Abstract
EFhd2 is a novel protein conserved from C. elegans to H. sapiens. This novel protein was originally identified in cells of the immune and central nervous systems. However, it is most abundant in the central nervous system, where it has been found associated with pathological forms of the microtubule-associated protein tau. The physiological or pathological roles of EFhd2 are poorly understood. In this study, a functional and structural analysis was carried to characterize the molecular requirements for EFhd2’s calcium binding activity. The results showed that mutations of a conserved aspartate on either EF-hand motif disrupted the calcium binding activity, indicating that these motifs work in pair as a functional calcium binding domain. Furthermore, characterization of an identified single-nucleotide polymorphisms (SNP) that introduced a missense mutation indicates the importance of a conserved phenylalanine on EFhd2 calcium binding activity. Structural analysis revealed that EFhd2 is predominantly composed of alpha helix and random coil structures and that this novel protein is thermostable. EFhd2’s thermo stability depends on its N-terminus. In the absence of the N-terminus, calcium binding restored EFhd2’s thermal stability. Overall, these studies contribute to our understanding on EFhd2 functional and structural properties, and introduce it into the family of canonical EF-hand domain containing proteins.
Keywords: EFHD2, calcium binding, EF-hand domain, coil-coiled domain, tau-associated protein, secondary structure
Introduction
EFhd2 is a novel calcium binding protein conserved from invertebrates to vertebrates, suggesting that this protein may play an important physiological role that withstands the forces of evolution [1]. EFhd2 has a polyalanine motif, two EF-hand motifs and a coiled-coil domain that are highly conserved across distant taxa [1, 2]. The polyalanine motif is located at the N-terminus, while the coiled coil domain is at the C-terminus [1]. Both, polyalanine and coiled-coil, are known to promote protein-protein interactions [3]. The EF-hand motifs are known to mediate calcium binding, and these are found close to the center of EFhd2’s protein sequence [1–2, 4].
EFhd2 expression is detected in most organs, but it is mainly abundant within the brain [2]. This protein was first identified in CD8 T cells, but it has also been detected in CD4 T cells and mast cells [5–7]. In B cells, it appears to serve as a modulator of B-cell receptor (BCR) proapototic signaling [7]. Previous work showed that EFhd2 protein influences cell survival by negatively modulating the NF-kB anti-apoptotic pathway and affected calcium efflux from the endoplasmic reticulum storage [7]. In the brain, EFhd2 was found associated with the microtubule-associated protein tau in a tauopathy mouse model (JNPL3) and humans with tauopathies, such as Alzheimer’s disease [2]. However, the physiological or pathological roles that EFhd2 plays in either the nervous or immune system remains poorly understood.
EF-hand domain containing proteins mediate calcium signaling pathways in the cell [3]. These calcium-binding proteins are mainly divided in two types: calcium sensors and calcium modulators. This classification is based on the structure and function of these proteins after calcium binding [8–9]. Calmodulin is the most classical example of a calcium “sensor”. Calcium binding induces a conformational change in calmodulin that leads to the exposure of specific residues that enable it to associate with other proteins, such as the calmodulin-dependent protein kinase II [10]. Other calcium-binding proteins, such as calbindin D9K, are classical calcium “buffers” or “modulators”. Calcium binding does not induce a significant conformational change in these proteins. Members of the EF-hand calcium buffers serve as modulators of cytoplasmic calcium ions [9, 11–13]. A more recent study on EFhd2’s calcium binding activity indicated that this novel protein influences the intracellular calcium concentration upon activation of the B cell receptor in WEHI231 cells [14]. However, it is still unclear if EFhd2 belongs to either of the classes of calcium binding proteins. In this study, a functional domain analysis was performed to characterize EFhd2’s calcium binding activity and structural stability.
Materials and Methods
Bioinformatics Analyses
Protein Sequence alignment was made using NCBI sequences of EFhd2 for various organisms at http://www.ncbi.nlm.nih.gov/sites/entrez. EF-hand domain prediction was made using the SMART server at http://smart.embl-heidelberg.de/. Coiled-coil domain prediction was made using the COILS algorithm at: http://www.ch.embnet.org/software/COILSform.html [15]. Prediction of EFhd2 secondary structure from its primary sequence was made using the SCRATCH protein prediction tool at: http://scratch.proteomics.ics.uci.edu/index.html [16]. Protein structure prediction was carried out using the Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2/) [17]. Mouse EFhd2 sequence was analyzed using the “Intensive Modeling Mode” on this server. The structure shown is the one obtained at a 99.9% confidence level.
Cloning of Recombinant EFhd2 Proteins
His-tagged EFhd2 proteins
The mouse EFhd2 cDNA encoding the wild type (753bps) and the C-terminus truncated mutant (594 bps) EFhd2 protein were subcloned from pIV-53 and pIV-54 respectively (constructs synthesized by IDT DNA Technologies, not shown in Table 1) to Vector pp-80L (5Prime Cat No. 2400850) between BamHI and HindIII restriction sites resulting in pIV-56 and pIV-57. Ligation reactions were transformed in E. coli XL-1 cells using standard procedures. Colonies were screened by colony PCR. Positive colonies were further confirmed by sequencing (Sequencing & Genotyping facility (SGf) UPR-RP and MCLAB Sequencing services).
Table 1.
List of constructs used to study EFhd2.
| Plasmid | Plasmid name | Description | Protein name |
|---|---|---|---|
| pIV-35 | pPP80 | Cloning vector encompassing N-terminus 6XHistidine tag. | |
| pIV-38 | pPP80::EFhd2ΔNT | EFhd2 gene sequence (567pbs) encoding for amino acids 52 to 241 cloned in to pIV-35 between BamHI and KpnI restriction sites. Encodes for His-EFhd2 ΔNT protein. | His-EFhd2ΔNT |
| pIV-56 | pPP80::EFhd2 | EFhd2 full length (723bps) sequence cloned into pIV-35 between BamHI and HindIII restriction sites. Encodes for His-EFhd2 full length protein. | His-EFhd2 |
| pIV-57 | pPP80::EFhd2ΔCC | EFhd2 gene sequence (609 bps) encoding amino acids 1 to 198 (Δ C-terminus) cloned in to pIV-35 between BamHI and HindIII restriction sites. Encodes for His-EFhd2 ΔCC protein. | His-EFhd2ΔCC |
| pIV-58 | pPP80::EFhd2F89L | Site directed mutagenesis of pIV-56. Point mutation from Phe to Leu at position 89. | His-EFhd2F89L |
| pIV-59 | pPP80::EFhd2D141A | Site directed mutagenesis of pIV-56. Point mutation from Asp to Ala at position 141. | His-EFhd2D141A |
| pIV-60 | pPP80::EFhd2D105A | Site directed mutagenesis of pIV-56. Point mutation from Asp to Ala at position 105. | His-EFhd2D105A |
Point mutants
His EFhd2 point mutants near and in the EF-Hand domains were generated by site directed mutagenesis (Stratagene) following manufacturer’s recomendations in pIV-56 using the following primer pairs: for D105A REV 5′ GCC ATC CCT GCC GGC GGC AGC ATA CTG CTT GAA CAT CTT CTC 3′ and FWD: 5′ GAG AAG ATG TTC AAG CAG TAT GCT GCC GCC GGC AGG GAT GGC 3′; for D141A FWD: 5′CTC AAG AGT ATG ATC CAG GAG GTG GCC GAG GAT TTC GAC AGC 3′ and REV: 5′ GCT GTC GAA ATC CTC GGC CAC CTC CTG GAT CAT ACT CTT GAG 3′; for F89L FWD 5′ CCC TAC ACC GAG TTC AAG GAG TTG TCC AGG AAG CAG ATC AAA GAC 3′ and REV 5′GTC TTT GAT CTG CTT CCT GGA CAA CTC CTT GAA CTC GGT GTA GGG 3′. All mutations were confirmed by sequencing (MCLAB sequencing services). The resulting plasmids were named pIV-60, pIV-59 and pIV-58, respectively.
The EFhd2ΔNT was amplified from plasmid pIV-5 by PCR using primers FWD 5′ GCC ATA GGA TCC ATG GCC ACG GAC GAG TTG GCC 3′ and REV 5′ GCC AAT GGT ACC CTA CTT GAA CGT GGA CTG 3′. PCR were performed in a 20 μl total volume, and the reaction mixtures contained 100 ng of DNA template, 150 ng of DNA primers, 200 μM dNTPs and 0.2 μl Phusion™ DNA polymerase. PCR conditions were as follows: Initial denaturation for 4 min. at 94 °C; denaturation at 94°C for 30 seconds, annealing at 55 °C for 30 seconds, and extension at 72 °C for 45 seconds (25 cycles total). Final extension was performed at 72 °C for 7 min. PCR products were resolved in a 2% agarose gel, and 567 bps DNA fragments were isolated using Qiagen’s Gel Extraction Kit (Qiagen Cat No. 28704) according to manufacturer’s recommendations. DNA was then digested with BamHI and Kpn1. Plasmid pIV-35 (PerfectPro cis-repress Vector Set from 5 Prime (Cat No. 2400850) was also digested with BamHI and KpnI. PCR products were ligated into linearized plasmid. Ligation reactions were transformed in E. coli BL-21 cells using standard procedures. Colonies were screened by colony PCR. Positive colonies were further confirmed by sequencing (Sequencing & Genotyping facility) UPR-RP.
Recombinant Protein Purification
His tagged proteins
E. coli bacteria overexpressed EFhd2 His-tagged proteins full length, point mutants, N-terminus truncation and coiled-coil truncation after the addition of 0.4 mM IPTG and induction for 1 hr. Bacteria were collected by centrifugation, resuspended in a buffer composed of 150 mM sodium chloride, 50 mM Tris Base, 5 mM imidazole, pH 7.4, and lysed by sonication. The cytoplasmic fraction of the lysate was incubated with a His-tag affinity nickel column and proteins were eluted using a buffer composed of 150 mM sodium chloride, 50 mM sodium phosphate, 250 mM imidazole, pH 8. To assess purification, eluted proteins were resolved by SDS-PAGE and visualized by coomassie blue staining.
Radioactive Calcium 45 Binding Assays
In vitro calcium45 binding assay
Recombinant His-EFhd2WT, His-EFhd2ΔNT, His-EFhd2ΔCC and EF-hand motif mutants (D105A and D141A) were affinity purified from bacterial lysate using a nickel affinity column as described above. Proteins bound to the column beads were extensively washed and equilibrated with binding buffer (10 mM Tris-HCl pH 7.5, 100 mM KCl). The same amount of beads was used for each reaction and these were incubated for 30 min. with 1.3 μCi 45CaCl2 at room temperature. After incubation, beads were washed five times with binding buffer to remove excess 45Ca and added to 10 mL scintillation counter liquid. The radioactivity associated to these beads was measured using a Beckman Coulter scintillation counter (LS6500 Multi-Purpose Scintillation Counter). The amount of 45CaCl2 bound to the beads was corrected for the amount of protein used on each reaction.
Circular Dichroism
Circular dichroism experiments were performed in an Olis™ DSM 10 CD spectrophotometer instrument, in a temperature controlled cell using a 0.2 mm quartz cuvette. Spectra of His-EFhd2WT, His-EFhd2ΔNT, His-EFhd2ΔCC and BSA secondary structure were obtained in the far-UV region (190–260 nm). All spectra were corrected against the solvent background. To study the effect of calcium binding in the secondary structure of EFhd2WT and EFhd2 mutants, recombinant His-tagged proteins were purified from bacteria using a nickel affinity column. These proteins were extensively washed, eluted, dialyzed against ddH2O and purity was assessed by SDS PAGE and coomassie staining. Protein solutions of 1.4 mg/mL for His-EFhd2 proteins (51 μM His-EFhd2WT, 62 μM His-EFhd2ΔNT, 59μM His-EFhd2ΔCC) were prepared in ddH2O to avoid absorbance of compounds in the far UV region and prevent the formation of unwanted salts when CaCl2 was added. Protein quantification using UV at 280 nm was made using the extinction coefficient: 2980 M−1cm−1 for all three EFhd2 proteins. Molecular weights used for each protein were the following: His-EFhd2WT: 27623.0 g/mol, His-EFhd2ΔNT: 22510.5 g/mol, His-EFhd2ΔCC: 23823.8 g/mol, as estimated by the Prot Param algorithm (http://www.expasy.ch/tools/protparam.html) [18]. Protein concentrations were also confirmed using a Bradford protein quantification assay and by SDS PAGE, resolving 1uL of purified proteins with concentration standards and staining with coomassie blue. In the BSA control experiments, purified lyophilized protein (MW 69323.4 g/mol) was resuspended in ddH2O and 1mg/mL (14 μM) of protein was used plus or minus 1mM CaCl2. For the thermal denaturation experiments, a step-wise 10°C increase was made from 25°C to 75°C, incubating 5 min. on each temperature. The spectra shown in all EFhd2 figures represent an average of 10 scans at 25°C and 75°C. These spectra were not smoothed. The secondary structural elements that composed these spectra were estimated using the CDNN software [19]. The units used to analyze the data were molar residue ellipticity (in degree cm2 dmol−1). The CD data was collected in milliabsorbance units (ΔA) and transformed to ellipticity θ using the relationship [θ = ΔA × 32.982]. The molar residue ellipticity was obtained using the following relationship: [Θ] = θ/(10 × n × l × c) where θ is the ellipticity (millidegrees), n is the number of amino acid residues in the protein, l is the cuvette pathlength in cm and c is the concentration in M.
A recent study has used the CDNN program to analyze a novel calcium-binding protein’s far UV-CD [20]. In this program, the secondary structure total content should be as close as possible to 100 %. If this value deviates more than 10%, possibly the analyzed spectra were not correctly matched to the CDNN’s protein structural database (i.e. the spectrum is very different from any other in the program database) or there is an error in the data unit conversion. The total sum is not presented on Tables 2–4. Some of the total percentages were above the 10% error threshold, mostly when analyzing spectra at 75°C (i.e. His-EFhd2ΔNT, 118.4%).
Table 2.
EFhd2 secondary structure prediction in the presence and absence of calcium.
| His EFhd2WT | His EFhd2ΔNT | His EFhd2ΔCC | ||||
|---|---|---|---|---|---|---|
| Calcium | − | + | − | + | − | + |
| α- helix | 23.8±0.6 | 22.7±1.2 | 22.2±1.3 | 22.4±1.7 | 21.5±1.2 | 21.7±1.1 |
| Antiparallel β-sheet | 15.7±5.4 | 15.9±4.5 | 15.5±3.7 | 14.7±2.7 | 16.8±4.9 | 16.9±5.1 |
| Parallel β-sheet | 11.6±0.6 | 12.2±1.1 | 12.7±1.1 | 12.7±1.2 | 12.8±1.2 | 12.6±1.2 |
| β-Turn | 18.8±0.3 | 19.1±0.3 | 19.0±0.4 | 18.9±0.5 | 19.3±0.3 | 19.3±0.3 |
| Random coil | 39.1±1.7 | 40.5±2.5 | 42.2±1.8 | 42.6±1.8 | 42.0±2.4 | 41.5±2.5 |
Table 4.
Thermal stability studies and secondary structure content prediction of BSA, His-Efhd2WT and His-EFhd2 mutants at 25°C and 75°C in the presence of calcium.
| BSA | His EFhd2WT | His EFhd2ΔNT | His EFhd2ΔCC | |||||
|---|---|---|---|---|---|---|---|---|
| Temperature | 25°C | 75°C | 25°C | 75°C | 25°C | 75°C | 25°C | 75°C |
| α- helix | 46.1±3.0 | 23.2±0.4 | 22.7±1.2 | 23.2±0.4 | 22.4±1.7 | 23.6±1.5 | 21.7±1.1 | 20.5±1.1 |
| Antiparallel β-sheet | 4.1±1.2 | 14.7±4.0 | 15.9±4.5 | 14.7±4.0 | 14.7±2.7 | 13.5±2.1 | 16.9±5.1 | 18.6±6.5 |
| Parallel β-sheet | 6.8±0.9 | 12.3±0.7 | 12.2±1.1 | 12.3±0.7 | 12.7±1.2 | 12.2±0.9 | 12.6±1.2 | 13.1±1.3 |
| β-Turn | 14.2±0.4 | 18.6±0.1 | 19.1±0.3 | 18.6±0.1 | 18.9±0.5 | 18.6±0.4 | 19.3±0.3 | 19.6±0.3 |
| Random coil | 29.8±4.5 | 42.0±0.8 | 40.5±2.5 | 42.0±0.8 | 42.6±1.8 | 42.0±1.1 | 41.5±2.5 | 42.2±2.9 |
Statistical analyses
Student’s T-test
To statistically analyze and compare the samples in the radioactive calcium binding assays was used a student’s T-test, paired, with a 2 tailed distribution. Using this test was determined whether there were significant differences between two samples that came from the same two underlying populations and had the same mean. The p values returned from the T-test algorithm were considered significant if p < 0.05.
Results
The N- and C-terminus are not required for EFhd2 Calcium Binding Activity
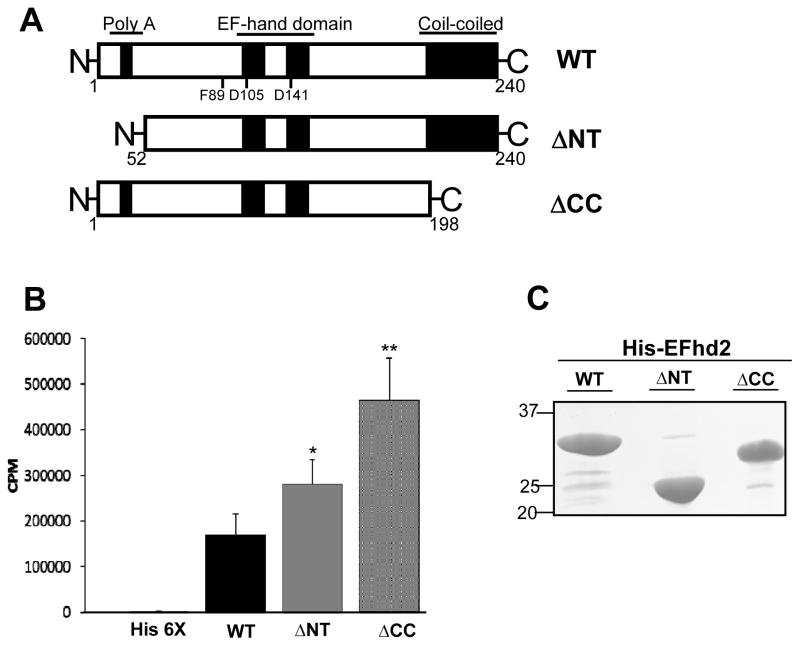
The EFhd2 protein of 240 amino acids could be divided in three principal regions: N-terminus, EF-hand domain and C-terminus (Fig. 1A). The N-terminus contains a polyalanine motif, found most frequently in nuclear proteins such as DNA-binding transcription regulators, but the specific function of this domain is still unclear [21]. EFhd2’s polyalanine motif is only conserved among mammals, from mice to humans (data not shown). In contrast to the polyalanine motif, the coiled-coil domain is located at the C-terminus is highly conserved (Fig. 1A). Coiled-coils are one of the most well-characterized protein domains and they are known to mediate protein-protein interactions [4]. Conversely, calcium-binding capacity of proteins with interaction domains could be affected while associating with other proteins or ligands because of sterical constraint or induced conformation changes. In order to determine whether the N- and C-terminus of EFhd2 have an effect on its calcium binding capacity, N- and C-terminal truncation mutants were constructed and subjected to calcium binding activity assays. As previously shown, the His-EFhd2WT protein binds calcium (Fig. 1B; p < 0.002). However, truncation of either the N- or C-terminus increased the calcium binding capacity of EFhd2 when compared to His-EFhd2WT (Fig. 1B; ΔNT, p<0.01 and ΔCC, p<0.001). Radioactive calcium binding was normalized based on the amount of recombinant protein purified (Fig. 1C). Thus, the results suggest that the N- and C-terminus may affect the accessibility of calcium ions to the EF-hand motifs or the deletion promote a conformational change that enhances calcium binding.
Fig. 1. His-EFhd2WT and truncation mutants His-EFhd2ΔNT and His-EFhd2ΔCC bind calcium in vitro.
A) Illustrated representation of EFhd2 domains under study, the N-terminus, the polyalanine domain, within the central region of the protein, the two EF-hand motifs and the coiled-coil domain at the C-terminus of the protein. The illustration is not at scale. B) Recombinant His-EFhd2WT, His-EFhd2ΔNT and His-EFhd2ΔCC were purified and incubated with radioactive 45CaCl2 to determine their calcium binding activity. Nickel-column beads were incubated with un-induced bacterial extract as negative control (His 6X). The radioactivity remaining in the beads was measured on a scintillation counter in counts per minute (CPM). C) A gel representative of the proteins used in this assay is illustrated. Protein amount was used to normalize the level of calcium binding detected. Significance was determined by a student’s T-test (two-tailed, paired). Statistical significance of * p < 0.01 and ** p < 0.001 is indicated.
The EF-hand Motifs are required for EFhd2 Calcium Binding
Based on sequence alignment, EFhd2 has two canonical EF-hand motifs (Fig. 2). EFhd2’s predicted EF-hand motifs are found from amino acids 95–123 and 131–159. The loop in the helix-loop-helix that forms the EF-hand motif (shown as EF-loop in Fig. 2) should mediate calcium binding. The canonical EF-loops on EFhd2 are: EF-loop1, 105aa DAGRDGFIDLME 116aa, and EF-loop 2, 141aa DEDFDSKLSFRE 152aa. The other two coordinating residues are provided by a bidentate carboxylate from an acidic amino acid located in the helix closest to the C-terminus of the motif. The first residue in this loop is always an aspartate that plays an essential role in the stereochemistry of the domain’s arrangement and the calcium ion positioning [3]. Mutation of this important residue has been shown to disrupt the calcium binding capacity of other EF-hand containing proteins, such as Tescalcin [22]. Additionally, mutation of the last glutamate residue (E116 and E152) on EFhd2’s EF-hand loops has shown to affect its calcium binding activity [14]. Thus, in order to corroborate EFhd2’s EF-hand motifs calcium-binding activity, a point mutation was made on the first aspartate residue of either EF-loop 1 (D105A) or EF-loop 2 (D141A), which are 100% conserved from human to nematodes (Fig. 2).
Fig. 2. EFhd2 EF-hand motifs are conserved throughout evolution.
Sequence alignment of EFhd2 calcium binding motifs (1st and 2nd EF-hand motif) was performed using sequences from various species. The aspartates (D) in bold represent the first amino acid within each EF-hand motif loop (EF-loop) that is essential for the domain’s calcium-binding activity. These aspartate residues were changed to alanine (A) by introducing a point mutation in the cDNA sequence. The phenylalanine residue (F) in the first EF-hand represents a conserved amino acid affected by the identified SNP rs12131549. The SNP introduces a missense mutation that changes the phenylalanine to a leucine (L).
EFhd2 point mutants, named HIS-EFhd2D105A and HIS-EFhd2D141A, were generated using site-directed mutagenesis. HIS-EFhd2WT was used as calcium-binding positive control, whereas nickel beads incubated with bacterial extract was used as negative control (Fig. 3A). Equal protein concentration of the purified recombinant proteins was used in the calcium-binding assay (Fig. 3B). Mutation of the first aspartate residue in either EF1 or EF2 significantly reduced the calcium binding capacity of EFhd2 (Fig. 3A). These results indicate that EFhd2 calcium binding activity depends on the integrity of both canonical EF-hand motifs.
Fig. 3. Both EF-hand domains are required for EFhd2 calcium-binding affinity.

A) Recombinant proteins HIS-EFhd2WT, HIS-EFhd2F89L, HIS-EFhd2D105A and HIS-EFhd2D141A were subjected to in vitro calcium binding assay using radioactive 45CaCl2. Nickel-column beads were incubated with un-induced bacterial extract as negative control (His 6X). The amount of radioactive calcium was measured with a scintillation counter in counts per minute (CPM). B) The amount of recombinant protein was used to normalize the level of calcium binding. Significance was determined by a student’s T-test (two-tailed, paired). Statistical significance of * p< 0.02 and **p<0.01 is indicated.
EFHD2 gene is located within chromosome 1 in humans and chromosome 4 in mice. Interestingly, the EFHD2 gene is found in chromosome 1 at the 1p36.21 locus. On the AlzGene database, two different studies showed that the chromosome region encompassing EFHD2 gene locus has been linked to (and in a third study associated to) late-onset Alzheimer’s disease [23–26]. Furthermore, chromosome 1 sequences were aligned and several single nucleotide polymorphisms (SNPs) were detected by the SNP Discovery Group, a consortium between NIH and Sanger Institute, UK (the NCBI Single Nucleotide Polymorphisms Reference Assembly at http://www.ncbi.nlm.nih.gov/SNP/index.html). Most SNPs were identified outside the coding region. However, the SNP rs12131549 was found to introduce a missense mutation on the EFHD2 gene’s coding sequence that changed a phenylalanine, at amino acid position 89, to a leucine (F89L). Previous studies using well-characterized calcium binding proteins such as Calbindin D9k and troponin C, have produced mutants with similar hydrophobic substitutions near or within the EF-hand domains to study its structural and functional consequences [8, 12, 27–28]. Thus, to evaluate whether the F89L mutation had an effect on the calcium binding activity of EFhd2, recombinant HIS-EFhd2F89L was generated. Recombinant HIS-EFhd2F89L mutant showed a noticeable reduction in radioactive calcium binding in comparison to HIS-EFhd2WT (Fig. 3A). These results suggest that the SNP that generates the point mutation F89L could compromise the calcium binding activity of EFhd2.
Random coil and alpha helix are the predominant secondary structure in EFhd2
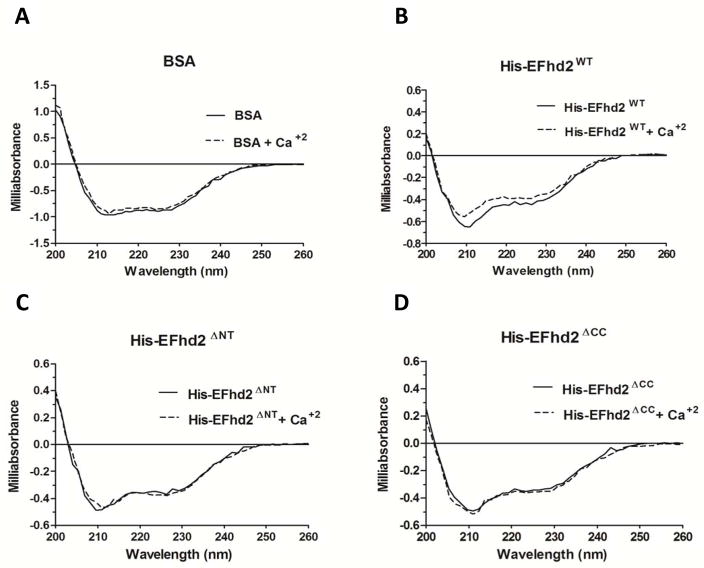
EFhd2 secondary structure is still unknown. All conserved domains in the EFhd2’s protein sequence are known to be predominantly alpha helical [3–4, 29]. These structural units comprise most of EFhd2’s protein sequence. Thus, it is very reasonable to hypothesize that EFhd2 may be a globular protein, consisting of alpha helical structures. Consistently, bioinformatics analyses using SCRATCH [16] and Phyre2 [17] protein prediction tools suggested that EFhd2 structure consisted mainly of alpha helices (Fig. 7). In order to validate the bioinformatics results, purified recombinant His-EFhd2WT was subjected to circular dichroism (CD) analyses (Fig. 4). The far-UV CD spectrum (far-UV region 190–260 nm) of purified proteins gives information about the presence of several types of secondary structure, such as alpha-helix, beta-sheet, β-turn, and random coil [30–31]. The shape of the far-UV CD spectrum for proteins with large amounts of alpha helical secondary structure is quite characteristic and can be exemplified using a well-known model protein, such as Bovine serum albumin (BSA). The spectrum of BSA shows two positive peaks at 200 nm and 222 nm, and two negative peaks at 210 nm and 230 nm, characteristics of predominantly alpha helical structure (Fig. 4A). Similar profile was observed for His-EFhd2WT (Fig. 4B), indicating that the secondary structure of this novel protein contains alpha helical elements.
Fig. 7. Structural prediction of the EFhd2 protein.
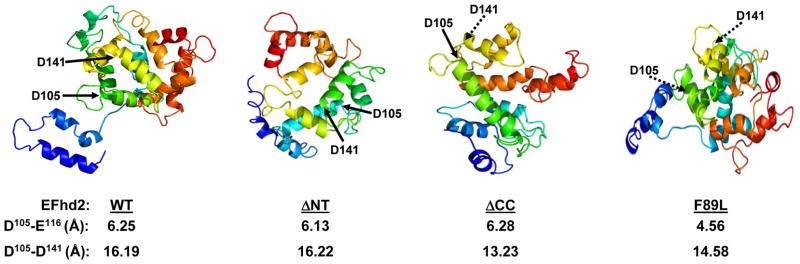
The mouse protein sequence of EFhd2 was analyzed using the protein structure prediction algorithms Phyre2. The predicted structures of EFhd2 wild-type (WT) protein, N- (ΔNT) and C-terminus (ΔCC) deletion mutants and EFhd2F89L mutant are illustrated. The position of the conserved aspartates (D105 and D141), is indicated (arrows). Dashed arrows indicate that D105 and D141 are behind the plane. The distance (Å) between amino acids in the same EF-hand loop (D105-D116) and adjacent EF-hand motifs (D105-D141) is illustrated.
Fig. 4. His-EFhd2WT, His-EFhd2ΔNT and His-EFhd2ΔCC secondary structure revealed by circular dichroism.
A–D) Circular dichroism was used to determine changes upon calcium binding in the secondary structure of recombinant His-EFhd2WT and truncation mutants at 25°C. The secondary structure of BSA (A), His-EFhd2WT (B), His-EFhd2ΔNT (C) and EFhd2ΔCC (D) was analyzed in the far-UV region (200–260 nm) without calcium (black solid line) and with 1mM of CaCl2 (dashed line). Bovine serum albumin was used as control for instrument precision and as a negative control for structural changes after addition of calcium.
To determine if the deletion of EFhd2’s N- and C-terminus induced a structural change when compared to the native protein, CD analyses were performed. Interestingly, none of these truncations generated spectra that significantly diverged from the full length protein’s secondary structure spectrum (Fig. 4C and 4D). The similarities between these mutants and the native protein spectra suggest that the absence of the N- or C-terminus fragments did not change the secondary structural organization of EFhd2, as detected by CD. Importantly, deconvolution of the CD spectra predicted that the composition of secondary structures in the EFhd2’s truncation mutant proteins did not significantly deviate from those on the EFhd2WT protein (Table 2).
Calcium binding can induce a conformational change as described for proteins that function as calcium sensors [10]. CD analyses were performed to determine if EFhd2 secondary structure changed upon calcium binding. Recombinant His-EFhd2WT and truncation mutants were subjected to CD analysis in the presence of calcium (Fig. 4, dashed line). As control, a CD spectrum of BSA in presence of calcium was also obtained. The CD spectra obtained did not show significant deviation from that of the proteins without calcium (Fig. 4, solid line). Deconvolution of the CD spectra showed no significant change in the secondary structures between untreated and treated with calcium (Table 2). The results suggest that calcium binding does not significantly induced secondary structure changes on EFhd2 protein.
EFhd2 N-terminus and Calcium Binding Confer Thermal Stability
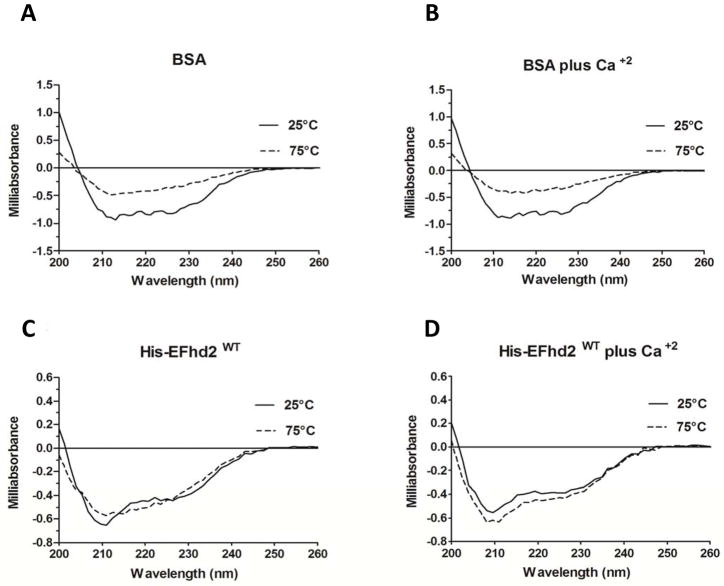
Calcium binding is known to confer thermal stability to some proteins [32–33]. Thus, the thermal stability of EFhd2’s secondary structure was determined in the presence and absence of calcium to further characterize the effects of calcium binding upon this protein. CD analyses were performed throughout a temperature gradient from 25°C to 75°C, in presence or absence of 1 mM CaCl2. The results showed that as the temperature increases to 75°C, His-EFhd2WT secondary structure did not deviated from that of the native at 25°C (Fig. 5A). The presence of calcium did not change this result, suggesting that EFhd2 is a thermostable protein (Fig. 5A and 5B). As expected, the secondary structure of BSA, used as a control in these experiments, was disrupted at 75°C, regardless of the absence (Fig. 5C) or presence (Fig. 5D) of calcium. Deconvolution of the CD spectra clearly showed a significant change in BSA’s secondary structure that was not observed for EFhd2WT protein (Tables 3 and 4). These results indicate that EFhd2 is a thermally stable protein that can withstand temperatures of at least 75°C, regardless of its calcium biding activity.
Fig. 5. Thermal stability of EFhd2WT.
A–D) Thermal stability studies on His-EFhd2WT were conducted using circular dichroism. Measurements of the protein in the far-UV region (200–260 nm presented) were taken every 10°C from 25°C to 75°C. Two representative temperature spectra at 25°C (solid line) and 75°C (dashed line) are shown. The thermal stability of BSA (used as positive denaturation control) was measured in absence (A) or presence (B) of 1mM CaCl2. His-EFhd2WT was analyzed in the absence (C) or presence (D) of 1mM CaCl2 using the same temperature range. The representative spectra at 25°C (solid line) and 75°C (dashed line) are shown.
Table 3.
Thermal stability studies and secondary structure content prediction of BSA, His-Efhd2WT and His-EFhd2 mutants at 25°C and 75°C.
| BSA | His EFhd2WT | His EFhd2ΔNT | His EFhd2ΔCC | |||||
|---|---|---|---|---|---|---|---|---|
| Temperature | 25°C | 75°C | 25°C | 75°C | 25°C | 75°C | 25°C | 75°C |
| α- helix | 47.2±3.2 | 24.4±1.1 | 23.7±0.5 | 22.3±1.3 | 22.2±1.3 | 17.8±1.6 | 21.5±1.2 | 21.0±0.9 |
| Antiparallel β-sheet | 3.8±1.3 | 11.1±4.2 | 15.7±5.4 | 18.6±8.8 | 25.5±3.7 | 20.4±6.4 | 16.8±4.9 | 17.9±6.0 |
| Parallel β-sheet | 6.6±1.0 | 11.7±0.5 | 11.6±0.6 | 11.9±0.6 | 12.7±1.1 | 15.1±1.9 | 12.8±1.2 | 12.9±1.2 |
| β-Turn | 14.0±0.4 | 18.4±0.2 | 18.8±0.3 | 19.3±0.6 | 19.0±0.4 | 20.2±0.5 | 19.3±0.3 | 19.5±0.3 |
| Random coil | 29.4±4.7 | 40.6±1.0 | 39.1±1.7 | 39.5±1.6 | 42.2±1.8 | 47.2±2.7 | 42.0±2.4 | 42.0±2.6 |
To determine whether the C- or N-terminus influenced EFhd2’s thermal stability, we performed the same experiments using the EFhd2 truncation mutants in presence or absence of 1mM CaCl2 (Fig. 6). The CD analyses showed that His-EFhd2ΔNT mutant is the most sensitive of all constructs when exposed to high temperatures of 75°C without calcium (Fig. 6A). Deconvolution of the CD spectra clearly demonstrated a significant deviation in secondary structure at 25°C versus 75°C (Table 3). Conversely, His-EFhd2ΔCC showed not significance change in the CD spectra at 25°C versus 75°C (Fig. 6C). Thus, the presence of the N-terminus confers the thermo stability detected for the EFhd2 protein. CD analysis of His-EFhd2ΔNT in presence of calcium showed no significant change in secondary structure upon temperature increase to 75°C (Fig. 6B). Interestingly, calcium binding restored the thermal stability to His-EFhd2ΔNT protein. The results suggest that EFhd2’s N-terminus confers structural stability, which in its absence is restored by calcium ions.
Fig. 6. The N-terminus and calcium binding are required for EFhd2’s thermal stability.
Recombinant His-EFhd2ΔNT and His-EFhd2ΔCC secondary structures were analyzed by circular dichroism to determine the contribution of the N- and C-terminus on EFhd2 thermal stability. Measurements were taken every 10°C from 25°C to 75°C in the far-UV region (200–260 nm). Two representative spectra, 25°C (solid line) and 75°C (dashed line) are shown. The effect of temperature on the structure of His-EFhd2ΔNT was determined in the absence (a) or presence (b) of 1mM CaCl2. The same analyses were performed for His-EFhd2ΔCC without calcium (c) or with (d) 1mM CaCl2. Deconvolution of the CD spectra obtained for His-EFhd2ΔNT and His-EFhd2ΔCC spectra are presented in Tables 3 (without calcium) and 4 (plus calcium)
EFhd2 Structural Modeling
Deletion of either the N- or C-terminus increases the calcium binding capability of EFhd2 protein. Thus, the N- and C-terminus may preclude calcium binding activity or deletion of these domains induces a conformational change that generates a more efficient calcium binding protein. Analysis of EFhd2’s protein sequence through the protein structure prediction program Phyre2 [17] modeled an extended structure at 99.9% confidence, formed predominantly of alpha helices and random coils (Fig. 7), which coincides with the CD analysis (Fig. 5). In this structure, the EF-hand motifs fold into a domain distant from the N- and C-terminus (Fig. 7, WT). Both conserved aspartate residues are located adjacent to each other (Fig. 7). The deletion of the N-terminus generates a more globular protein, maintaining the EF-hand motifs almost at the same distance as the full-length protein (Fig. 7, compare WT vs. ΔNT). The distance between the two EF-hand motifs was reduced in the C-terminus deletion mutant (Fig. 7). However, the distance between important negative charge amino acids (D105 and E116) in the same EF-hand loop did not deviate from that of the full-length protein (Fig. 7). Therefore, deletion of N- and C-terminus mutants may render the EF-hand motifs more accessible for calcium binding. In contrast, the F89L mutation induced a conformational change that reduces the distance between negative amino acids in the same EF-hand loop and between both EF-hand motifs. This conformational change may explain the HIS-EFhd2F89L mutant reduced calcium binding activity. Further experiments are required to validate these structural models.
Discussion
EFhd2’s EF-hand motifs are conserved from humans to nematodes, suggesting that EFhd2’s calcium binding activity may be essential for its physiological role. Recently, Hagen et al. (2012) showed that EFhd2 may play a role in BCR-induced calcium flux in WEHI231 cells [14]. They tested different EF-hand loops point mutants (E116A and E152A) than the one shown here, but obtained the same results [14]. As shown here, their data also indicated that both EF-hand motifs are functional and necessary for EFhd2’s calcium binding activity [14]. They concluded that one EFhd2 molecule binds two calcium ions based on equilibrium centrifugation assays [14]. Furthermore, in vivo assays using excitable ratiometric calcium indicator to monitor the BCR-induced calcium flux suggested that overexpression of EFhd2 enhances significantly the amount of free calcium detected in WEHI213 cells after stimulation with IgM antibodies [14]. Hagen et al. (2012) also showed that this phenomenon was primarily mediated by EFhd2’s N-terminus domain and the first EF-hand motif [14]. Although calcium binding activity of the N-terminus deletion mutant was not measured, the results suggested that expression of N- and C-terminal deletions failed to restore the BCR-induced calcium flux in WEHI231 cells [14]. Based on our results, N-terminal and C-terminal truncations enhanced the calcium binding capability of EFhd2. Thus, it is possible that the reduced level of free calcium detected by Hagen et al. (2012) upon stimulation of WEHI213 cells could be due to the enhanced calcium binding capability of the overexpressed EFhd2 N- and C-terminus mutant. On the other hand, it is also plausible to suggest that N- and C-terminus deletion could disrupt the association of EFhd2 with other proteins required to mediate or facilitate the BCR-induced calcium flux.
EFhd2 may function as a calcium sensor involved in signal transduction pathways induced by cellular cues or insults. In cells of the immune system, EFhd2 has been associated to signaling pathways triggered by the B-cell receptor. Additionally, its expression level in B-cells modulates the apoptotic signaling pathway [5]. EFhd2 has been suggested to function as a scaffolding protein that brings together spleen tyrosine kinase, SLP-65 and PLCγ2 to mediate BCR-induced calcium flux [7]. In the central nervous system, EFhd2 has been shown to associate with the microtubule-associated protein tau [2]. The association between EFhd2 and tau was detected in the course of tau-mediated neurodegeneration [2]. In both systems where EFhd2 has been studied, calcium ions play an important signaling role. Calcium plays a crucial role in B-cell activation and fate [34]. In addition, calcium dysregulation (i.e. increased intracellular calcium levels) has been linked to the pathophysiology of neurodegenerative disorders such as Alzheimer’s disease [35]. Thus, EFhd2 main physiological function and putative pathological role may be directly linked to calcium signaling pathways involved in regulating cellular fate. Further studies are required to decipher the physiological and pathological role of the novel calcium binding protein EFhd2.
Acknowledgments
The authors are grateful for the help provided by Dr. Julie Dutil in the identification of EFhd2 SNP. This work was supported by NIH-NINDS grant 1SC1NS066988 to IEV. YFA and ENRC were supported by NIH-NIGMS training grant 5R25GM061151. ACV was supported by NIH-NIGMS training grant 5T34GM007821.
Abbreviations
- BCR
B-cell receptor
- BSA
Bovine serum albumin
- CD
Circular dichroism
- Efhd2
EF-hand domain 2
- GST
Glutathione-S-transferase
- SNP
Single nucleotide polymorphism
Footnotes
Conflict of Interest: The authors declare that there are no conflicts of interest.
References
- 1.Dütting S, Brachs S, Mielenz D. Fraternal twins: Swiprosin-1/EFhd2 Swiprosin-2/EFhd1 two homologous EF-hand containing calcium binding adaptor proteins with distinct functions. Cell Commun Signal. 2011;9:2–14. doi: 10.1186/1478-811X-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vega IE, Traverso EE, Ferrer-Acosta Y, Matos E, Colón M, González J, Dickson D, Hutton M, Lewis J, Yen SH. A novel calcium binding protein is associated with tau proteins in tauopathy. J Neurochem. 2008;106:96–106. doi: 10.1111/j.1471-4159.2008.05339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson MR, Thulin E, Fagan PA, Forsén S, Chazin WJ. The EF-hand domain: A globally cooperative structural unit. Protein Sci. 2002;11:198–205. doi: 10.1110/ps.33302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woolfson DN. The Design of Coiled-Coiled Structures and assemblies. Adv Prot Chem Rev. 2005;70:79–111. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- 5.Avramidou A, Kroczek C, Lang C, Schuh W, Jäck HM, Mielenz D. The novel adaptor protein Swiprosin 1 enhances BCR signals and contributes to BCR-induced apoptosis. Cell Death Differ. 2007;14:1936–1947. doi: 10.1038/sj.cdd.4402206. [DOI] [PubMed] [Google Scholar]
- 6.Thylur RP, Kim YD, Kwon MS, Oh HM, Kwon HK, Kim SH, Im SH, Chun JS, Park ZY, Jun CD. Swiprosin-1 is expressed in mast cells and up-regulated through the protein kinase C beta I/eta pathway. J Cell Biochem. 2009;108:705–715. doi: 10.1002/jcb.22307. [DOI] [PubMed] [Google Scholar]
- 7.Kroczek C, Lang C, Brachs S, Grohman M, Dütting S, Schweizer A, Nitschke L, Feller SM, Jäck HM, Mielenz D. Swiprosin-1/Efhd2 Controls B Cell Receptor Signaling through the Assembly of the B Cell Receptor Syk and Phospholipase C gamma2 in Membrane Rafts. J Immunol. 2010;184:3665–3676. doi: 10.4049/jimmunol.0903642. [DOI] [PubMed] [Google Scholar]
- 8.Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem J. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- 9.Schwaller B. Cytosolic Ca2+ buffers. Cold Spring Harb Perspect Biol. 2010;2:1–21. doi: 10.1101/cshperspect.a004051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meador WE, Means AR, Quiocho FA. Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science. 1993;262:1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- 11.Ababou A, Desjarlais JR. Solvation energetics and conformational change in EF-hand proteins. Protein Sci. 2001;10:301–312. doi: 10.1110/ps.33601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skelton NJ, Kördel J, Akke M, Forsén S, Chazin WJ. Signal transduction versus buffering activity in Ca(2+)-binding proteins. Nat Struct Biol. 1994;1:239–245. doi: 10.1038/nsb0494-239. [DOI] [PubMed] [Google Scholar]
- 13.Wendt B, Hofmann T, Martin SR, Bayley P, Brodin P, Grundström T, Thulin E, Linse S, Forsén S. Eur J Biochem. 1988;175:439–445. doi: 10.1111/j.1432-1033.1988.tb14214.x. [DOI] [PubMed] [Google Scholar]
- 14.Hagen S, Brachs S, Kroczek C, Fürnrohr BG, Lang C, Mielenz D. The B cell receptor-induced calcium flux involves a calcium mediated positive feedback loop. Cell Calcium. 2012;51:411–417. doi: 10.1016/j.ceca.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 16.Cheng J, Randall AZ, Sweredoski MJ, Baldi P. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 2005;33:W72–6. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 18.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 19.Böhm G, Muhr R, Jaenicke R. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 1992;5:191–195. doi: 10.1093/protein/5.3.191. [DOI] [PubMed] [Google Scholar]
- 20.Bucholc M, Ciesielski A, Goch G, Anielska-Mazur A, Kulik A, Krzywińska E, Dobrowolska G. SNF1-related protein kinases 2 are negatively regulated by a plant-specific calcium sensor. J Biol Chem. 2011;286:3429–3441. doi: 10.1074/jbc.M110.115535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albrecht A, Mundlos S. The other trinucleotide repeat: polyalanine expansion disorders. Curr Opin Genet Dev. 2005;15:285–293. doi: 10.1016/j.gde.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Gutierrez-Ford C, Levay K, Gomes AV, Perera EM, Som T, Kim YM, Benovic JL, Berkovitz GD, Slepak VZ. Characterization of tescalcin, a novel EF-hand protein with a single Ca2+-binding site: metal-binding properties, localization in tissues and cells, and effect on calcineurin. Biochemistry. 2003;42:14553–14565. doi: 10.1021/bi034870f. [DOI] [PubMed] [Google Scholar]
- 23.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 24.Myers A, Wavrant De-Vrieze F, Holmans P, Hamshere M, Crook R, Compton D, Marshall H, Meyer D, Shears S, Booth J, Ramic D, Knowles H, Morris JC, Williams N, Norton N, Abraham R, Kehoe P, Williams H, Rudrasingham V, Rice F, Giles P, Tunstall N, Jones L, Lovestone S, Williams J, Owen MJ, Hardy J, Goate A. Full genome screen for Alzheimer disease: stage II analysis. Am J Med Genet. 2002;114:235–244. doi: 10.1002/ajmg.10183. [DOI] [PubMed] [Google Scholar]
- 25.Holmans P, Hamshere M, Hollingworth P, Rice F, Tunstall N, Jones S, Moore P, Devrieze FW, Myers A, Crook R, Compton D, Marshall H, Meyer D, Shears S, Booth J, Ramic D, Williams N, Norton N, Abraham R, Kehoe P, Williams H, Rudrasingham V, O’donovan M, Jones L, Hardy J, Goate A, Lovestone S, Owen M, Williams J. Genome screen for loci influencing age at onset rate of decline in late onset Alzheimer’s disease. Am J Med Genet B Neuropsychiatr Genet. 2005;135:24–32. doi: 10.1002/ajmg.b.30114. [DOI] [PubMed] [Google Scholar]
- 26.Hiltunen M, Mannermaa A, Thompson D, Easton D, Pirskanen M, Helisalmi S, Koivisto AM, Lehtovirta M, Ryynänen M, Soininen H. Genome-wide linkage disequilibrium mapping of late-onset Alzheimer’s disease in Finland. Neurology. 2001;57:1663–1668. doi: 10.1212/wnl.57.9.1663. [DOI] [PubMed] [Google Scholar]
- 27.Julenius K, Thulin E, Linse S, Finn BE. Hydrophobic core substitutions in Calbindin D9k: effects on stability and structure. Biochemistry. 1998;37:8915–8925. doi: 10.1021/bi972642d. [DOI] [PubMed] [Google Scholar]
- 28.Kragelund BB, Jönsson M, Bifulco G, Chazin WJ, Nilsson H, Finn BE, Linse S. Hydrophobic core substitutions in calbindin D9k: effects on Ca2+ binding and dissociation. Biochemistry. 1998;37:8926–8937. doi: 10.1021/bi9726436. [DOI] [PubMed] [Google Scholar]
- 29.Giri K, Ghosh U, Bhattacharyya NP, Basak S. Caspase 8 mediated apoptotic cell death induced by beta-sheet forming polyalanine peptides. FEBS Lett. 2003;55:380–384. doi: 10.1016/s0014-5793(03)01294-8. [DOI] [PubMed] [Google Scholar]
- 30.Kelly SM, Price NC. The use of circular dichroism in the investigation of protein structure and function. Curr Protein Pept Sci. 2000;1:349–384. doi: 10.2174/1389203003381315. [DOI] [PubMed] [Google Scholar]
- 31.Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochim, Biophys Acta. 2005;1751:119–39. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Kataeva IA, Uversky VN, Ljungdahl LG. Calcium and domain interactions contribute to the thermostability of domains of the multimodular cellobiohydrolase, CbhA, a subunit of the Clostridium thermocellum cellulosome. Biochem J. 2003;M15 372(Pt 1):151–61. doi: 10.1042/BJ20021621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wijeyesakere SJ, Gafni AA, Raghavan M. Calreticulin is a thermostable protein with distinct structural responses to different divalent cation environments. J Biol Chem. 2011;286:8771–8785. doi: 10.1074/jbc.M110.169193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scharenberg AM, Humphries LA, Rawlings DJ. Calcium signalling and cell-fate choice in B cells. Nat Rev Immunol. 2007;7:778–789. doi: 10.1038/nri2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thibault O, Gant JC, Landfield PW. Expansion of the calcium hypothesis of brain aging and Alzheimer’s disease: minding the store. Aging Cell. 2007;6:307–317. doi: 10.1111/j.1474-9726.2007.00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]