Abstract
Immunization with a recombinant yeast-expressed Plasmodium falciparum merozoite surface protein 3 (MSP3) protected Aotus nancymai monkeys against a virulent challenge infection. Unfortunately, the production process for this yeast-expressed material was not optimal for human trials. In an effort to produce a recombinant MSP3 protein in a scaleable manner, we expressed and purified near-full-length MSP3 in Escherichia coli (EcMSP3). Purified EcMSP3 formed non-globular dimers as determined by analytical size-exclusion HPLC with in-line multi-angle light scatter and quasi-elastic light scatter detection and velocity sedimentation (Rh 7.6 ± 0.2 nm and 6.9 nm, respectively). Evaluation by high-resolution atomic force microscopy revealed non-linear asymmetric structures, with beaded domains and flexible loops that were recognized predominantly as dimers, although monomers and larger multimers were observed. The beaded substructure corresponds to predicted structural domains, which explains the velocity sedimentation results and improves the conceptual model of the protein. Vaccination with EcMSP3 in Freund's adjuvant-induced antibodies that recognized native MSP3 in parasitized erythrocytes by an immunofluorescence assay and gave delayed time to treatment in a group of Aotus monkeys in a virulent challenge infection with the FVO strain of P. falciparum. Three of the seven monkeys vaccinated with EcMSP3 had low peak parasitemias. EcMSP3, which likely mimics the native MSP3 structure located on the merozoite surface, is a viable candidate for inclusion in a multi-component malaria vaccine.
Keywords: Malaria, Plasmodium falciparum, MSP3, Vaccine, Aotus, Escherichia coli
1. Introduction
Malaria is a disease caused by infection of red blood cells by protozoan parasites of the genus Plasmodium and is transmitted by Anopheles mosquitoes. It is a leading cause of morbidity and mortality in human populations in tropical countries. There were an estimated 300–660 million episodes of clinical malaria and over one million deaths in 2002 [1]. Among the four species of malaria parasites infecting humans, P. falciparum is the focus of most of the research because it accounts for almost all mortality, especially in children under 5 years of age living in sub-Saharan Africa.
Merozoite surface protein 3 (MSP3 or previously identified as SPAM [2]) is a ∼43-kilodalton (kDa) soluble protein associated with the merozoite membrane surface. MSP3 was first identified using human hyperimmune serum from Papua New Guinea [2] and with antibodies that inhibit P. falciparum growth by cooperating with human monocytes in an antibody-dependent cellular inhibition (ADCI) in vitro assay [3].
Current evidence supports MSP3 as a vaccine candidate. The initial observations of ADCI dependent on naturally occurring human antibodies have been extended [4,5] and genotyping data suggests that MSP3 is under active immune selection pressure [6]. An Aotus monkey trial using full-length recombinant MSP3 expressed from Pichia pastoris gave significant protection against a virulent challenge of P. falciparum [7]. Prechallenge antibody titers to MSP3 correlated with time to treatment, consistent with antibodies to MSP3 protecting from the challenge infection. The immunogenicity and protective efficacy of various MSP3 fragment-adjuvant formulations have also been evaluated in Saimiri sciureus monkeys and were able to control parasitemia upon experimental P. falciparum blood-stage infection [8]. A Phase 1 human study using a long synthetic peptide which encompassed a 70 amino acid conserved region showed that the formulation was safe, well tolerated with alum and Montanide ISA 720 [9], and antibodies from vaccine-induced volunteers were able to kill parasites in vitro and in vivo in a mouse severe combined immunodeficiency disease (SCID) model [10].
The most remarkable features of the primary structure of MSP3 are three blocks of alanine-rich repeats with a seven amino acid motif (AXXAXXX), which are characteristic of parallel left-handed coiled coils [11,12] (Fig. 1). The msp3 gene and the predicted MSP3 protein have considerable diversity, especially in the N-terminal segment prior to these alanine repeats. The conserved C-terminal half of the protein contains a 50 amino acid sequence rich with 35 glutamic and aspartic acid residues (Fig. 1). The C-terminus of MSP3 also contains a heptad “leucine zipper-like” region responsible for dimerization [13] (Fig. 1). An Escherichia coli expressed full-length MSP3, which included the N-terminal signal sequence fused with a His6 tag also at the N-terminus, appeared as an extended dimer or tetramer in solution [13].
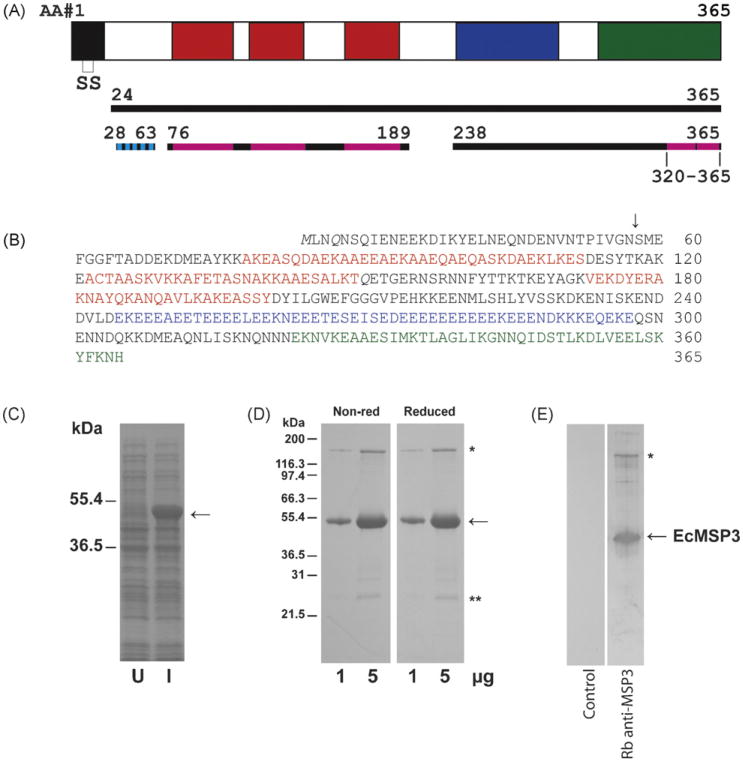
Fig. 1.
Production of EcMSP3. A schematic of the MSP3 protein (solid black line represents expressed region of MSP3, broken black line represents portions of the sequence used for structure modeling, cyan indicates beta strands and magenta indicates alpha helices) (A), amino acid sequence of EcMSP3 including non-native M (B), Coomassie blue stained SDS-PAGE gels with uninduced (U) and induced (I) solubilized cells produced by shake flask fermentation (C) and purified EcMSP3 protein (D), and analysis by immunoblot with rabbit anti-EcMSP3 antiserum or control (E). Blocks within the schematic as well as amino acids highlighted by colors are the three blocks (H1–H3) of Ala-X-X-Ala-X-X-X heptad repeats (red), the glutamic acid-rich region (blue), and the leucine zipper-like domain (green). Two glutamines (Q) shown in italics in panel B are N to Q point mutations introduced into the synthetic gene (AF440682).
MSP3 is found to associate with the merozoite surface, although this association is not mediated by a transmembrane domain or a glycophosphatidylinositol lipid anchor. Truncation of the C-terminal coil region disrupted localization of both MSP3 and another parasite protein, the acidic–basic repeat antigen (ABRA) to the merozoite surface in transfected parasites. Thus this region of the protein may be involved with protein–protein interactions important for targeting and localization of the protein to the merozoite surface [14].
Although MSP3 is initially detected as a full-length protein during early schizogony [2], it is subsequently processed by an unknown protease [15]. The processed form is associated with the merozoite surface at the time of schizont rupture [15]. In this paper, we report the expression in E. coli and purification of an untagged MSP3 with a short N-terminal deletion (EcMSP3) that may more closely resemble the structure of the native processed MSP3 found on the surface of the merozoite. We describe its biochemical and biophysical properties and its ability to elicit an immune response that attenuates a virulent P. falciparum infection in Aotus monkeys.
2. Materials and methods
2.1. Expression plasmid
A synthetic gene based on a yeast codon preference of msp3 FVO strain (GenBank accession no. AF440682) was used as a template for PCR cloning. The synthetic msp3 gene corresponding to amino acids 25–365 was cloned with the primer set 5′-AGGAGGATATTCATATGT-TGAACCAAAACTCCC-3′ and 5′-CCTGCAGGTCGACTCTAGACTCGAGT-TAGTGG-3′ (restriction sites are underlined) into expression vector pET-24a(+), which was used to transform E. coli BL21 DE3 (Invitrogen, Carlsbad, CA). The sequence of the msp3 gene insert was then verified by sequencing. The synthetic gene template contained three asparagines mutated to glutamines; one of these was reverted from glutamine to asparagine at amino acid position 234 using standard techniques. The transformant was designated as pMSP3 n21/pET. EcMSP3 transformants were selected on agar plates containing 30 μg/mL kanamycin.
2.2. Fermentation and purification
Fermentation was performed using defined media in 5 L bioreactors using BioFlo 3000 fermenter units (New Brunswick Scientific) as described [16]. In brief, the scalable purification process, which will be detailed elsewhere (Shimp RL, Zhang Y, Narum DL, manuscript in preparation) was as follows. Cells containing soluble EcMSP3 were harvested by centrifugation, resuspended in lysis buffer, passed thrice through a microfluidizer with a 100 μm interaction chamber (Microfluidics Corp., Newton, MA) prior to depth filtration and purification. Recombinant EcMSP3 was purified using standard column chromatography methods with step gradients. The column chromatography steps were as follows: (1) capture on ceramic hydroxyapatite column (80 μm bead size, BioRad, Hercules, CA), (2) purification using MEP HyperCel (Pall Life Sciences, East Hills, NY), (3) ceramic hydroxyapatite to reduce endotoxin, (4) Q Sepharose HP (GE Healthcare, Piscataway, NJ) for purification, endotoxin removal and concentration, and (5) size-exclusion using Superdex 200 (GE Healthcare). The final bulk antigen was diluted to 1.0 mg/mL in PBS pH 7.4, sterile filtered and stored at −80 °C.
2.3. SDS-PAGE and Western blot
SDS-PAGE was performed on 4–20% gradient Tris-glycine polyacrylamide gels as described by the manufacturer (Invitrogen). For reducing conditions, samples were mixed with 5% (v/v) β-mercaptoethanol (β-ME) before electrophoresis. For Western blot analysis, proteins were transferred electrophoretically onto nitrocellulose membranes and Western blotting was performed as described [17].
2.4. Analytical reverse phase HPLC, N-terminal sequencing and MALDI-TOF
Purified EcMSP3 protein was acidified with 10% (v/v) trifluoroacetic acid (TFA, Fisher Scientific) to a final concentration of 0.1% TFA (v/v) and then microfuged at 13,000 rpm for 2 min. Approximately, 100 μg of EcMSP3 was analyzed on a Vydac (Hesperia, CA) C4 reverse phase column (2.1 mm i.d. × 250 mm). The initial mobile phase combined 99% mobile phase A (0.1% (w/v) TFA in water) and 1% mobile phase B (0.1% (w/v) TFA in acetonitrile). EcMSP3 was eluted with increasing concentrations of mobile phase B with the flow rate maintained at 0.2 mL/min over 60 min. The primary peak was collected and vacuum dried in organic mode (Eppendorf vacufuge) and submitted to the NIAID Research Technology Branch (RTB/NIAID/NIH, Rockville, MD) for mass spectrometry measurements and N-terminal sequencing. MALDI-TOF spectra (in the positive mode) were acquired on an Axima CFR Plus mass spectrometer (Shimadzu Biotech, Columbia, MD) equipped with a pulsed nitrogen laser emitting at 337 nm. Samples were analyzed in a linear mode using an accelerating voltage operating in a positive ion mode of 20 kV. Each chromatographic fraction was loaded on the target by the dried droplet method. Sinapinic acid was used as matrix. Spectra were externally calibrated using the [M+H]+ and [M+2H]++/2 ions of apomyoglobin (SIGMA). The amount of protein deposited on each spot was typically 1–5 pmol. Around 200 scans were averaged for each spectrum acquisition, using Kompact Shimadzu acquisition software.
2.5. Analytical SEC-MALS-HPLC and QELS
The purity, identity, and solution aggregation profile of purified EcMSP3 was analyzed using size-exclusion chromatography (SEC) with in-line multi-angle light scattering (MALS), refractive index, and ultraviolet detection [16]. A Waters (Milford, MA) 2695 HPLC, Waters 2996 PDA detector, Wyatt (Santa Barbara, CA) Dawn EOS Light Scattering detector, Wyatt Quasi Elastic Light Scattering (QELS) detector and Wyatt Optilab DSP refractive index detector were in series for acquiring the data and the Wyatt Astra V software suite was used for data processing. For separation, a Tosoh Bioscience (Montgomeryville, PA) TSK gel G3000SWxl 7.8 mm i.d. × 30 cm, 5 μm particle size analytical column was used with a TSK gel Guard SWxl 6.0 mm i.d. × 4cm, 7 μm particle size guard column. The column was equilibrated in mobile phase (1.04 mM KH2PO4, 2.97 mM Na2HPO4, with 308 mM NaCl, 0.02% azide) for at least 60 min at 0.5 mL/min prior to sample injection. Prior to analysis, EcMSP3 was filtered through a 0.45 μm PVDF centrifugal filtration device (Ultrafree-MC 0.45 μm, PVDF, Durapore, Millipore (Bedford, MA)) spun at 2000 rpm for 2 min. Approximately, 75 μg of sample was injected using an isocratic run at 0.50 mL/min in mobile phase for 60 min. Instrument calibration was performed with 10 μL of Bio-Rad (Hercules, CA) Gel Filtration Standard HPLC standards for size comparisons and a 75 μL injection of 5 mg/mL bovine serum albumin for MALS detector normalization.
2.6. Analytical ultracentrifugation characterization of EcMSP3
Boundary sedimentation velocity (SV) and sedimentation equilibrium measurements were made in Optima XLI analytical ultracentrifuge (Beckman-Coulter Instruments). SV analysis was performed at 20 °C at a rotor speed of 58,000 rpm. The centrifuge cell was filled with 400 μL of protein solution at a concentration of 1.0 mg/mL in PBS. Absorbance scans were obtained at 280 nm using the continual scan mode. Sedimentation coefficient distribution analysis to deconvolute the boundary velocity data into sedimenting species was performed as previously described [18] using the public domain software Sedfit developed by Peter Schuck (http://www.analyticalultracentrifugation.com/). The resolution parameter was set at 200 for the analysis. This computational analysis resolves sedimenting species by eliminating the effect of diffusion. The results of this analysis are presented in the form of a c(s) vs. s plot, where the y-axis c(s) equals dc/ds(1/S), where c is the concentration or absorbance, and the x-axis is the sedimentation coefficient range of the analysis, i.e., the experimental time scale converted to sedimentation coefficient S units. In this computational treatment, the sedimentation boundary velocity data are subjected to maximum entropy regularization statistical analysis for the most parsimonious distribution of sedimenting species [18] that best fits the data. The software program Sednterp developed by Hayes, Laue, and Philo (http://www.bbri.org/RASMB/rasmb.html) [19] was used to calculate the partial specific volume of the EcMSP3 and the viscosity (η) and density (ρ) for the buffer/salt solution used in the sedimentation analyses. This software was also used for hydrodynamic characterization of EcMSP3.
Sedimentation equilibrium measurements were used to determine molar mass of MSP3. Lebowitz et al. [20] have reviewed the biophysical foundation of this methodology. Cells were loaded with 180 μL of the protein solution at the protein concentrations indicated in the figure legend. Sedimentation equilibrium absorbance data at radial increments of 0.001 cm with 10 repeats were obtained at rotor speeds indicated in the figure legends. The SE experiment was performed at 4 °C. The obtainment of equilibrium was evaluated by subtracting 6 h scans and observing whether the difference data points randomly distributed around the zero base line. The public domain software program Sedphat, developed by Peter Schuck, was used for the analysis of the sedimentation equilibrium data to (http://www.analyticalultracentrifugation.com/).
2.7. Atomic force microscopy
Gentle tapping-mode atomic force microscopy (AFM) studies were performed using a Nanoscope® IV or V controller, a PicoForce® Multimode platform, and a type E scanner head (Veeco, CA) basically as reported elsewhere [21,22]. AFM imaging both in fluid and in air was pursued to investigate aspects of this protein product. To achieve a suitable MSP3 attachment to the substrate for high-resolution imaging in air, freshly peeled mica substrates (of 9 or 15 mm diameter glued on a metal disk) were either used directly or modified by incubation under CaCl2 or MgCl2 solutions of up to 20 mM for up to 10 min at room temperature [21]. To evaluate the sensitivity of the observed EcMSP3 structure to the conditions of the AFM sample preparation, we diluted the PBS buffer-based purified EcMSP3 protein product with variable amount of deionized water (dH2O) with and without addition of variable amount of EDTA or EGTA for reduction of multivalent ions. Typically, 2–7 μL of such a solution, with the final protein concentration under 1 μg/mL, the total ionic strength ranging from 10 mM to 1.1 M and the pH ranging from 4 to 8.5, were spread over the entire mica surface to deposit EcMSP3 proteins for several minutes under the fluid or while drying under a gentle flow of argon or N2 gas. The sample surface was then washed with ca. 0.5 mL dH2O, dried completely under the inert gas flow, and placed into the Multimode compartment filled with Drierite® particles and sealed with a piece of Parafilm® for further humidity reduction and equilibration. Gentle tapping mode imaging in air was then carried out with a small tapping amplitude set-point reduction of 5–10% and a tapping frequency lower than the free-oscillation resonance. In high-resolution experiments, preselected AFM probes having a sharpened tip of a radius less than 5 nm were used. AFM images were evaluated within the Nanoscope software (Veeco, CA), and exported to Image J (NIH, Bethesda, MD) and Mathcad (Mathsoft, MA) for further analyses and display.
2.8. Indirect immunofluorescence assay
P. falciparum FVO strain parasitized erythrocytes were assayed essentially as previously described [7]. Briefly, methanol fixed parasitized thin blood films were reacted with primary antibodies at a serum dilution of 1/100 to 1/500 in buffer, washed and reacted with anti-human IgG conjugated with Alexa 488 (Molecular Probes, Eugene, OR).
2.9. Aotus challenge study
Aotus nancymai monkeys were housed at the National Institutes of Health Primate Research Facility in compliance with an NIH Animal Care and Use Committee-approved protocol (ASP MVDB8E). Fourteen naïve monkeys were randomly assigned to two groups of seven. Seven monkeys were vaccinated with 100 μg of EcMSP3, seven with 100 μg of control protein Pfs25, a parasite protein expressed during the mosquito stage of the life cycle [17,23]. Each monkey received 0.125 mL of antigen emulsified in complete Freund's adjuvant at four sites, for a total of 0.5 mL, followed by two booster vaccinations with the same dose of antigen in a Montanide ISA51 (SEPPIC) formulation at 3-week intervals. Seventeen days after the third vaccination, the monkeys were challenged by intravenous infusion of 5 × 104 P. falciparum FVO strain parasitized RBCs collected from a naïve donor monkey. Parasitemias were monitored daily by Giemsa-stained thin blood films for 28 days after challenge. Hematocrits were monitored every third day. Monkeys were treated with 25 mg/kg of mefloquine when the parasitemia reached 5% or the hematocrit fell below 25%.
2.10. ELISA
Serum antibodies to MSP3, AMA1 and MSP142 were assayed by a standardized enzyme-linked immunosorbent assay (ELISA) [24]. Briefly, flat-bottom 96-well ELISA plates were coated with 100 μL per well of a 1.0 μg/mL recombinant antigen solution of 15 mM sodium carbonate and 35 mM sodium bicarbonate, pH 9.6 and then incubated overnight at 4°C. Plates were washed with 0.1% Tween 20 in Tris-buffered saline (TBS) and then blocked with 5% skim milk (Difco, Detroit, MI) in TBS for 2h at room temperature. After the plates were again washed, test sera (primary antibodies) were diluted in 0.1% bovine serum albumin-0.05% Tween 20 in TBS, added to antigen-coated wells in triplicate, and incubated for 2 h at room temperature. A duplicate control dilution series of a reference Aotus standard antiserum to the plate antigen was also added to each plate. After extensive washing, plates were incubated with alkaline phosphatase-labeled goat anti-human IgG heavy-plus-light chain conjugate diluted in the same buffer for 2 h. Bound antibodies were visualized by addition of the substrate solution (p-nitrophenyl phosphate; Sigma Chemical Co.). Absorbance at 405 nm was read with a SpectraMAX 340P ELISA reader (Molecular Devices Co, Sunnyvale, CA). The undiluted reference serum was assigned an antibody unit value equal to the reciprocal of the dilution giving an OD405 = 1.0 in this assay. Duplicates of the reference sera were included on each test plate and used to generate a standard curve. A four-parameter hyperbolic curve was generated from the standard curve values and this curve was used to convert the absorbance of individual test sera in to antibody units (SOFTmax PRO version 3; Molecular Devices Co.).
2.11. Statistical analysis
Non-parametric, unpaired one-tailed Mann–Whitney U-tests were performed to compare test group vaccinated with EcMSP3 to the control group that received Pfs25. The primary end-points for the Aotus challenge study only included data until the first monkey was treated for low hematocrit rather than for parasitemia. On that day, all monkeys were ranked using the following criteria: (1) monkeys that were treated for parasitemia prior to the primary endpoint day ranked first, in order of their day of treatment, (2) if two or more monkeys were treated on the same day, then these monkeys were ranked by their cumulative parasitemia (the sum of that monkey's daily parasite burden) and (3) monkeys not treated for parasitemia up until that point were ranked in order of their cumulative parasitemias.
2.12. Structure prediction
Secondary structure predictions were obtained using the SAM, JUFO, and PSIPRED algorithms [25–27], which provide amino acid-specific confidence values. Regions of the sequence that were predicted to have significant secondary structure were excised and used for de novo structure prediction in Rosetta [28] (see Table 2). The resulting decoys were clustered by the root-mean-square similarity in the positions of their alpha carbons. The highest scoring decoy for each region was chosen for fitting to the AFM data. Controls included Rosetta and LOOPP [29] predictions on orthologous sequences of variant strains of MSP3 (MSP3-K1: GenBank accession no. AAC47831; MSP3-NF54: GenBank accession no. AAC09377). AFM images were converted to a pseudo-electron density mesh by creating a cubic voxel field based on the resolution of the image and its corresponding volume. Each voxel was assigned a density value based on the pixel value from an AFM topological trace, but scaled in the Z direction by the normal distribution so that the density would be greatest inside the AFM-derived envelope. After initial manual placement of the predicted regions into the pseudodensity, the fit was refined with the colacor package of the Situs suite [30].
3. Results
3.1. Cloning and expression of EcMSP3
A synthetic msp3 gene [7] was used as a template for PCR cloning as discussed in Section 2. The expression product from pMSP3 n21/pET, identified here as EcMSP3, consists of a 40 amino acid deletion from the N-terminal methionine of K1 or D10 parasites [31]. This construct was selected due to its integrity of the expressed protein (Fig. 1C). EcMSP3 was observed as a single dominant band with an apparent molecular weight of 47 kDa (Fig. 1C). The expression clone was fermented in a 2.5 L fermentation run in defined media and EcMSP3 expressed as a soluble protein was purified from the cell pellet as discussed in Section 2. The integrity and purity of the recombinant protein is shown by Coomassie blue stained SDS-PAGE gel (Fig. 1D) and immunoblot (Fig. 1E). An EcMSP3 protein complex (Fig. 1D and E, asterisks) consistently migrated at approximately 160 kDa, which was resistant to significant reduction by DTT or β-ME and was also detected in solution (see details below). A C-terminal EcMSP3 fragment was also identified (NT: YVSSKDK) (Fig. 1D, double asterisks), which was not removed by size exclusion column chromatography.
3.2. Aotus challenge study
Two groups of seven monkeys were immunized with a control antigen Pfs25 or with EcMSP3. Analysis of sera taken 2 weeks after the last vaccination showed EcMSP3 elicited specific antibody responses by ELISA (Table 1) and by immunofluorescent antibody analysis using P. falciparum FVO infected erythrocytes (data not shown).
Table 1.
Summary of Aotus challenge study.
| Immunogen | Aotus ID | Pre-challenge EcMSP3 ELISA titera | Peak parasitemia (%) | Day to treatment | |
|---|---|---|---|---|---|
|
| |||||
| Parasitemia | Anemia | ||||
| Pfs25H | 3110 | 18 | 5.7 | 10 | |
| T1360 | 4 | 6.5 | 10 | ||
| 3137 | 9 | 12.3 | 11 | ||
| T1327 | 6 | 6.9 | 11 | ||
| 3098 | 11 | 6. 5 | 11 | ||
| 3100 | 6 | 5.3 | 11 | ||
| 3111 | 21 | 5 .7 | 14 | ||
| Mean | 11.1 | ||||
| EcMSP3 | 3128 | 17,992 | 5.1 | 12 | |
| 3126 | 28,105 | 6.9 | 13 | ||
| 3073 | 16,085 | 10.0 | 14 | ||
| 3075 | 25,146 | 5.9 | 14 | ||
| 3135 | 6,078 | 3.6 | 18 (2.2)b | ||
| 3130 | 33,013 | 1.4 | 18 (0.65)b | ||
| 3123 | 36,651 | 0.1 | 15 (0.00)b | ||
| Mean | 14.9 | 17.0 | |||
ELISA titer represents the reciprocal of the dilution which gives an OD = 1 in a standardized ELISA assay.
Percent parasitemias on day of treatment.
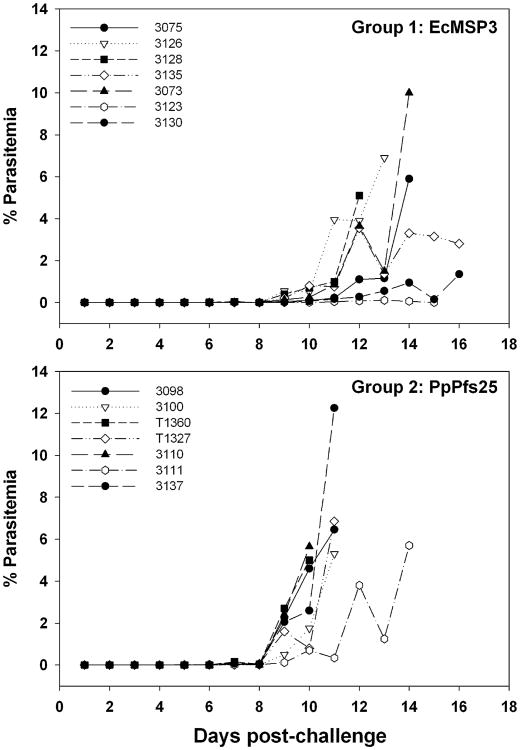
Three weeks after the third immunization, the monkeys were challenged with P. falciparum FVO parasitized erythrocytes. By day 11 post-challenge, the parasitemia in all but one monkey in the control group had reached the predetermined upper limit and were treated In contrast, no animals in the EcMSP3-vaccinated group required treatment by this time (Fig. 2). Four animals in the MSP3 group subsequently developed high parasitemia and were treated between days 12 and 14 post-challenge. The remaining three animals maintained relatively low levels of parasitemia but were eventually treated for anemia (days 15, 17 and 18).
Fig. 2.
Aotus challenge study. Parasitemia curves in Aotus monkeys challenged intravenously with P. falciparum-infected Aotus erythrocytes after immunization with EcMSP3 or PpPfs25, a control protein.
All monkeys were ranked on the first day a monkey was treated for a low hematocrit (Table 1, day 15) using the criteria described in Section 2. An evaluation of the rankings by use of an one-tailed Mann–Whitney U-test showed that the MSP3-vaccinated group differed from the Pfs25 control group (P=0.0013). Additional analysis was completed on the cumulative day 10 parasitemia (the day when the first monkey was treated). The cumulative parasitemia of the MSP3 group differed significantly from the Pfs25 group (P=0.006 by one tailed Mann–Whitney U-test) (Table 1). Clearly EcMSP3 elicited an immune response that controlled the parasite infection in some animals in the test group compared to the control group and is therefore consistent with the recombinant protein having the correct structure. Within the test group, there was a trend to a milder infection with higher pre-challenge EcMSP3 antibody titers (Spearman rank correlation of 0.46), but this was not significant (P =0.147).
3.3. Biochemical and biophysical analysis of EcMSP3
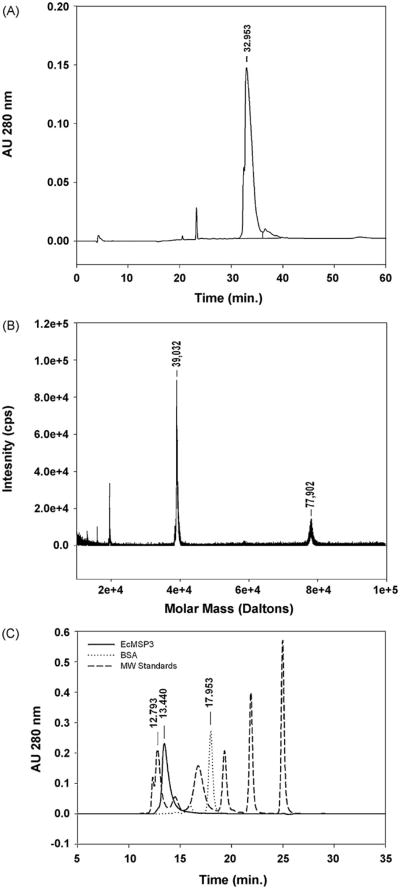
Purified EcMSP3 was characterized biochemically and biophysically. The purity of EcMSP3 was >95% based on RP-HPLC under non-reduced conditions (Fig. 3A). The RP-HPLC elution peak was analyzed by matrix-assisted laser adsorption ionization-mass spectrometry. The observed mass for the non-reduced form was 39,032 Da (Fig. 3B) which is within 6.9 Da (≤0.02%) of the non-reduced theoretical mass 39038.9 Da. The observed mass is within the expected range of error (±0.2%). A second peak with a mass of 77,902 Da, which likely represents a dimer, was observed (Fig. 3B). The identity of EcMSP3 was confirmed by N-terminal sequence analysis, which yielded the expected N-terminal sequence (MLNQNSQIENEE). The endotoxin concentration ranged between 35 and 70 EU/mg of protein.
Fig. 3.
Biochemical analysis of EcMSP3. EcMSP3 was characterized by RP-HPLC (A), matrix assisted laser desorption ionization mass spectrometry (B) and SEC-HPLC (solid line) in comparison with molecular weight standards (dashed line) and bovine serum albumin (dash-dot-dash line) (C). Molecular weight standards used: thyroglobulin (670 kDa), immunoglobulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa) and vitamin B12 (1350 Da).
Analysis of EcMSP3 by SEC-HPLC indicated that the protein was comprised of >99% as a single peak (Fig. 3C) although the retention time (RT: 13.440 min) was significantly less than albumin (67 kDa, RT: 17.953 min) (Fig. 3C, see legend). EcMSP3 retention time was close to thyroglobulin (RT: 12.793) that has a mass of 630 kDa. Analysis by in-line SEC-MALS-HPLC showed that the peak at 13.440 min had a weight average molar mass of 92,630 ± 278 (Fig. 4, (a)), which is ∼14.6 kDa larger than the theoretical mass if EcMSP3 formed a dimer, as reported for another E. coli expressed MSP3 protein [13]. The weight average mass of the front shoulder (Fig. 4, (b)) ranged from ∼160 to 300 kDa. The larger than expected weight average mass (92,630 Da vs. 78,064 Da) is likely due to the population of multimers derived from the front shoulder that are included in the mass analysis (Fig. 4, (b)) as has been described [16]. Consistent with an earlier report [13], evidence that the C-terminus is responsible for dimer formation was observed during purification (data not shown).
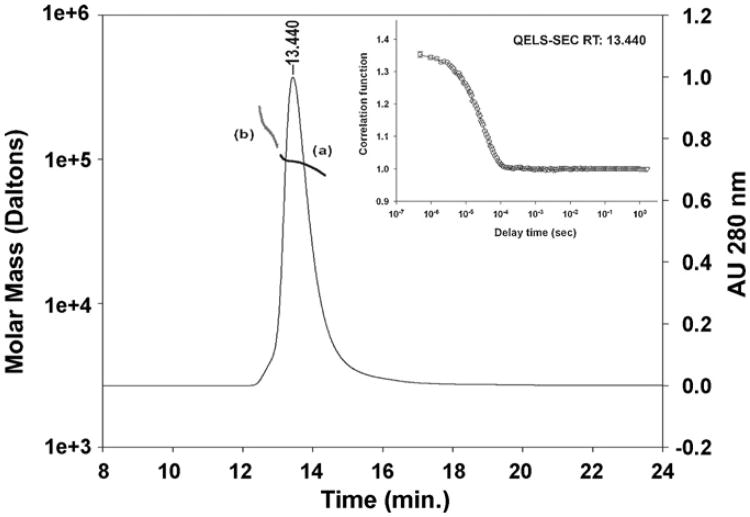
Fig. 4.
Biophysical characterization by inline SEC-MALS and QELS. Analysis of EcMSP3 by in-line MALS-SEC yielded the molar mass distribution of main peak (a, dark line, RT 13.440 min) and front shoulder (b, gray line) compared to the absorbance 280 nm. The insert shows SEC-QELS-HPLC result for the goodness of fit of the hydrodynamic radius at the apex of the primary peak (RT 13.440).
SEC-MALS-QELS-HPLC was used to analyze the hydrodynamic radius (Rh) of the dimeric EcMSP3 given the significant increase in the apparent weight average mass. The results of the QELS analysis demonstrated that the hydrodynamic radius at the peak (RT: 13.440 min) was 7.6 ± 0.2nm (Fig. 4 inset), which indicates that the dimeric form of EcMSP3 is highly extended compared to a more typical globular protein such as BSA (Rh ≈ 3.4 nm). To confirm our interpretation we analyzed the purified EcMSP3 by sedimentation velocity studies.
3.4. Boundary sedimentation velocity, sedimentation equilibrium, and hydrodynamic characterization of EcMSP3
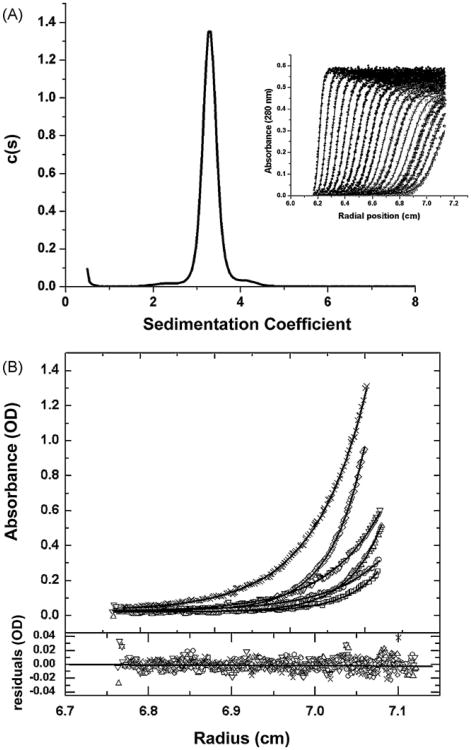
Boundary sedimentation velocity was used to obtain the sedimentation coefficient distribution of the EcMSP3 sample. The absorbance scans of the sedimenting boundary vs. radial position are shown in the inset of Fig. 5A. This data was analyzed using the software program Sedfit to deconvolute the sedimenting species of EcMSP3 that compose the sedimenting boundary (Fig. 5A). It is evident from the c(s) vs. s plot that EcMSP3 appears to sediment as a single peak with an uncorrected weight average sedimentation coefficient of 3.30S. This is corrected to an s20,W value of 3.41S. The weight average frictional ratio (f/f0) is optimized in the fitting process to account for boundary diffusion. An f/f0 of 2.50 was obtained and this value in combination with the weight average s value of the peak gives a molar massfor the peak of 103 kDa (results not shown). Although the latter value is close to the weight average molecular weight obtained from SEC-MALS it is of importance to obtain a molar mass determination using a sedimentation approach that is independent of the shape of the protein. To accomplish this we used sedimentation equilibrium, a thermodynamically rigorous method for molar mass determination.
Fig. 5.
Biophysical characterization by sedimentation analyses. Panel A: sedimentation velocity characterization of EcMSP3. The inset of this figure shows the sedimentation boundary movement (time independent noise removed) of the EcMSP3 sample vs. radial position. The solid lines through the data points are the fit generated by Sedfit software analysis. For clarity only one half of the optical scans are shown, but data analysis was performed for all the scans. The deconvolution of the boundary data into resolved sedimenting components is shown as a c(s) vs. s plot (see Section 2). The essentially single peak was integrated to give a weight average sedimentation coefficient of 3.29S. Panel B: sedimentation equilibrium determination of the molar mass of MSP3. The top panel shows the overlay of the sedimentation equilibrium distributions, obtained at 4°C, absorbance vs. radius scans at 280 nm for EcMSP3 at the following concentrations: 0.22, 0.44, and 1.09 mg/mL at centrifugal speeds of 12,000 and 15,000 rpm. The × and diamond symbols show the data for the 1.09 mg/mL sample at 12,000 and 15,000 rpm, respectively. The inverted and upright triangles show the data for the 0.44 mg/mL sample at 12,000 and 15,000 rpm, respectively. The open circles and open squares show the data for the 0.22 mg/mL sample at 12,000 and 15,000 rpm, respectively. The solid line for each data set is the best fit to the data from the global analysis of all the scans using a single species model (see text). This non-linear regression analysis gives a molar mass 100,199 for EcMSP3. Residuals of the fitted lines to the experimental data are displayed in the lower panel with the corresponding symbols listed above. The best-fit rms error is 0.0058 and the global reduced χ2 was 2.0169.
Sedimentation equilibrium measurements were made using three concentrationsof EcMSP3 and two centrifugal speeds in order to perform global non-linear regression analysis of different models for the species present in solution. A single species model gave a very good fit to the data as shown in Fig. 5B. The molar mass from this fit was 100,191. All other multiple species models tested gave poor fits to the data (results not shown). The molar mass results from sedimentation equilibrium, SEC-MALS-HPLC and the c(s) vs. s analysis are in good agreement regarding the weight average molar mass.
Using the average weight average molar mass of 96,411 obtained from SEC-MALS- and sedimentation equilibrium we obtain a frictional ratio (f/f0) of 2.38 and a Stokes radius of 7.17 nm. This value for f/f0 strongly supports a non-globular shape for EcMSP3. The simplest models are elongated prolate and oblate ellipsoids. For the prolate model, using a hydration value of 0.3g water/protein, the major semi-axis is 27.5 nm and the minor semi-axis is 1.19 nm. For a comparable hydrated oblate model, the major semi-axis is 11.1 nm and the minor semi-axis is 0.32 nm. The volume of both hydrated ellipsoids is about 162 nm3. The volume without hydration would be 114 nm3. These ellipsoid models are used for convenience in calculation, but do not present a high-resolution view of the protein shape. Atomic force microscopy measurements were made as described below to better understand the unusual structure that causes such a high degree of translation friction.
3.5. AFM structures of EcMSP3
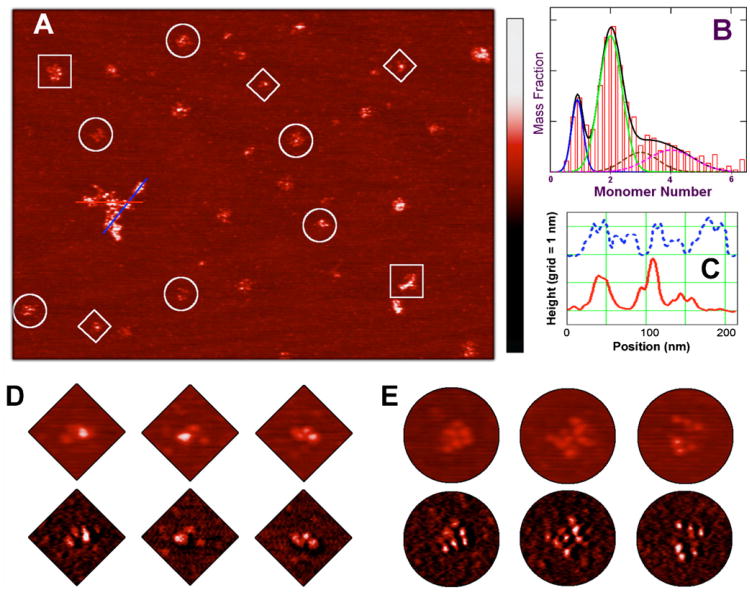
EcMSP3 and various oligomers of it can be highly resolved after in-solution deposition over a flat mica surface and low humidity AFM imaging (see Section 2). The most striking feature in both topological and phase contrast maps were large, asymmetric filamentous structures with a topological height of about 1.5 nm, and a variable size range of about 5 nm in full width at half maximum (Fig. 6A). At higher protein concentrations, these structures even formed flattened micrometer-sized meshes (data not shown). We interpret these structures as aggregated protein.
Fig. 6.
Atomic force microscopy structures of EcMSP3 and oligomers. EcMSP3 and oligomers are highly resolved, after adsorption initiated under 20 mM PBS buffer (pH 7.4) over a flat mica surface pretreated with a CaCl2 solution, revealing large filamentous structures and smaller, isolated particles. Both structures have a consistent topological height of around 1.5 nm, but their sizes range from tens of nanometers to several hundred nanometers (A). The isolated particles can be grouped according to their volumes and identified as monomers, dimers, and higher order oligomers. The mass distribution derived from over 2000 EcMSP3 particles of high-resolution AFM images reveals discrete populations of monomer, dimer, and larger oligomers (B). The mass ratio of the dominant dimer population (under the green curve) to the monomer population (blue) is approximately four. The distribution of larger oligomers, totaling about 30% in the overall mass, can be attributed mainly as trimers (brown), tetramers (magenta) and a few sporadic higher oligomers. The solid black curve is the sum of the four components from monomer to tetramers. Each feature in the topological trace displays a beaded substructure as plotted on the line chart (C), showing the topological height along two cross sections of the largest structure in (A). The bead separation averaged about 14 nm. The number of resolved beads in the isolated particles corresponded with their oligomeric state. EcMSP3 monomers (diamonds) typically had three beads, dimers (circles) had up to seven to nine beads, and higher order oligomers (squares) had up to 12–15 beads. The color scale bar from dark to red to bright is universal with a range of 0–4 nm for all height images and of 0–15 degrees for all phase images.
Smaller, isolated particles were also observed with a topological height similar to the larger structures, but that can be grouped according to their apparent molecular volume. The smallest particles have an AFM volume of about 70 nm3. A more prevalent group has an AFM volume of about 140 nm3. Larger particles have an AFM volume mostly ranging from 150 to 400 nm3. The molecular weight of EcMSP3 is about 39 kDa, so the protein volume should be about 60 nm3, assuming a common specific volume value for proteins of 0.75 cm3 per gram. Because buffer hydration typically adds the equivalent of one or two water layers to the protein volume and because the AFM tip-broadening effect tends to overestimate the apparent molecular volume, it is reasonable to assume that the smallest particles of about 70 nm3 are monomeric EcMSP3. The most prevalent group of particles has a volume twice that of the monomeric EcMSP3, which also agrees with the ellipsoid volumes calculated from the sedimentation studies and suggests identifying this group as the dimeric form of EcMSP3. Larger particles have volumes corresponding to higher order oligomers (Fig. 6A and B).
Both the aggregated protein and the isolated particles clearly exhibit a complicated, non-linear, beaded substructure (Fig. 6A and C–E). The height of these beads ranged from about 0.8 to 1.8 nm, with a mean of about 1.2 nm. The bead separation ranged from 9 to 18 nm, with a mean of about 14 nm. The number of resolved beads in the isolated particles corresponded with their oligomeric state. EcMSP3 monomers typically had three beads, dimers had up to 7–9 beads, and higher order oligomers had as many as 12–15 beads. We interpret these beads as structural domains of the protein corresponding to the domains identified in the amino acid sequence (Fig. 1). To develop a working structural model for the observed EcMSP3 domains molecular computational modeling was performed as described in the next section.
3.6. Structure predictions of EcMSP3
We used computational techniques to test how the tertiary structure of EcMSP3 might correspond with the domains identified in the sequence and if that tertiary structure could be reconciled with the AFM data. Traditional homology modeling would be ineffective for EcMSP3 because little sequence homology exists between it and any known protein structure. De novo tertiary structure predictions in Rosetta are aided by secondary structure predictions. The three long helices previously identified by sequence repeats in the N-terminal half of the protein were detected by secondary structure predictions. A COILS [32–34] prediction suggested a loose coiled coil structure: the first and third helices have a nearly 100% probability of forming a coiled coil, the second had only a 50% probability. The two short C-terminal leucine zipper-like helices were also detected by secondary structure predictions. The acid-rich middle domain was predicted to have some secondary structure near the leucine zipper-like domain,so the two domains were submitted for tertiary structure predictions together. This C-terminal domain is separated from the coiled coil domain by 48 amino acids of no secondary structure. For this reason, predictions of tertiary structure were attempted for the coiled coil (76–189) and the C-terminal (238–365) domains individually. Other regions of the sequence were also submitted for tertiary structure prediction.
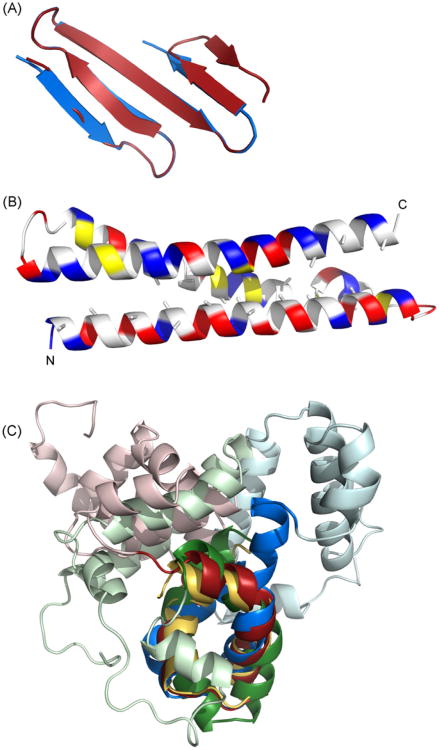
The coiled coil and the leucine zipper-like domains yielded consistent and reproducible tertiary structures with Rosetta and LOOPP no matter which sequence variant was used (Fig. 7), suggesting a robust and reasonably accurate prediction for these domains. The N-terminal residues 28–63 of EcMSP3 and MSP3-K1 also folded neatly into an antiparallel beta sheet and are separated from the coiled coil domain by a linker of 12 amino acids. This beta sheet likely does not exist in MSP3-NF54 because there is little secondary structure or homology in the corresponding sequence. In contrast, tertiary structure predictions for the acid-rich middle domain were not consistent for any of the variant sequences or between any of them. This inconsistency suggests that the tertiary structure for the majority of the C-terminal domain should not be considered to be accurate. Tertiary structure predictions for the linker between the coiled coil and C-terminal domains were also inconsistent (see Fig.7 and Table 2).
Fig.7.
Computational prediction of MSP3 structure. Structural models of MSP3 were generated by homology from sequence threading (LOOPP) or de novo with Rosetta. (A) Rosetta predicts that the N-terminal domain forms a small anti-parallel beta-sheet; two alternative models are shown in blue and red. (B) The alanine repeat region forms a coiled coil of three helices. These alanines face the interior of the bundle. Charged residues are shown in blue and red, hydrophobic residues in yellow (C) Rosetta predicts a leucine zipper-like domain at the C-terminus (shown in darker colors). This leucine zipper-like domain is a structural anchor around which four helices of the D/E-rich middle domain pack (shown in lighter colors). The top-scoring decoys from nearly 200,000 trials using the EcMSP3 sequence have little internal agreement and the top-scoring decoys from another 100,000 trials with closely related homologues do not agree between themselves or with the EcMSP3 decoys, suggesting that the orientation of these helices cannot be assigned with any confidence. The leucine zipper-like domain alone is shown in gold, the EcMSP3 C-terminus in red, the MSP3-K1 C-terminus in green, and the MSP3-NF54 C-terminus in blue.
Table 2.
Computational prediction and fitting results.
| Domaina | N-terminal | Coiled-coil | C-terminal | |||
|---|---|---|---|---|---|---|
| Strainb | EcMSP3-FVOc | EcMSP3-FVO | MSP3-NF54 | EcMSP3-FVO | MSP3-K1 | MSP3-NF54 |
| Modeled amino acidsc | 28–63 | 76–189 | 76–186 | 238–365 | 254–379 | 228–354 |
| LOOPP templated | None | 1EZ3 | 1EZ3 | None | None | None |
| LOOPP score | – | 1.91 (“high”) | 1.56 (“good”) | – | – | – |
| Rosetta scoree | −87.5 (3.2) | −94.8 (2.7) | −87.1 (3.4) | −230.3 (4.3) | −190.6 (2.6) | −199.4 (4.3) |
| Rosetta cluster sizef | 1431 (2.9) | 39 (3.4) | 198 (5.7) | 89 (6.8) | 39 (4.0) | 52 (4.8) |
| Rosetta decoysg | 20 (2) | 40 (4) | 20 (2) | 190 (4.5) | 50 (2) | 50 (2) |
| AFM fittingh | 0.09 | 0.19 | – | 0.22 | – | – |
The protein was divided into domains to aid prediction and fitting. Where strain variants differed, additional predictions with those sequences were made to verify the robustness of the solution.
There is no difference between the sequences of EcMSP3 and MSP3-K1 in the coiled-coil domain.
EcMSP3 amino acid numbering refers to GenBank accession no. AF440682 and with the exception of two point mutations corresponds to the MSP3 derived from the FVO strain. EcMSP3 comprises amino acids 25–365.
No suitable template was found with LOOPP for the N- and C-terminal regions. 1EZ3: neuronal t-SNARE syntaxin-1A.
The top score from the largest cluster is reported with the Z value in parentheses. The N-terminal domain produced two equally viable solutions; see Fig. 7A.
Z value is in parentheses.
Total number of decoys generated in thousands (the number of highest scoring decoys used for clustering is in parentheses).
A dimer envelope was used for fitting; correlation scores were identical for both monomers at the reported precision.
Tertiary structures were predicted for 85% of the full-length sequence. About 60% of the full-length sequence was predicted with a reasonable confidence. Assuming random coil secondary structure for the uncertain and missing portions of the structure, the protein secondary structure composition is predicted to be 41% alpha helix, 7% beta strand and 52% turn and random coil. These values correspond well with circular dichroism studies, which indicated a secondary structure composition of about 47% or 50% alpha helix, and 6% or beta-strand ([13] and data not shown, respectively).
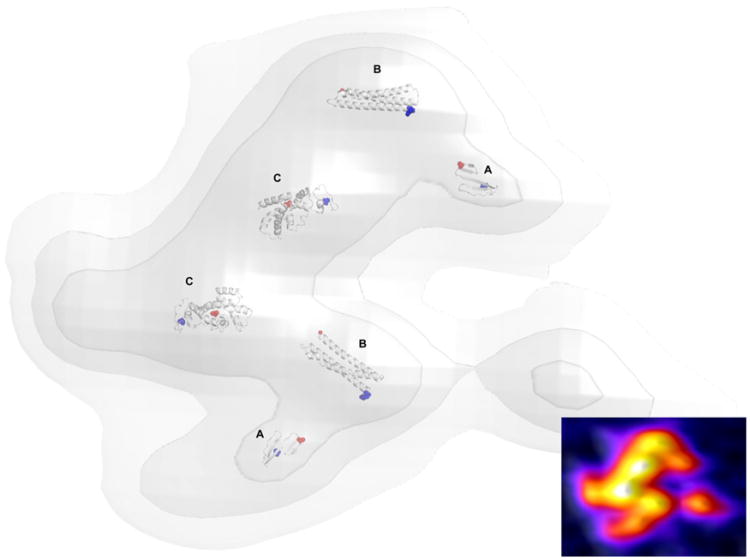
We chose to fit the predicted domains of EcMSP3 into an AFM topographic trace for a dimer because it is the solution state of the protein (Fig.8). Although the majority of the C-terminal domain was predicted with little confidence, we fit the entirety of this largest domain into the largest bead structures of the dimer to illustrate the concept of dimerization between their leucine zipper-like domains. We restrict the level of uncertainty in the model by not modeling the linkers between domains. The correlation values for the fit of the best tertiary structures into the AFM pseudodensity were low (see Table 2), primarily because (1) the predicted domains are only fragments of the complete structure, (2) the predicted domains do not include the hydration shells measured by AFM, and (3) the protein pseudodensity is only approximated by the AFM topographic envelope since the AFM traces contain some effects of protein surface charge and compressibility distribution and finite AFM tip size.
Fig. 8.
A structural model of dimeric MSP3. The N-terminus and coiled coil regions are predicted with high confidence (A and B, respectively). The larger C-terminus contains leucine zipper-like domain responsible for dimerization (C). The top-scoring predictions for EcMSP3 were chosen for fitting to the protein pseudodensity map derived from high-resolution AFM topological imaging. The N-termini of each domain are colored blue and the C-termini of each domain are colored red. A heat map of the corresponding AFM image data is inset.
4. Discussion
A preclinical study in which Aotus monkeys were immunized with a P. pastoris expressed MSP3 protein showed that MSP3 provided significant protection against a virulent P. falciparum FVO challenge [7]. Unfortunately, this particular yeast-expressed MSP3 protein was highly fragmented and not optimal for scaled production for phase 1 human vaccine trials. As an alternative, we have developed a scalable E. coli fermentation and purification process to produce a recombinant, soluble form of MSP3 (EcMSP3) that has been extensively characterized both biochemically and biophysically and evaluated for its protective efficacy. The details of the production process will be described elsewhere [2].
Biochemical characterization of purified EcMSP3 showed that the N-terminal sequence and mass spectrum were comparable to the expected results. Evaluation of the impurity profiles from the bench scale production indicated that the concentration of endotoxin and E. coli host cell proteins were at acceptable levels for human studies (data not shown). Analytical biophysical studies of the solution state of EcMSP3 by SEC-MALS and QELS, as well as velocity sedimentation showed that EcMSP3 is predominantly an extended dimer (Figs. 2C, 3 and 4). Both QELS and sedimentation velocity data agree on a value of the hydrodynamic radius of EcMSP3, which gives it an unusually high translational frictional coefficient, strongly supporting a highly non-globular protein. This was also observed qualitatively from the elution behavior on SEC chromatograms. A recent study [13] of a different form of E. coli-expressed MSP3 that contained an N-terminal His6 tag and the first 21 additional amino acids of the unprocessed protein showed different sedimentation velocity behavior compared to EcMSP3, using a similar sedimentation methodology (see Section 2). For their construct, a resolvable equilibrium between dimers and tetramers was observed in the boundary sedimentation velocity. The s20,w of this protein was 4.52S and the f/f0 was 1.64 compared to the s20,w of EcMSP3 which is 3.4S and the f/f0 is 2.5. The difference in the association properties and shape of these different MSP3 constructs based on sedimentation analysis is significant. Furthermore, circular dichroism measurements for EcMSP3 were comparable to those reported by Burgess et al. [13] (data not shown). Thus it is difficult to explain the striking differences observed for the sedimentation velocity behavior for these two recombinant forms of MSP3 other than to suggest that the presence of the N-terminal His6 tag and/or the additional 21 amino acids in their product are responsible for the different biophysical characteristics. We believe that EcMSP3 more closely mimics the native structure of the processed form of MSP3 [15].
To further resolve the structural features that give EcMSP3 such an unusually high translational frictional coefficient, we characterized the protein using high-resolution AFM. These measurements identified various non-linear molecular structures for EcMSP3 recognizable predominantly as dimers, but also as monomers, tetramers, and other multimeric forms. The detection of these alternative forms of EcMSP3 by AFM, which were not resolved in solution by SEC-MALS and sedimentation studies, may be promoted by the particular AFM substrate, but the distribution of these forms is fairly constant over the range of AFM observations. In each of these forms, a beaded substructure is apparent and commonly reveals three beads per monomer. We conclude that these beads are actually structural domains of EcMSP3 connected by flexible loops, giving rise to the asymmetry observed in the substructure and accounting for the considerable hydrodynamic friction and large hydrodynamic radius. It is also possible that the self association of the flexible arms of the alanine-rich repeats of EcMSP3 observed here by AFM contribute to protein–protein interactions with other malaria surface proteins such as ABRA [14]. Such high-resolution single-protein studies provide information that is not obtainable from bulk solution studies. It is clear that the structures of EcMSP3 observed here by AFM are not extended rod-like molecules, as is proposed for the full-length recombinant MSP3 protein containing the N-terminal His6 tag plus 21 amino acids [13].
The Aotus challenge results reported here, as well as those of Hisaeda et al. [7] demonstrate the protective efficacy of MSP3 protein in this stringent primate model. Both the yeast- and E. coli-expressed recombinant forms of MSP3-induced protective responses. Overall the levels of protection appeared similar [7]. Because the Aotus study was not designed to compare the potency of the two recombinant forms of MSP3, which require a head-to-head comparison,no statistical comparison of the two independent outcomes was performed. Analysis of the antibody responses in the previous Aotus challenge study conducted with the yeast-expressed MSP3 showed that some of the pre-challenge sera from the MSP3 group cross-reacted with an E. coli-expressed MSP142 and P. pastoris expressed AMA1 [7]. Such cross-reactivity was not observed in this study (data not shown) and the reason(s) for this difference is unclear.
Our results show that MSP3 expressed in a soluble form by E. coli can induce a protective immune response in Aotus monkeys when it is formulated with Freund's adjuvant. Future vaccine studies should be directed toward the identification, development, and characterization of adjuvants that will be suitable for human use alone or, more importantly, in combination with other malaria vaccine antigens.
Acknowledgments
We are grateful to Drs. David Garboczi and Brandt Burgess (Structural Biology Section, Laboratory of Immunogenetics, NIAID/NIH) for discussions on the design and expression of MSP3 constructs in E. coli; to Jacqueline Glen, Vu Nguyen, and Dr. Yanling Zhang for assistance with production of recombinant EcMSP3; to Karine Reiter for assistance with characterization and to Olga Muratova for performing the ELISAs. We thank Dr. Paul Smith of NIBIB/NIH for encouragement and support. Part of this work was carried out by using the resources of the Computational Biology Service Unit from Cornell University, which is partially funded by Microsoft Corporation. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov). This research is supported by intramural funding of NIAID/NIH.
References
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434(7030):214–7. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McColl DJ, Silva A, Foley M, et al. Molecular variation in a novel polymorphic antigen associated with Plasmodium falciparum merozoites. Mol Biochem Parasitol. 1994;68(1):53–67. doi: 10.1016/0166-6851(94)00149-9. [DOI] [PubMed] [Google Scholar]
- 3.Oeuvray C, Bouharoun-Tayoun H, Gras-Masse H, et al. Merozoite surface protein-3: a malaria protein inducing antibodies that promote Plasmodium falciparum killing by cooperation with blood monocytes. Blood. 1994;84(5):1594–602. [PubMed] [Google Scholar]
- 4.Singh S, Soe S, Mejia JP, et al. Identification of a conserved region of Plasmodium falciparum MSP3 targeted by biologically active antibodies to improve vaccine design. J Infect Dis. 2004;190(5):1010–8. doi: 10.1086/423208. [DOI] [PubMed] [Google Scholar]
- 5.Roussilhon C, Oeuvray C, Muller-Graf C, et al. Long-term clinical protection from falciparum malaria is strongly associated with IgG3 antibodies to merozoite surface protein 3. PLoS Med. 2007;4(11):e320. doi: 10.1371/journal.pmed.0040320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polley SD, Tetteh KK, Lloyd JM, et al. Plasmodium falciparum merozoite surface protein 3 is a target of allele-specific immunity and alleles are maintained by natural selection. J Infect Dis. 2007;195(2):279–87. doi: 10.1086/509806. [DOI] [PubMed] [Google Scholar]
- 7.Hisaeda H, Saul A, Reece JJ, et al. Merozoite surface protein 3 and protection against malaria in Aotus nancymai monkeys. J Infect Dis. 2002;185(5):657–64. doi: 10.1086/339187. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho LJ, Oliveira SG, Theisen M, et al. Immunization of Saimiri sciureus monkeys with Plasmodium falciparum merozoite surface protein-3 and glutamate-rich protein suggests that protection is related to antibody levels. Scand J Immunol. 2004;59(4):363–72. doi: 10.1111/j.0300-9475.2004.01409.x. [DOI] [PubMed] [Google Scholar]
- 9.Audran R, Cachat M, Lurati F, et al. Phase I malaria vaccine trial with a long synthetic peptide derived from the merozoite surface protein 3 antigen. Infect Immun. 2005;73(12):8017–26. doi: 10.1128/IAI.73.12.8017-8026.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Druilhe P, Spertini F, Soesoe D, et al. A malaria vaccine that elicits in humans antibodies able to kill Plasmodium falciparum. PLoS Med. 2005;2(11):e344. doi: 10.1371/journal.pmed.0020344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulhern TD, Howlett GJ, Reid GE, et al. Solution structure of a polypeptide containing four heptad repeat units from a merozoite surface antigen of Plasmodium falciparum. Biochemistry. 1995;34(11):3479–91. doi: 10.1021/bi00011a001. [DOI] [PubMed] [Google Scholar]
- 12.Cohen C, Parry DA. Alpha-helical coiled coils and bundles: how to design an alpha-helical protein. Proteins. 1990;7(1):1–15. doi: 10.1002/prot.340070102. [DOI] [PubMed] [Google Scholar]
- 13.Burgess BR, Schuck P, Garboczi DN. Dissection of merozoite surface protein 3, a representative of a family of Plasmodium falciparum surface proteins, reveals an oligomeric and highly elongated molecule. J Biol Chem. 2005;280(44):37236–45. doi: 10.1074/jbc.M506753200. [DOI] [PubMed] [Google Scholar]
- 14.Mills KE, Pearce JA, Crabb BS, Cowman AF. Truncation of merozoite surface protein 3 disrupts its trafficking and that of acidic-basic repeat protein to the surface of Plasmodium falciparum merozoites. Mol Microbiol. 2002;43(6):1401–11. doi: 10.1046/j.1365-2958.2002.02834.x. [DOI] [PubMed] [Google Scholar]
- 15.Pearce JA, Hodder AN, Anders RF. The alanine-rich heptad repeats are intact in the processed form of Plasmodium falciparum MSP3. Exp Parasitol. 2004;108(3–4):186–9. doi: 10.1016/j.exppara.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 16.Shimp RL, Jr, Martin LB, Zhang Y, et al. Production and characterization of clinical grade Escherichia coli derived Plasmodium falciparum 42 kDa merozoite surface protein 1 (MSP1(42)) in the absence of an affinity tag. Protein Expr Purif. 2006;50(1):58–67. doi: 10.1016/j.pep.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 17.Tsai CW, Duggan PF, Shimp RL, Jr, Miller LH, Narum DL. Overproduction of Pichia pastoris or Plasmodium falciparum protein disulfide isomerase affects expression, folding and O-linked glycosylation of a malaria vaccine candidate expressed in P. pastoris. J Biotechnol. 2006;121(4):458–70. doi: 10.1016/j.jbiotec.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 18.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78(3):1606–19. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S, Rowe A, editors. Analytical ultracentrifugation in biochemistry and polymer science. Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 20.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 2002;11(9):2067–79. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin AJ, Prasad K, Smith PD, Lafer EM, Nossal R. Measuring the elasticity of clathrin-coated vesicles via atomic force microscopy. Biophys J. 2006;90(9):3333–44. doi: 10.1529/biophysj.105.068742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tokumasu F, Jin AJ, Feigenson GW, Dvorak JA. Nanoscopic lipid domain dynamics revealed by atomic force microscopy. Biophys J. 2003;84(4):2609–18. doi: 10.1016/S0006-3495(03)75066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou L, Miles AP, Wang J, Stowers AW. Expression of malaria transmission-blocking vaccine antigen Pfs25 in Pichia pastoris for use in human clinical trials. Vaccine. 2003;21(15):1650–7. doi: 10.1016/s0264-410x(02)00701-6. [DOI] [PubMed] [Google Scholar]
- 24.Miura K, Orcutt A, Muratova O, Miller LH, Saul A, Long CA. Development and characterization of a standardized ELISA including a reference serum on each plate to detect antibodies induced by experimental malaria vaccines. Vaccine. 2008;26(2):193–200. doi: 10.1016/j.vaccine.2007.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krogh A, Brown M, Mian IS, Sjolander K, Haussler D. Hidden Markov models in computational biology. Applications to protein modeling J Mol Biol. 1994;235(5):1501–31. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- 26.Meiler J, Baker D. Coupled prediction of protein secondary and tertiary structure. Proc Natl Acad Sci USA. 2003;100(21):12105–10. doi: 10.1073/pnas.1831973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292(2):195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 28.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 29.Tobi D, Elber R. Distance-dependent, pair potential for protein folding: results from linear optimization. Proteins. 2000;41(1):40–6. [PubMed] [Google Scholar]
- 30.Chacon P, Wriggers W. Multi-resolution contour-based fitting of macromolecular structures. J Mol Biol. 2002;317(3):375–84. doi: 10.1006/jmbi.2002.5438. [DOI] [PubMed] [Google Scholar]
- 31.McColl DJ, Anders RF. Conservation of structural motifs and antigenic diversity in the Plasmodium falciparum merozoite surface protein-3 (MSP-3) Mol Biochem Parasitol. 1997;90(1):21–31. doi: 10.1016/s0166-6851(97)00130-8. [DOI] [PubMed] [Google Scholar]
- 32.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252(5010):1162–4. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 33.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–25. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 34.Parry DA. Coiled-coils in alpha-helix-containing proteins: analysis of the residue types within the heptad repeat and the use of these data in the prediction of coiled-coils in other proteins. Biosci Rep. 1982;2(12):1017–24. doi: 10.1007/BF01122170. [DOI] [PubMed] [Google Scholar]