol. 2007;17:204–213. doi: 10.1016/j.semcancer.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Hill RP. Idemtifying cancer stem cells in solid tumors: case not proven. Can Res. 2006;66:1891–1895. doi: 10.1158/0008-5472.CAN-05-3450. [DOI] [PubMed] [Google Scholar]
- 13.Charles N, Ozawa T, Squatrito M, Bleau AM, Brennan CW, et al. Perivascular Nitric Oxide Activates Notch Signaling and Promotes Stem-like Character in PDGF-Induced Glioma Cells. Cell Stem Cell. 2010;6:141–152. doi: 10.1016/j.stem.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taussig DC, Vargaftig J, Miraki-Moud F, Griessinger E, Sharrock K, et al. Leukemia-initiating cells from some acute myeloid leukemia patients with mutated nucleophosmin reside in the CD34- fraction. Blood. 2010;115:1976–1984. doi: 10.1182/blood-2009-02-206565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo T, Raff M. Oligodendrocyte precursor cells reprogrammed to become multipotential CNS stem cells. Science. 2000;289:1754–1757. doi: 10.1126/science.289.5485.1754. [DOI] [PubMed] [Google Scholar]
- 16.Sharif A, Legendre P, Prévot V, Allet C, Romao L, et al. Transforming growth factor alpha promotes sequential conversion of mature astrocytes into neural progenitors and stem cells. Oncogene. 2007;26:2695–2706. doi: 10.1038/sj.onc.1210071. [DOI] [PubMed] [Google Scholar]
- 17.Pepper J, Sprouffske K, Maley C. Animal cell differentiation patterns suppress somatic evolution. PLoS Comput Biol. 2007;3:e250. doi: 10.1371/journal.pcbi.0030250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michor F, Hughes TP, Iwasa Y, Branford S, Shah NP, Sawyers CL, et al. Dynamics of chronic myeloid leukaemia. Nature. 2005;435:1267–1270. doi: 10.1038/nature03669. [DOI] [PubMed] [Google Scholar]
- 19.Frank, Iwasa Y, Nowak MA. Patterns of cell division and the risk of cancer. Genetics. 2003;163:1527–1532. doi: 10.1093/genetics/163.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michor F, Frank May RM, Iwasa Y, Nowak MA. Somatic selection for and against cancer. J Theor Biol. 2003;225:377–382. doi: 10.1016/s0022-5193(03)00267-4. [DOI] [PubMed] [Google Scholar]
- 21.Nowak MA, Michor F, Iwasa Y. The linear process of somatic evolution. Proc Natl Acad Sci USA. 2003;100:14966–14969. doi: 10.1073/pnas.2535419100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dingli D, Traulsen A, Pacheco JM. Stochastic dynamics of hematopoietic tumor stem cells. Cell Cycle. 2007;6:461–466. doi: 10.4161/cc.6.4.3853. [DOI] [PubMed] [Google Scholar]
- 23.Maley C, Galipeau P, Finley J, Wongsurawat V, Li X, Sanchez C, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 24.Michor F, Nowak MA, Iwasa Y. Stochastic dynamics of metastasis formation. J Theor Biol. 2006;240:521–530. doi: 10.1016/j.jtbi.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 25.Nagy JD. Competition and natural selection in a mathematical model of cancer. Bull Math Biol. 2004;66:663–687. doi: 10.1016/j.bulm.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Goldie JH, Coldman AJ. Quantitative model for multiple levels of drug resistance in clinical tumors. Cancer Treatment Reports. 1983;67:923–931. [PubMed] [Google Scholar]
- 27.Komarova NL, Wodarz D. Evolutionary dynamics of mutator phenotypes in cancer: implications for chemotherapy. Cancer Research. 2003;63:6635–6642. [PubMed] [Google Scholar]
- 28.Michor F, Nowak MA, Iwasa Y. Evolution of resistance to cancer therapy. Curr Pharm Des. 2006;12:261–271. doi: 10.2174/138161206775201956. [DOI] [PubMed] [Google Scholar]
- 29.Gatenby RA, Vincent TL. An evolutionary model of carcinogenesis. Cancer Research. 2003;63:6212–6220. [PubMed] [Google Scholar]
- 30.Magurran AE. Measuring Biological Diversity. Blackwell; Malden, Massachusetts: 2004. [Google Scholar]
- 31.Park SY, Gonen M, Kim HJ, Michor F, Polyak F. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120:636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merlo LMF, Shah NA, Li X, et al. A comprehensive survey of clonal diversity measures in Barrett’s esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prev Res. 2010;3:XXX. doi: 10.1158/1940-6207.CAPR-10-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]