Abstract
Emerging data in myeloma and other cancers indicates that heparan sulfate proteoglycans promote tumor progression by enhancing their growth and metastasis. By acting as key regulators of cell signaling via their interactions with multiple growth and angiogenic factors, heparan sulfates mediate a shift in the microenvironment that supports the tumor as an ‘organ’ and promotes an aggressive tumor phenotype. In addition, enzymatic remodeling of heparan sulfate proteoglycans provides a mechanism for rapid, localized and dynamic modulation of proteoglycan function thereby tightly regulating activities within the tumor microenvironment. New data from animal models demonstrates that heparan sulfate or the enzymes that regulate heparan sulfate are viable targets for cancer therapy. This strategy of targeting heparan sulfate may be particularly effective for attacking cancers like myeloma where extensive genetic chaos renders them unlikely to respond well to agents that target a single signaling pathway.
Keywords: Syndecan-1, Myeloma, Growth, Metastasis, Tumor Microenvironment, Heparan Sulfate, Proteoglycan
Introduction
Heparan sulfate proteoglycans are ubiquitous, being present on the cell surface (e.g., syndecans, glypicans), within the extracellular matrix (e.g., perlecan), or as soluble molecules present in connective tissue fluids and in the circulation [1, 2]. The soluble proteoglycans are derived from molecules shed from the cell surface or from secreted proteoglycans. Heparan sulfate can bind to, and assemble, extracellular matrix proteins and can mediate both cell-cell and cell-extracellular matrix interactions [3]. Additionally, because of their unique structural features, heparan sulfates bind a wide range of bioactive molecules (e.g., growth factors, chemokines) that regulate cell behaviors important in normal and pathological processes [1, 4, 5]. This binding of heparan sulfate to growth factors is not merely passive, but can decisively regulate growth factor function. For example, binding of heparan sulfate to the chemokine IL-8 stabilizes it in an active conformation [6]. Also, in some cell types, heparan sulfate is an obligate partner within the fibroblast growth factor-2 receptor complex (FGF-2/Heparan Sulfate/FGFR1) [7, 8]. Thus, heparan sulfate proteoglycans are not simply passive structural components of cells and extracellular matrices, rather, they are multifunctional molecules that regulate cell behavior by fine tuning the function of many regulatory proteins.
These unique features place heparan sulfate proteoglycans at the intersection of signaling pathways that become disregulated during disease, including those central to cancer growth and progression. An important concept is that by targeting heparan sulfate proteoglycans, many signaling pathways can be attenuated. This provides a distinct advantage over therapies that target a single signaling pathway particularly in cancer, where multiple pathways are often disregulated.
In this review we provide a general introduction to the relationship between heparan sulfate proteoglycans and cancer (for more comprehensive reviews of proteoglycan expression and function in cancer and disease see [9–12]). Emphasis is placed on newly emerging data demonstrating that enzymatic remodeling of proteoglycans can lead to modulation of the tumor microenvironment with resulting impact on tumor behavior. In addition, we discuss the role of syndecan-1 in multiple myeloma and how this disease is being used as a model to study heparan sulfate proteoglycan regulation of cancer with the goal of developing therapies that target heparan sulfate or the enzymes that modify heparan sulfate.
Heparan sulfate proteoglycans and cancer
Heparan sulfate proteoglycans regulate tumor cell signaling, growth and metastasis
Evidence is mounting that heparan sulfate can act as an important and potent promoter of tumor growth and metastasis. For example, unlike their heparan sulfate-expressing counterparts, Chinese hamster ovary cells deficient in heparan sulfate fail to form tumors in vivo [13]. More recently, it was demonstrated that expression of syndecan-1 is required for Wnt-1-induced mammary tumorigenesis in mice [14]. These studies indicated a strong genetic interaction between syndecan-1 and Wnt-1, as do numerous studies in Drosophila, showing that heparan sulfate proteoglycans play important roles in growth factor gradients during development [15]. Similarly, glypican-3 promotes growth of hepatocellular carcinoma via Wnt signaling [16]. However, heparan sulfate proteoglycans do not always act as promoters of tumor growth. Perlecan, an extracellular matrix proteoglycan, promotes the growth of colon carcinoma and prostate cancer cells, but suppresses growth of fibro carcinoma cells [17–19]. Thus, as might be expected due to its heterogeneity in structure and function, heparan sulfate can play diverse roles in regulating tumor cell behavior.
Promotion of metastasis by heparan sulfate is generally attributed to heparan sulfate proteoglycans within the extracellular matrix. These proteoglycans are either secreted by cells or initially expressed as cell surface molecules that are proteolytically clipped from the cell surface (e.g., syndecans). Shed proteoglycans can accumulate within the extracellular matrix or diffuse into the circulation where they remain soluble and can reach high levels in some cancer patients [20, 21]. Shedding may also be elevated in some tumors such as breast and pancreatic cancers that are reported to overexpress heparan sulfate proteoglycans [22, 23]. Within the extracellular matrix, heparan sulfate facilitates metastasis by sequestering chemokines or growth factors within close proximity to migrating tumor cells [24]. In this way, haptotactic gradients are established that drive cells toward specific stimuli. Other potential mechanisms for heparan sulfate in promoting metastasis include aiding in protection and localization of proteases, and blocking interactions between tumor cells and other adhesion molecules [15, 25–27].
At least part of the sinister nature of heparan sulfate proteoglycans in cancer is due to both quantitative and qualitative changes in heparan sulfate that occur on transformed cells as compared to normal cells. The multifunctional nature of heparan sulfate proteoglycans is related in part to the extensive structural heterogeneity of heparan sulfate chains [28, 29]. Considerable effort is underway to determine how heparan sulfate fine structure is regulated and how specific sequences within heparan sulfate define its function [9, 28, 30]. In simplistic terms, fine structural heterogeneity results predominantly from variations in the density and pattern of sulfation within a heparan sulfate chain. More specifically, heparan sulfates are composed of a linear chain of 10–200 disaccharide units, each disaccharide unit being formed by N-acetyl-D-glucosamine linked to D-glucuronic acid. Structural heterogeneity stems from five potential modifications that can occur to each disaccharide: N-sulfation, 6-O sulfation to glucosamine, 3-O sulfation to glucosamine, 2-O sulfation to uronic acid, and epimerization of D-glucuronic acid to L-glucuronic acid. These five different modifications give rise to 25=32 possible structural combinations for each disaccharide along the chain. Thus, even a short heparan sulfate such as an octasaccharide can have over a million possible sequences, thereby making heparan sulfate the most information-dense biopolymer found in nature [32].
In some tumors, there are diminished amounts of heparan sulfate and reduced sulfation of the heparan sulfate chain, changes that can clearly effect heparan sulfate function [33–36]. There is mounting evidence that important differences in heparan sulfate structure exist among individual tumors, even tumors of the same type, and that these differences in structure underlie differences in heparan sulfate function. For example, as a direct consequence of having distinct heparan sulfate signatures, we discovered that two myeloma cell lines exhibit distinct cell-adhesive properties [37]. In another study, comparison of a colon-derived adenoma and carcinoma revealed substantial structural differences, including distinct modifications in the domains high in sulfation and domains flanking these regions [38]. When S115 mammary carcinoma cells are induced to undergo malignant transformation, the level of sulfation of heparan sulfate decreases due to a 40% decrease in 6-O-sulfate units, while no significant change in 2-O-sulfate or N-sulfate is detected [39]. These studies indicate that the transition of normal cells to malignancy is accompanied by specific changes in heparan sulfate structure, and these changes will not be identical among patients, even patients with the same tumor type. Given the fact that heparan sulfates are essential partners in many signaling pathways, it is likely that understanding heparan sulfate structure/function signatures of tumors will lead to a new level of patient stratification that will help guide and determine diagnosis, treatment, and prognosis.
Enzymatic remodeling of heparan sulfate can dramatically impact tumor cell behavior
Once displayed on the cell surface or secreted into the extracellular matrix, the structure of heparan sulfate proteoglycans does not remain static. Rather, they are subjected to remodeling by a number of enzymes that attack either the core proteins or heparan sulfate chains. These events can dramatically alter proteoglycan function and thus impact tumor behavior.
Human heparanase
In humans, there appears to be a single dominant functional heparanase enzyme (heparanase-1) that can act on extracellular heparan sulfate [40–43]. Importantly, active heparanase does not completely digest the heparan sulfate chains it attacks; rather, it cleaves the glycosidic bonds of heparan sulfate chains at only a few sites, producing fragments that are 10–20 sugar residues long that retain their biological activity [44–46].
Heparanase expression is rare in normal tissues, but becomes evident in many human tumors where it significantly increases both the angiogenic and metastatic potential of tumor cells [47, 48]. The mechanism by which heparanase facilitates cancer progression appears to involve several distinct activities. First, cleavage of heparan sulfate within the extracellular matrix may be required for migration of metastasizing tumor cells and for remodeling of the vasculature during angiogenesis. Second, heparanase directly promotes angiogenesis by releasing heparin-binding angiogenic growth factors such as FGF-2 and VEGF that are trapped within the extracellular matrix [4, 49]. Third, of great importance is the emerging concept that heparanase does more than simply remodel extracellular matrix. There is evidence that the fragments of heparan sulfate generated by heparanase are more biologically active than the native heparan sulfate chain from which they are derived. [50]. For example, degradation of syndecan-1 heparan sulfate with heparanase yields fragments that activate FGF-2 mitogenicity better than does intact syndecan-1 [46]. Importantly, these “activating fragments” of heparan sulfate are present in wound fluids [46] and probably cancer as well. Heparanase produced by tumor cells may also directly impact other cells within the microenvironment to enhance the growth permissive environment. It was recently demonstrated that the inactive form of heparanase (proheparanase) is secreted by cells and then can be taken back up by cells via a panel of cell surface receptors (lipoprotein related receptor protein, mannose-6-phosphate receptor, heparan sulfate proteoglycan) where it is transported to the lysosomal compartment and activated [51]. This raises the possibility that tumor cell derived heparanase could act in a paracrine fashion once taken up by other cells within the tumor microenvironment such as endothelial cells. This would provide a unique mechanism by which tumor cells can enlist surrounding cells to partner in the process of tumor progression.
Although heparanase and heparan sulfate proteoglycans have multiple effects within the tumor microenvironment, the most substantial impact of the heparanase/heparan sulfate axis in cancer and other diseases may be related to its role in angiogenesis. Heparan sulfate positively modulates VEGF/VEGFR signaling by potentiating VEGF binding to its receptors and possibly by directly binding to VEGF receptors as well [52–56]. Additional important functions of heparan sulfate include its role in binding to and establishing gradients of VEGF required to promote proper endothelial branching [57] and the ability of heparan sulfate to salvage VEGF by reactivating it following oxidative damage [58]. Modulation by heparanase of at least some of these functions of heparan sulfate apparently further stimulate the angiogenic response.
Sulfatases
The extracellular endosulfatases (Sulf-1 and Sulf-2) attack disulfated GlcA-GlcNS6S and trisulfated IdoA2S-GlcNS6S of heparan sulfate to remove 6-O sulfate moieties [59], thereby altering signaling events that control cell behavior [60, 61]. For example, expression of sulfatase-1 is required for regulation of Wnt signaling and patterning in quail embryos [60]. Evidence suggests that heparan sulfate in the absence of modulation by sulfatase binds to Wnt with high affinity and blocks interaction with Frizzled receptors. In the presence of sulfatase and following removal of specific 6-O sulfates, the heparan sulfate-Wnt interaction enters a low affinity state, thereby allowing Wnt binding to Frizzled with subsequent signal transduction [59]. In contrast, another study suggests that sulfatase-1 acts to inhibit growth of head and neck tumors by decreasing activation of cellular growth signaling pathways [62]. A second human sulfatase, sulfatase-2, was recently discovered and, like sulfatase-1, has enzymatic activity for 6-O sulfates and can differentially modulate growth factor interactions with heparan sulfate [61, 63]. What has become clear from a number of reports is that the 6-O endosulfatases act to either promote or inhibit tumor growth and this is likely due to the growth factor pathways that are regulating a specific tumor and how the sulfatase impacts those pathways [64–68].
Sheddases
Like many cell surface molecules, proteoglycans can be readily cleaved from the cell surface by proteases (sheddases). For example, the extracellular core protein of syndecan-1 is cleaved near the cell membrane resulting in release of the intact extracellular domain with the attached heparan sulfate chains [69]. This released form of the proteoglycan can remain soluble within the stromal compartment or bind to the stromal matrix via heparan sulfate interactions with molecules such as fibronectin and collagens. Shed syndecans retain biological activity and can, for example, continue to facilitate growth factor interactions or act as reservoirs by holding and concentrating effector molecules within a defined area adjacent to tumor cells [70, 71]. Thus, heparan sulfate proteoglycans shed by tumor cells can provide a mechanism for conditioning the tumor microenvironment to support growth. In addition to their biological functions, shed proteoglycans can also serve as serum markers for tumor load and prognosis suggesting their potential as markers of early detection of some cancers [72–75].
Bacterial heparinases
In addition to the endogenous enzymes described above, recent work has tested the effects of exogenous heparan sulfate degrading enzymes on tumor behavior. It was discovered that the action of bacterial heparinase I (which cleaves heparan sulfate in regions similar to those cleaved by human heparanase) yields biologically active fragments of heparan sulfate that, when injected into mice bearing melanoma or lung carcinoma tumors, promote growth and metastasis of melanoma cells [76]. In contrast, bacterial heparinase III cuts heparan sulfate in regions that differ from heparinase I, and these heparinase III-generated fragments of heparan sulfate inhibit the growth and metastasis of melanoma and lung carcinoma tumors [76, 77]. Similar inhibitory effects were seen when the enzyme itself was infused into animals bearing tumor indicating that the anti-tumor heparan sulfate fragments can be generated by enzymatic modulation of heparan sulfate in vivo. Thus, encoded within intact heparan sulfate chains are cryptic structural elements that have the power to either positively or negatively impact the behavior of cancer cells, and the action of heparan sulfate-degrading enzymes can liberate these cryptic fragments with resulting biological consequences. This presents an exciting new avenue for therapy employing either the heparan sulfate fragments produced by heparinase III, or the enzyme itself, which can liberate tumor-inhibiting fragments of heparan sulfate in vivo. Moreover, it underscores the critical regulatory role of heparan sulfate proteoglycans in cancer.
Role of syndecan-1 heparan sulfate proteoglycan in myeloma
Multiple myeloma
Multiple myeloma is the second most prevalent hematologic malignancy and accounts for over 10% of all hematologic cancers in the United States [78]. At any one time, over 50,000 people suffer from this cancer. It is estimated that in 2006 over 16,500 new cases of myeloma were diagnosed and over 11,000 died from this disease. Myeloma is a devastating cancer that resides predominantly within the bone and is marked by fatigue, intractable bone pain, renal failure and recurrent infections [79]. These effects result from high tumor burden with accompanying cytokine dysregulation, osteolytic bone disease and from the deposition in some patients of high levels of immunoglobulin light chain. The etiology of myeloma remains a mystery.
Although progress has been made in the treatment of myeloma over the last decade, the overall outlook for patients is grim. High dose chemotherapy (e.g., melphalan, doxorubicin) followed by hematopoietic stem cell support has improved 5-year survival rates, however this therapeutic approach has to be repeated on relapse and in some patients this occurs multiple times before eventual death [79]. Thus, even though median survival has been extended, the quality of life for patients is often very poor. Much of the resistance of myeloma to chemotherapy is likely due to the high level of genomic instability prevalent in most tumors [80]. This is further complicated by the host response to tumor, including survival signals provided by the stroma that help ensure tumor growth as well as eventual relapse following therapy. Over the last few years, promising biologically-based therapies such as the proteasome inhibitor Velcade, bisphosphonates and thalidomide among others, have shown success in myeloma. The effectiveness of these agents, at least in part, is due to their impact on the myeloma tumor microenvironment, particularly the bone cell components and the microvasculature [81–84]. These initial successes provide evidence that targeting the microenvironment is a ripe opportunity for the discovery of new and better drugs for myeloma.
Syndecan-1 expression and function in myeloma
Work over the last decade has provided evidence that syndecan-1 is a major regulator of the bone marrow microenvironment that supports myeloma growth and metastasis. We first reported syndecan-1 expression of the surface of myeloma cells [85], and over time, syndecan-1 (CD138) expression has become a standard marker used by many labs for identification and purification of myeloma cells [86–89]. Syndecan-1 is not expressed on B lymphocytes but becomes present at the onset of plasma cell differentiation [90]. Levels of syndecan-1 expression are high on myeloma cells but, interestingly, similar high levels of syndecan-1 are also expressed on normal plasma cells within the bone marrow [91]. This implies that the bone marrow microenvironment promotes high level syndecan-1 expression on both normal and tumor cells and that this expression may contribute to the organ selectivity of myeloma.
Several groups have taken advantage of this high level of syndecan-1 expression to target cytotoxic drugs such as doxorubicin and naytansinoid to the myeloma tumor cell surface [92, 93]. Others have taken advantage of syndecan-1 in attempts to stimulate immune-mediated killing of tumor cells [94–96]. For example, coating myeloma cells with anti-syndecan-1 antibody enhanced cross presentation of cellular antigens by dendritic cells to autologous T cells from healthy donors. This led to dendritic cell generation of myeloma-specific killer T cells [96]. Together, these studies indicate that the syndecan-1 present on the myeloma cell surface can serve as a convenient address for delivering and enhancing therapies targeted to the myeloma cell.
Because syndecan-1 is the dominant, and in most cases may be the only abundant heparan sulfate proteoglycan expressed on the surface of myeloma cells, this cancer lends itself to examining the role of a single predominant heparan sulfate proteoglycan in regulating tumor behavior. Syndecan-1 is composed of a core protein and covalently attached heparan sulfate chains. The amino acid sequence of the core protein distinguishes it from the other three members of the syndecan family of proteoglycans (syndecans-2, -3 and -4) and from other heparan sulfate proteoglycans (e.g., perlecan, glypicans) [1]. As discussed above, the heparan sulfate chains of proteoglycans serve important biological functions, but the core proteins also have functions independent of their heparan sulfate chains. The syndecan core proteins are composed of three major domains; a short cytoplasmic domain, a plasma membrane-spanning hydrophobic domain and a long extracellular domain. The cytoplasmic domains can transmit signals and they also bind to anchoring molecules including PDZ family members [97]. The extracellular domains bear the attached heparan sulfate chains but also interact with, and regulate, other cell adhesion molecules and cell surface receptors independent of their heparan sulfate chains [98]. For example, it was recently demonstrated that a domain within the syndecan-1 core protein is required for activation of αvβ3 and αvβ5 integrins [99, 100]. Thus, both heparan sulfate chains and core proteins can perform functions that impact tumor behavior.
Cell surface syndecan-1 mediates adhesion of myeloma cells to collagen, inhibits invasion through collagen gels and also can mediate myeloma cell-cell adhesion [26, 37, 85, 101, 102]. Shedding of syndecan-1 from the cell surface occurs actively via proteolytic sheddases that mediate a low-level constitutive shedding as well as an enhanced activated shedding that can result in response to external stimuli or from myeloma cell apoptosis [103–105]. Shed syndecan-1 is present in high levels in the serum of some myeloma patients and, based on clinical data, we speculated that this was an indicator of poor prognosis [20]. This hypothesis was confirmed by Siedel et al. who demonstrated that a high level of syndecan-1 in the serum is an independent predictor of poor prognosis in myeloma [75]. Subsequent studies by a number of groups have confirmed the value of serum syndecan-1 as a prognostic indicator including a recent 324 patient study demonstrating its power as an independent prognostic factor both at diagnosis and at the plateau phase of disease [106]. In addition, we found that in myeloma patients, shed syndecan-1 becomes trapped within the bone marrow extracellular matrix and within regions of marrow fibrosis [88]. Because shed syndecan-1 is composed of the entire extracellular domain, including biologically active heparan sulfate chains [20], we wondered if the shed proteoglycan actively promoted myeloma tumor growth. This notion was confirmed using an in vivo model of myeloma in which we found that soluble syndecan-1 substantially enhances growth and metastasis of myeloma cells within the bone [107]. In recent studies we also found that soluble syndecan-1 correlates with increased tumor microvessel density (unpublished data), consistent with a previous report demonstrating a positive correlation between myeloma patient microvessel density and serum syndecan-1 [108]. Interestingly, soluble syndecan-1 from tumor stroma has recently been shown to promote growth of breast cancer in vivo where it may also be responsible for enhancing angiogenesis [109, 110]. Although the sheddases responsible for shedding syndecan-1 in myeloma have not been identified, MMP-7, MT1-MMP, MMP-9 and thrombin have been shown to shed syndecan-1 from several cell types, although one study on myeloma cell lines suggests that shedding occurs via a non-MMP activity [71, 103, 111–113].
Similar to the soluble syndecan-1 findings, enzymatically active heparanase is present in the plasma of myeloma patient bone marrow and high heparanase activity correlates with high microvessel density within the tumor [114]. Enhanced expression of heparanase in a myeloma cell line increased the growth and angiogenesis of tumors formed by these cells in mice [115]. Moreover, these tumors spontaneously metastasized at a high rate from subcutaneous sites to bone, indicating that heparanase is a critical determinant of the propensity of myeloma tumors to home selectively to bone [115]. Interestingly, when heparanase expression is enhanced in a breast cancer cell line and these cells are implanted in mammary fat pads, bone turnover is stimulated even before metastatic tumor cells are detected in bone [116]. This suggests that the expression of heparanase at a distal site results in signals to the bone that initiate the osteolytic process, perhaps as a mechanism to precondition the bone microenvironment to accept metastatic cells. Recent findings also have established that expression of heparanase promotes enhanced expression and shedding of syndecan-1 [117, 118]. Since we have demonstrated that shed syndecan-1 in itself promotes myeloma growth and metastasis, the finding that heparanase promotes syndecan-1 shedding suggests that one mechanism by which heparanase augments tumor aggressiveness is via upregulated shedding of syndecan-1.
How does syndecan-1 promote myeloma growth? There is substantial evidence from a number of labs strongly supporting the notion that growth and development of myeloma is dependent on signals coming from the bone marrow microenvironment [119]. Given the enormous genetic and molecular heterogeneity of myeloma, it is clear that multiple signaling molecules contribute to the survival and progression of most myeloma tumors [120], a fact supported by analysis of the transcriptome of myeloma patients [121, 122]. Because many of these signaling or signaling-related molecules resident in the bone marrow milieu (e.g., IL-6, IL-7, IL-8, HGF, FGF-2, VEGF, EGF-family ligands) have the capacity to bind to, and be regulated by, heparan sulfate, and because there are high levels of heparan sulfate in the myeloma marrow, heparan sulfate acts as a key mediator of signaling activity in myeloma. For example, HGF, an important myeloma growth factor, binds to the heparan sulfate chains of syndecan-1. This syndecan-1/HGF interaction promotes signaling via the cMET receptor with resulting stimulation of myeloma growth [123, 124]. In addition, syndecan-1 interacts with a number of heparan sulfate binding factors (e.g., VEGF, FGF2, IL-8) that together direct the generation and maintenance of the myeloma microenvironment. One example of how syndecan-1 can modulate the myeloma microenvironment is through its binding to OPG. After binding to syndecan-1 on the myeloma cell surface, OPG, a known osteoclast inhibitor, is taken up by the cell and degraded [125, 126]. Thus, the syndecan-1 mediated uptake and degradation of OPG may contribute to the enhanced osteoclastogenesis and bone destruction seen in myeloma. Further supporting this role for syndecan-1 regulation of growth factor activity in myeloma is the recent finding that overexpression of either HSulf-1 or HSulf-2 (6-O endosulfatases) in myeloma tumor cells dramatically inhibits their growth in vivo [68]. This effect is likely due to disruption of the activity of factors regulating myeloma growth.
Syndecan-1 as a therapeutic target for myeloma
These findings lead us to conclude that a key function of syndecan-1 is to facilitate integration of cross-talk between myeloma tumor and host cells thereby helping to establish and maintain the tumor as an “organ.” Figure 1 shows a model that includes several ways (but there are likely many more) in which syndecan-1 positively impacts multiple signaling pathways within the tumor microenvironment leading to aggressive tumor growth. From this conclusion we hypothesized that targeting syndecan-1 in the myeloma tumor microenvironment will shut down or attenuate multiple pathways critical to rampant tumor growth thereby inhibiting the growth and progression of this cancer. Blocking syndecan-1 function has a higher likelihood of impacting tumor growth than do therapies targeting a single pathway. This is particularly relevant to myeloma where disease progression is driven by a high incidence of genetic chaos leading to aberrant regulation of multiple signaling pathways [80, 127]. Therefore, interfering with a single signaling pathway will not likely be an effective therapy for this cancer.
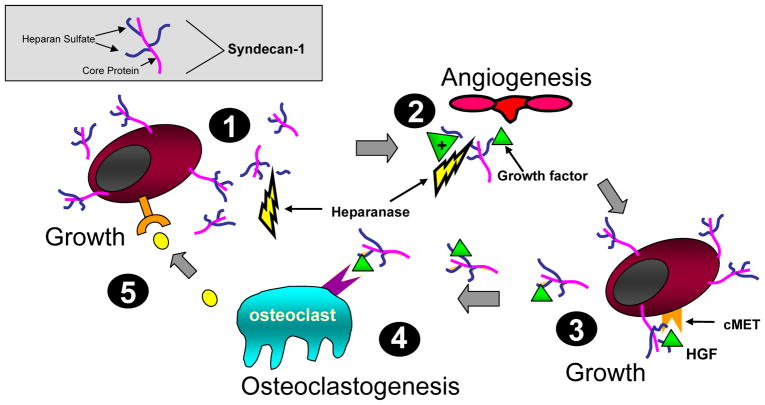
Figure 1. Model showing syndecan-1 expression and function in the myeloma microenvironment.
(1) Myeloma cells shed syndecan-1 that accumulates adjacent to cells and within bone marrow stroma (2), where it traps and concentrates growth and chemotactic factors and stimulates angiogenesis. Heparanase fuels the process by enhancing syndecan-1 shedding and releasing matrix-bound growth factors. This growth factor enriched microenvironment supports further seeding of myeloma cells in the stroma (3) that proliferate in response to signaling via the HGF/syndecan-1/cMET complex and others. These proliferating myeloma cells release factors stimulating osteoclastogenesis (4). Once activated, osteoclasts secrete additional unknown factors that further stimulate myeloma growth (5). This model depicts only a fraction of interactions between syndecan-1 and heparan sulfate in the myeloma microenvironment, it is likely that many others exist. The gray box illustrates syndecan-1 structure.
To test our hypothesis, animal models of myeloma were utilized to test three different strategies including degradation of heparan sulfate, inhibition of heparanase and knockdown of syndecan-1 expression. The results of these experiments were recently published [128]. To degrade heparan sulfate chains in vivo, bacterial heparinase III was used to treat tumors growing in the SCID-hu model. Fragments of human bone were implanted in SCID mice and freshly harvested cells from the bone marrow of myeloma patients were injected into the human bone. After it was determined that the human tumors were growing in the bone, animals were treated by daily injection of recombinant heparinase III. This treatment blocked tumor growth within the bone by over 80% as compared to controls. Dramatic growth inhibitory effects were also seen when tumor-bearing mice were injected with fragments of heparan sulfate generated by treatment of tumor cells ex vivo with bacterial heparinase III. This indicates that heparinase III liberates heparan sulfate fragments having anti-myeloma activity. To examine the effect of heparanase inhibitors we utilized chemically modified heparin that was 100% N-acetylated and 25% glycol-split. This modified form of heparin is a potent inhibitor of heparanase enzyme activity that is non-anticoagulant, binds tightly to heparanase but is not readily degraded by the enzyme [129]. Treatment of tumor-bearing animals with glycol-split heparin blocked the growth of myeloma tumors in vivo in a dose-dependent manner. When animals bearing an established subcutaneous myeloma tumor were treated with 36 mg/kg/day of glycol-split heparin (the highest concentration tested) only one of the six animals treated had tumor following 28 days of treatment. In comparison, all of the animals treated with PBS as a control had large, well-established tumors. Lastly, to directly assess the tumor promoting effect of syndecan-1 in myeloma, we produced stable knockdowns of syndecan-1 expression in the CAG myeloma cell line. Following infection with lentiviral vectors containing syndecan-1 shRNA, syndecan-1 expression was reduced by 90% as compared to normal levels of the proteoglycan. When injected into SCID mice, 70% of the animals injected with cells having low syndecan-1 expression remained tumor free after 8 weeks. In contrast, only 20% of control animals were tumor free. Taken together, these results confirm the importance of syndecan-1 in myeloma pathobiology and provide strong evidence that disruption of the normal function or amount of syndecan-1 or its heparan sulfate chains is a valid therapeutic approach for this cancer. Additional thereaputic targets for myeloma may include the endosulfatases, the core protein of syndecan-1 (to block integrin activation) or the enzymes that control heparan sulfate synthesis. Further exploration of the role of heparan sulfate porteoglycans in cancer will likely lead to novel proteoglycan-based therapies directed against myeloma and other cancer types.
Conclusions
Heparan sulfate proteoglycans modulate many growth factor signaling pathways by binding to and promoting the activity of these factors. Heparan sulfate – ligand interactions play important roles in regulating the tumor microenvironment thereby controling tumor growth, angiogenesis and metastasis. Syndecan-1 is present on both the myeloma cell surface and in the myeloma microenvironment and acts to promote aggressive tumor behavior. Therapeutic targeting of syndecan-1 or its heparan sulfate blocks myeloma growth in vivo, thus providing a ripe opportunity for development of new therapeutics directed against this deadly cancer.
Acknowledgments
This work was supported by NIH CA103054 and CA55819 to RDS. The authors apologize for the inability, due to space limitations, to reference all studies relevant to this review.
Abbreviations
- EGF
epidermal growth factor
- FGF
fibroblast growth factor
- FGFR
fibroblast growth factor receptor
- HGF
hepatocyte growth factor
- MMP
matrix metalloproteinase
- OPG
osteoprotegerin
- VEGF
vascular endothelial growth factor
- VEGFR
vascular endothelial growth factor receptor
References
- 1.Bernfield M, Gotte M, Park PW, et al. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–77. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 2.Lander AD, Selleck SB. The elusive functions of proteoglycans: in vivo veritas. J Cell Biol. 2000;148(2):227–32. doi: 10.1083/jcb.148.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woods A, Couchman JR. Syndecans: synergistic activators of cell adhesion. Trends Cell Biol. 1998;8(5):189–92. doi: 10.1016/s0962-8924(98)01244-6. [DOI] [PubMed] [Google Scholar]
- 4.Iozzo RV, San Antonio JD. Heparan sulfate proteoglycans: heavy hitters in the angiogenesis arena. J Clin Invest. 2001;108(3):349–55. doi: 10.1172/JCI13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446(7139):1030–7. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 6.Goger B, Halden Y, Rek A, et al. Different affinities of glycosaminoglycan oligosaccharides for monomeric and dimeric interleukin-8: a model for chemokine regulation at inflammatory sites. Biochemistry. 2002;41(5):1640–6. doi: 10.1021/bi011944j. [DOI] [PubMed] [Google Scholar]
- 7.Filla MS, Dam P, Rapraeger AC. The cell surface proteoglycan syndecan-1 mediates fibroblast growth factor-2 binding and activity. J Cell Physiol. 1998;174(3):310–21. doi: 10.1002/(SICI)1097-4652(199803)174:3<310::AID-JCP5>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 8.Perrimon N, Bernfield M. Cellular functions of proteoglycans--an overview. Semin Cell Dev Biol. 2001;12(2):65–7. doi: 10.1006/scdb.2000.0237. [DOI] [PubMed] [Google Scholar]
- 9.Sasisekharan R, Shriver Z, Venkataraman G, et al. Roles of heparan-sulphate glycosaminoglycans in cancer. Nat Rev Cancer. 2002;2(7):521–8. doi: 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- 10.Blackhall FH, Merry CL, Davies EJ, et al. Heparan sulfate proteoglycans and cancer. Br J Cancer. 2001;85(8):1094–8. doi: 10.1054/bjoc.2001.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fears CY, Woods A. The role of syndecans in disease and wound healing. Matrix Biol. 2006;25(7):443–56. doi: 10.1016/j.matbio.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Beauvais DM, Rapraeger AC. Syndecans in tumor cell adhesion and signaling. Reprod Biol Endocrinol. 2004;2(1):3. doi: 10.1186/1477-7827-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esko JD, Rostand KS, Weinke JL. Tumor formation dependent on proteoglycan biosynthesis. Science. 1988;241(4869):1092–6. doi: 10.1126/science.3137658. [DOI] [PubMed] [Google Scholar]
- 14.Alexander CM, Reichsman F, Hinkes MT, et al. Syndecan-1 is required for Wnt-1-induced mammary tumorigenesis in mice. Nat Genet. 2000;25(3):329–32. doi: 10.1038/77108. [DOI] [PubMed] [Google Scholar]
- 15.Perrimon N, Bernfield M. Specificities of heparan sulphate proteoglycans in developmental processes. Nature. 2000;404(6779):725–8. doi: 10.1038/35008000. [DOI] [PubMed] [Google Scholar]
- 16.Capurro MI, Xiang YY, Lobe C, et al. Glypican-3 promotes the growth of hepatocellular carcinoma by stimulating canonical Wnt signaling. Cancer Res. 2005;65(14):6245–54. doi: 10.1158/0008-5472.CAN-04-4244. [DOI] [PubMed] [Google Scholar]
- 17.Sharma B, Handler M, Eichstetter I, et al. Antisense targeting of perlecan blocks tumor growth and angiogenesis in vivo. J Clin Invest. 1998;102(8):1599–608. doi: 10.1172/JCI3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathiak M, Yenisey C, Grant DS, et al. A role for perlecan in the suppression of growth and invasion in fibrocarcinoma cells. Cancer Res. 1997;57(11):2130–6. [PubMed] [Google Scholar]
- 19.Savore C, Zhang C, Muir C, et al. Perlecan knockdown in metastatic prostate cancer cells reduces heparin-binding growth factor responses in vitro and tumor growth in vivo. Clin Exp Metastasis. 2005;22(5):377–90. doi: 10.1007/s10585-005-2339-3. [DOI] [PubMed] [Google Scholar]
- 20.Dhodapkar MV, Kelly T, Theus A, et al. Elevated levels of shed syndecan-1 correlate with tumor mass and decreased matrix metalloproteinase-9 activity in the serum of patients with multiple myeloma. Br J Hematol. 1997;99:368–71. doi: 10.1046/j.1365-2141.1997.3893203.x. [DOI] [PubMed] [Google Scholar]
- 21.Joensuu H, Anttonen A, Eriksson M, et al. Soluble syndecan-1 and serum basic fibroblast growth factor are new prognostic factors in lung cancer. Cancer Res. 2002;62(18):5210–7. [PubMed] [Google Scholar]
- 22.Kleeff J, Ishiwata T, Kumbasar A, et al. The cell-surface heparan sulfate proteoglycan glypican-1 regulates growth factor action in pancreatic carcinoma cells and is overexpressed in human pancreatic cancer. J Clin Invest. 1998;102(9):1662–73. doi: 10.1172/JCI4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuda K, Maruyama H, Guo F, et al. Glypican-1 is overexpressed in human breast cancer and modulates the mitogenic effects of multiple heparin-binding growth factors in breast cancer cells. Cancer Res. 2001;61(14):5562–9. [PubMed] [Google Scholar]
- 24.Sanderson RD. Heparan sulfate proteoglycans in invasion and metastasis. Semin Cell Dev Biol. 2001;12(2):89–98. doi: 10.1006/scdb.2000.0241. [DOI] [PubMed] [Google Scholar]
- 25.Yu WH, Woessner JF., Jr Heparan sulfate proteoglycans as extracellular docking molecules for matrilysin (matrix metalloproteinase 7) J Biol Chem. 2000;275(6):4183–91. doi: 10.1074/jbc.275.6.4183. [DOI] [PubMed] [Google Scholar]
- 26.Liebersbach BF, Sanderson RD. Expression of syndecan-1 inhibits cell invasion into type I collagen. J Biol Chem. 1994;269:20013–9. [PubMed] [Google Scholar]
- 27.Borsig L, Wong R, Feramisco J, et al. Heparin and cancer revisited: Mechanistic connections involving platelets, P-selectin, carcinoma mucins, and tumor metastasis. Proc Natl Acad Sci U S A. 2001;98(6):3352–7. doi: 10.1073/pnas.061615598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turnbull J, Powell A, Guimond S. Heparan sulfate: decoding a dynamic multifunctional cell regulator. Trends Cell Biol. 2001;11(2):75–82. doi: 10.1016/s0962-8924(00)01897-3. [DOI] [PubMed] [Google Scholar]
- 29.Selleck SB. Proteoglycans and pattern formation: sugar biochemistry meets developmental genetics. Trends Genet. 2000;16(5):206–12. doi: 10.1016/s0168-9525(00)01997-1. [DOI] [PubMed] [Google Scholar]
- 30.Gallagher JT. Heparan sulfate: growth control with a restricted sequence menu. J Clin Invest. 2001;108(3):357–61. doi: 10.1172/JCI13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindahl U, Kusche-Gullberg M, Kjellen L. Regulated diversity of heparan sulfate. J Biol Chem. 1998;273(39):24979–82. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- 32.Sasisekharan R, Venkataraman G. Heparin and heparan sulfate: biosynthesis, structure and function. Curr Opin Chem Biol. 2000;4(6):626–31. doi: 10.1016/s1367-5931(00)00145-9. [DOI] [PubMed] [Google Scholar]
- 33.Underhill CB, Keller JM. A transformation-dependent difference in the heparan sulfate associated with the cell surface. Biochem Biophys Res Commun. 1975;63:448–54. doi: 10.1016/0006-291x(75)90708-1. [DOI] [PubMed] [Google Scholar]
- 34.Winterbourne DJ, Mora PT. Altered metabolism of heparan sulphate in simian virus 40 transformed cloned mouse cells. J Biol Chem. 1978;253:5109–20. [PubMed] [Google Scholar]
- 35.David G, Van Den Berghe H. Transformed mouse mammary epithelial cells synthesize undersulfated basement membrane proteoglycan. J Biol Chem. 1983;258(12):7338–44. [PubMed] [Google Scholar]
- 36.Robinson J, Viti M, Höök M. Structure and properties of an under-sulfated heparan sulfate proteoglycan synthesized by a rat hepatoma cell line. J Cell Biol. 1984;98:946–53. doi: 10.1083/jcb.98.3.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanderson RD, Turnbull JE, Gallagher JT, et al. Fine structure of heparan sulfate regulates syndecan-1 function and cell behavior. J Biol Chem. 1994;269(18):13100–6. [PubMed] [Google Scholar]
- 38.Jayson GC, Lyon M, Paraskeva C, et al. Heparan sulfate undergoes specific structural changes during the progression from human colon adenoma to carcinoma in vitro. J Biol Chem. 1998;273(1):51–7. doi: 10.1074/jbc.273.1.51. [DOI] [PubMed] [Google Scholar]
- 39.Safaiyan F, Lindahl U, Salmivirta M. Selective reduction of 6-O-sulfation in heparan sulfate from transformed mammary epithelial cells. Eur J Biochem. 1998;252(3):576–82. doi: 10.1046/j.1432-1327.1998.2520576.x. [DOI] [PubMed] [Google Scholar]
- 40.Vlodavsky I, Friedmann Y, Elkin M, et al. Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nat Med. 1999;5(7):793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- 41.Kussie PH, Hulmes JD, Ludwig DL, et al. Cloning and functional expression of a human heparanase gene. Biochem Biophys Res Commun. 1999;261(1):183–7. doi: 10.1006/bbrc.1999.0962. [DOI] [PubMed] [Google Scholar]
- 42.Hulett MD, Freeman C, Hamdorf BJ, et al. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis. Nat Med. 1999;5(7):803–9. doi: 10.1038/10525. [DOI] [PubMed] [Google Scholar]
- 43.Toyoshima M, Nakajima M. Human heparanase. Purification, characterization, cloning, and expression. J Biol Chem. 1999;274(34):24153–60. doi: 10.1074/jbc.274.34.24153. [DOI] [PubMed] [Google Scholar]
- 44.Vlodavsky I, Friedmann Y. Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J Clin Invest. 2001;108(3):341–7. doi: 10.1172/JCI13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bitan M, Polliack A, Zecchina G, et al. Heparanase expression in human leukemias is restricted to acute myeloid leukemias. Exp Hematol. 2002;30(1):34–41. doi: 10.1016/s0301-472x(01)00766-4. [DOI] [PubMed] [Google Scholar]
- 46.Kato M, Wang H, Kainulainen V, et al. Physiological degradation converts the soluble syndecan-1 ectodomain from an inhibitor to a potent activator of FGF-2. Nat Med. 1998;4(6):691–7. doi: 10.1038/nm0698-691. [DOI] [PubMed] [Google Scholar]
- 47.Vlodavsky I, Goldshmidt O, Zcharia E, et al. Mammalian heparanase: involvement in cancer metastasis, angiogenesis and normal development. Semin Cancer Biol. 2002;12(2):121–9. doi: 10.1006/scbi.2001.0420. [DOI] [PubMed] [Google Scholar]
- 48.Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem Cell Biol. 2006;38(12):2018–39. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 49.Whitelock JM, Murdoch AD, Iozzo RV, et al. The degradation of human endothelial cell-derived perlecan and release of bound basic fibroblast growth factor by stromelysin, collagenase, plasmin, and heparanases. J Biol Chem. 1996;271(17):10079–86. doi: 10.1074/jbc.271.17.10079. [DOI] [PubMed] [Google Scholar]
- 50.Elkin M, Ilan N, Ishai-Michaeli R, et al. Heparanase as mediator of angiogenesis: mode of action. Faseb J. 2001;15(9):1661–3. doi: 10.1096/fj.00-0895fje. [DOI] [PubMed] [Google Scholar]
- 51.Vreys V, Delande N, Zhang Z, et al. Cellular uptake of mammalian heparanase precursor involves low density lipoprotein receptor-related proteins, mannose 6-phosphate receptors, and heparan sulfate proteoglycans. J Biol Chem. 2005;280(39):33141–8. doi: 10.1074/jbc.M503007200. [DOI] [PubMed] [Google Scholar]
- 52.Cohen T, Gitay-Goren H, Sharon R, et al. VEGF121, a vascular endothelial growth factor (VEGF) isoform lacking heparin binding ability, requires cell-surface heparan sulfates for efficient binding to the VEGF receptors of human melanoma cells. J Biol Chem. 1995;270(19):11322–6. doi: 10.1074/jbc.270.19.11322. [DOI] [PubMed] [Google Scholar]
- 53.Gitay-Goren H, Soker S, Vlodavsky I, et al. The binding of vascular endothelial growth factor to its receptors is dependent on cell surface-associated heparin-like molecules. J Biol Chem. 1992;267(9):6093–8. [PubMed] [Google Scholar]
- 54.Chiang MK, Flanagan JG. Interactions between the Flk-1 receptor, vascular endothelial growth factor, and cell surface proteoglycan identified with a soluble receptor reagent. Growth Factors. 1995;12(1):1–10. doi: 10.3109/08977199509003208. [DOI] [PubMed] [Google Scholar]
- 55.Dougher AM, Wasserstrom H, Torley L, et al. Identification of a heparin binding peptide on the extracellular domain of the KDR VEGF receptor. Growth Factors. 1997;14(4):257–68. doi: 10.3109/08977199709021524. [DOI] [PubMed] [Google Scholar]
- 56.Fuster MM, Wang L, Castagnola J, et al. Genetic alteration of endothelial heparan sulfate selectively inhibits tumor angiogenesis. J Cell Biol. 2007;177(3):539–49. doi: 10.1083/jcb.200610086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruhrberg C, Gerhardt H, Golding M, et al. Spatially restricted patterning cues provided by heparin-binding VEGF-A control blood vessel branching morphogenesis. Genes Dev. 2002;16(20):2684–98. doi: 10.1101/gad.242002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gengrinovitch S, Berman B, David G, et al. Glypican-1 is a VEGF165 binding proteoglycan that acts as an extracellular chaperone for VEGF165. J Biol Chem. 1999;274(16):10816–22. doi: 10.1074/jbc.274.16.10816. [DOI] [PubMed] [Google Scholar]
- 59.Ai X, Do AT, Lozynska O, et al. QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J Cell Biol. 2003;162(2):341–51. doi: 10.1083/jcb.200212083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dhoot GK, Gustafsson MK, Ai X, et al. Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science. 2001;293(5535):1663–6. doi: 10.1126/science.293.5535.1663. [DOI] [PubMed] [Google Scholar]
- 61.Morimoto-Tomita M, Uchimura K, Werb Z, et al. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J Biol Chem. 2002;277(51):49175–85. doi: 10.1074/jbc.M205131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lai JP, Chien J, Strome SE, et al. HSulf-1 modulates HGF-mediated tumor cell invasion and signaling in head and neck squamous carcinoma. Oncogene. 2004;23(7):1439–47. doi: 10.1038/sj.onc.1207258. [DOI] [PubMed] [Google Scholar]
- 63.Uchimura K, Morimoto-Tomita M, Bistrup A, et al. HSulf-2, an extracellular endoglucosamine-6-sulfatase, selectively mobilizes heparin-bound growth factors and chemokines: effects on VEGF, FGF-1, and SDF-1. BMC Biochem. 2006;7:2. doi: 10.1186/1471-2091-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morimoto-Tomita M, Uchimura K, Bistrup A, et al. Sulf-2, a proangiogenic heparan sulfate endosulfatase, is upregulated in breast cancer. Neoplasia. 2005;7(11):1001–10. doi: 10.1593/neo.05496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Narita K, Chien J, Mullany SA, et al. Loss of HSulf-1 expression enhances autocrine signaling mediated by amphiregulin in breast cancer. J Biol Chem. 2007;282(19):14413–20. doi: 10.1074/jbc.M611395200. [DOI] [PubMed] [Google Scholar]
- 66.Staub J, Chien J, Pan Y, et al. Epigenetic silencing of HSulf-1 in ovarian cancer:implications in chemoresistance. Oncogene. 2007 doi: 10.1038/sj.onc.1210300. [DOI] [PubMed] [Google Scholar]
- 67.Lai JP, Chien JR, Moser DR, et al. hSulf1 Sulfatase promotes apoptosis of hepatocellular cancer cells by decreasing heparin-binding growth factor signaling. Gastroenterology. 2004;126(1):231–48. doi: 10.1053/j.gastro.2003.09.043. [DOI] [PubMed] [Google Scholar]
- 68.Dai Y, Yang Y, MacLeod V, et al. HSulf-1 and HSulf-2 are potent inhibitors of myeloma tumor growth in vivo. J Biol Chem. 2005;280(48):40066–73. doi: 10.1074/jbc.M508136200. [DOI] [PubMed] [Google Scholar]
- 69.Jalkanen M, Rapraeger A, Saunders S, et al. Cell surface proteoglycan of mouse mammary epithelial cells is shed by cleavage of its matrix-binding ectodomain from its membrane-associated domain. J Cell Biol. 1987;105:3087–96. doi: 10.1083/jcb.105.6.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanderson RD, Yang Y, Suva LJ, et al. Heparan sulfate proteoglycans and heparanase--partners in osteolytic tumor growth and metastasis. Matrix Biol. 2004;23(6):341–52. doi: 10.1016/j.matbio.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 71.Li Q, Park PW, Wilson CL, et al. Matrilysin shedding of syndecan-1 regulates chemokine mobilization and transepithelial efflux of neutrophils in acute lung injury. Cell. 2002;111(5):635–46. doi: 10.1016/s0092-8674(02)01079-6. [DOI] [PubMed] [Google Scholar]
- 72.Capurro M, Wanless IR, Sherman M, et al. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125(1):89–97. doi: 10.1016/s0016-5085(03)00689-9. [DOI] [PubMed] [Google Scholar]
- 73.Nakatsura T, Kageshita T, Ito S, et al. Identification of glypican-3 as a novel tumor marker for melanoma. Clin Cancer Res. 2004;10(19):6612–21. doi: 10.1158/1078-0432.CCR-04-0348. [DOI] [PubMed] [Google Scholar]
- 74.Nakatsura T, Nishimura Y. Usefulness of the novel oncofetal antigen glypican-3 for diagnosis of hepatocellular carcinoma and melanoma. BioDrugs. 2005;19(2):71–7. doi: 10.2165/00063030-200519020-00001. [DOI] [PubMed] [Google Scholar]
- 75.Seidel C, Sundan A, Hjorth M, et al. Serum syndecan-1: a new independent prognostic marker in multiple myeloma. Blood. 2000;95(2):388–92. [PubMed] [Google Scholar]
- 76.Liu D, Shriver Z, Venkataraman G, et al. Tumor cell surface heparan sulfate as cryptic promoters or inhibitors of tumor growth and metastasis. Proc Natl Acad Sci U S A. 2002;99(2):568–73. doi: 10.1073/pnas.012578299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ernst S, Langer R, Cooney CL, et al. Enzymatic degradation of glycosaminoglycans. Crit Rev Biochem Mol Biol. 1995;30(5):387–444. doi: 10.3109/10409239509083490. [DOI] [PubMed] [Google Scholar]
- 78.Bergsagel D. The incidence and epidemiology of plasma cell neoplasms. Stem Cells. 1995;13(Suppl 2):1–9. [PubMed] [Google Scholar]
- 79.Barlogie B, Shaughnessy J, Epstein J, et al. Plasma Cell Myeloma. In: Lichtman MA, Beutler E, Kipps TJ, et al., editors. Williams Hematology. 7. New York: McGraw-Hill; 2006. [Google Scholar]
- 80.Kuehl WM, Bergsagel PL. Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer. 2002;2(3):175–87. doi: 10.1038/nrc746. [DOI] [PubMed] [Google Scholar]
- 81.Anderson KC. Targeted therapy of multiple myeloma based upon tumor-microenvironmental interactions. Exp Hematol. 2007;35(4 Suppl 1):155–62. doi: 10.1016/j.exphem.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 82.Mundy GR. Myeloma bone disease. Eur J Cancer. 1998;34(2):246–51. doi: 10.1016/s0959-8049(97)10133-2. [DOI] [PubMed] [Google Scholar]
- 83.Roodman GD. Pathogenesis of myeloma bone disease. Blood Cells Mol Dis. 2004;32(2):290–2. doi: 10.1016/j.bcmd.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 84.Mundy GR. Metastasis to bone: causes, consequences and therapeutic opportunities. Nat Rev Cancer. 2002;2(8):584–93. doi: 10.1038/nrc867. [DOI] [PubMed] [Google Scholar]
- 85.Ridley RC, Xiao H, Hata H, et al. Expression of syndecan regulates human myeloma plasma cell adhesion to type I collagen. Blood. 1993;81(3):767–74. [PubMed] [Google Scholar]
- 86.Wijdenes J, Vooijs WC, Clement C, et al. A plasmocyte sleective monoclonal antibody (B-B4) recognizes syndecan-1. British J Haematol. 1996;94(2):318–23. doi: 10.1046/j.1365-2141.1996.d01-1811.x. [DOI] [PubMed] [Google Scholar]
- 87.Chilosi M, Adami F, Lestani M, et al. CD138/syndecan-1: a useful immunohistochemical marker of normal and neoplastic plasma cells on routine trephine bone marrow biopsies. Mod Pathol. 1999;12(12):1101–6. [PubMed] [Google Scholar]
- 88.Bayer-Garner IB, Sanderson RD, Dhodapkar MV, et al. Syndecan-1 (CD138) immunoreactivity in bone marrow biopsies of multiple myeloma: shed syndecan-1 accumulates in fibrotic regions. Mod Pathol. 2001;14(10):1052–8. doi: 10.1038/modpathol.3880435. [DOI] [PubMed] [Google Scholar]
- 89.Witzig TE, Kimlinger T, Stenson M, et al. Syndecan-1 expression on malignant cells from the blood and marrow of patients with plasma cell proliferative disorders and B-cell chronic lymphocytic leukemia. Leuk Lymphoma. 1998;31(1–2):167–75. doi: 10.3109/10428199809057596. [DOI] [PubMed] [Google Scholar]
- 90.Sanderson RD, Lalor P, Bernfield M. B lymphocytes express and lose syndecan at specific stages of differentiation. Cell Regulation. 1989;1(1):27–35. doi: 10.1091/mbc.1.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhan F, Tian E, Bumm K, et al. Gene expression profiling of human plasma cell differentiation and classification of multiple myeloma based on similarities to distinct stages of late-stage B-cell development. Blood. 2003;101(3):1128–40. doi: 10.1182/blood-2002-06-1737. [DOI] [PubMed] [Google Scholar]
- 92.Tassone P, Goldmacher VS, Neri P, et al. Cytotoxic activity of the maytansinoid immunoconjugate B-B4-DM1 against CD138+ multiple myeloma cells. Blood. 2004;104(12):3688–96. doi: 10.1182/blood-2004-03-0963. [DOI] [PubMed] [Google Scholar]
- 93.Post J, Vooijs WC, Bast BJ, et al. Efficacy of an anti-CD138 immunotoxin and doxorubicin on drug-resistant and drug-sensitive myeloma cells. Int J Cancer. 1999;83(4):571–6. doi: 10.1002/(sici)1097-0215(19991112)83:4<571::aid-ijc21>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 94.Ragnarsson L, Stromberg T, Wijdenes J, et al. Multiple myeloma cells are killed by syndecan-1-directed superantigen-activated T cells. Cancer Immunol Immunother. 2001;50(7):382–90. doi: 10.1007/s002620100211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.von Strandmann EP, Hansen HP, Reiners KS, et al. A novel bispecific protein (ULBP2-BB4) targeting the NKG2D receptor on natural killer (NK) cells and CD138 activates NK cells and has potent antitumor activity against human multiple myeloma in vitro and in vivo. Blood. 2006;107(5):1955–62. doi: 10.1182/blood-2005-05-2177. [DOI] [PubMed] [Google Scholar]
- 96.Dhodapkar KM, Krasovsky J, Williamson B, et al. Antitumor monoclonal antibodies enhance cross-presentation of cellular antigens and the generation of myeloma-specific killer T cells by dendritic cells. J Exp Med. 2002;195(1):125–33. doi: 10.1084/jem.20011097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zimmermann P, Tomatis D, Rosas M, et al. Characterization of syntenin, a syndecan-binding PDZ protein, as a component of cell adhesion sites and microfilaments. Mol Biol Cell. 2001;12(2):339–50. doi: 10.1091/mbc.12.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Couchman JR, Chen L, Woods A. Syndecans and cell adhesion. Int Rev Cytol. 2001;207:113–50. doi: 10.1016/s0074-7696(01)07004-8. [DOI] [PubMed] [Google Scholar]
- 99.Beauvais DM, Burbach BJ, Rapraeger AC. The syndecan-1 ectodomain regulates alpha V beta 3 integrin activity in human mammary carcinoma cells. J Cell Biol. 2004;167(1):171–81. doi: 10.1083/jcb.200404171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McQuade KJ, Beauvais DM, Burbach BJ, et al. Syndecan-1 regulates alpha V beta5 integrin activity in B82L fibroblasts. J Cell Sci. 2006;119(Pt 12):2445–56. doi: 10.1242/jcs.02970. [DOI] [PubMed] [Google Scholar]
- 101.Sanderson RD, Hinkes MT, Bernfield M. Syndecan-1, a cell-surface proteoglycan, changes in size and abundance when keratinocytes stratify. Journal of Investigative Dermatology. 1992;99(4):390–6. doi: 10.1111/1523-1747.ep12616103. [DOI] [PubMed] [Google Scholar]
- 102.Stanley MJ, Liebersbach BF, Liu W, et al. Heparan sulfate-mediated cell aggregation: Syndecans-1 and -4 mediate intercellular adhesion following their transfection into human B lymphoid cells. J Biol Chem. 1995;270:5077–83. doi: 10.1074/jbc.270.10.5077. [DOI] [PubMed] [Google Scholar]
- 103.Subramanian SV, Fitzgerald ML, Bernfield M. Regulated shedding of syndecan-1 and -4 ectodomains by thrombin and growth factor receptor activation. J Biol Chem. 1997;272(23):14713–20. doi: 10.1074/jbc.272.23.14713. [DOI] [PubMed] [Google Scholar]
- 104.Fitzgerald ML, Wang Z, Park PW, et al. Shedding of syndecan-1 and -4 ectodomains is regulated by multiple signaling pathways and mediated by a TIMP-3-sensitive metalloproteinase. J Cell Biol. 2000;148(4):811–24. doi: 10.1083/jcb.148.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jourdan M, Ferlin M, Legouffe E, et al. The myeloma cell antigen syndecan-1 is lost by apoptotic myeloma cells. Br J Haematol. 1998;100(4):637–46. doi: 10.1046/j.1365-2141.1998.00623.x. [DOI] [PubMed] [Google Scholar]
- 106.Lovell R, Dunn JA, Begum G, et al. Soluble syndecan-1 level at diagnosis is an independent prognostic factor in multiple myeloma and the extent of fall from diagnosis to plateau predicts for overall survival. Br J Haematol. 2005;130(4):542–8. doi: 10.1111/j.1365-2141.2005.05647.x. [DOI] [PubMed] [Google Scholar]
- 107.Yang Y, Yaccoby S, Liu W, et al. Soluble syndecan-1 promotes growth of myeloma tumors in vivo. Blood. 2002;100(2):610–7. doi: 10.1182/blood.v100.2.610. [DOI] [PubMed] [Google Scholar]
- 108.Andersen NF, Standal T, Nielsen JL, et al. Syndecan-1 and angiogenic cytokines in multiple myeloma: correlation with bone marrow angiogenesis and survival. Br J Haematol. 2005;128(2):210–7. doi: 10.1111/j.1365-2141.2004.05299.x. [DOI] [PubMed] [Google Scholar]
- 109.Su G, Blaine SA, Qiao D, et al. Shedding of syndecan-1 by stromal fibroblasts stimulates human breast cancer cell proliferation via FGF2 activation. J Biol Chem. 2007;282(20):14906–15. doi: 10.1074/jbc.M611739200. [DOI] [PubMed] [Google Scholar]
- 110.Maeda T, Desouky J, Friedl A. Syndecan-1 expression by stromal fibroblasts promotes breast carcinoma growth in vivo and stimulates tumor angiogenesis. Oncogene. 2006;25(9):1408–12. doi: 10.1038/sj.onc.1209168. [DOI] [PubMed] [Google Scholar]
- 111.Endo K, Takino T, Miyamori H, et al. Cleavage of syndecan-1 by membrane-type matrix metalloproteinase-1 stimulates cell migration. J Biol Chem. 2003;278:40764–70. doi: 10.1074/jbc.M306736200. [DOI] [PubMed] [Google Scholar]
- 112.Brule S, Charnaux N, Sutton A, et al. The shedding of syndecan-4 and syndecan-1 from HeLa cells and human primary macrophages is accelerated by SDF-1/CXCL12 and mediated by the matrix metalloproteinase-9. Glycobiology. 2006;16(6):488–501. doi: 10.1093/glycob/cwj098. [DOI] [PubMed] [Google Scholar]
- 113.Holen I, Drury NL, Hargreaves PG, et al. Evidence of a role for a non-matrix-type metalloproteinase activity in the shedding of syndecan-1 from human myeloma cells. Br J Haematol. 2001;114(2):414–21. doi: 10.1046/j.1365-2141.2001.02963.x. [DOI] [PubMed] [Google Scholar]
- 114.Kelly T, Miao HQ, Yang Y, et al. High heparanase activity in multiple myeloma is associated with elevated microvessel density. Cancer Res. 2003;63(24):8749–56. [PubMed] [Google Scholar]
- 115.Yang Y, Macleod V, Bendre M, et al. Heparanase promotes the spontaneous metastasis of myeloma cells to bone. Blood. 2005;105(3):1303–9. doi: 10.1182/blood-2004-06-2141. [DOI] [PubMed] [Google Scholar]
- 116.Kelly T, Suva LJ, Huang Y, et al. Expression of heparanase by primary breast tumors promotes bone resorption in the absence of detectable bone metastases. Cancer Res. 2005;65(13):5778–84. doi: 10.1158/0008-5472.CAN-05-0749. [DOI] [PubMed] [Google Scholar]
- 117.Yang Y, Macleod V, Miao HQ, et al. Heparanase enhances syndecan-1 shedding: A novel mechanism for stimulation of tumor growth and metastasis. J Biol Chem. 2007;282(18):13326–33. doi: 10.1074/jbc.M611259200. [DOI] [PubMed] [Google Scholar]
- 118.Mahtouk K, Hose D, Raynaud P, et al. Heparanase influences expression and shedding of syndecan-1, and its expression by the bone marrow environment is a bad prognostic factor in multiple myeloma. Blood. 2007 doi: 10.1182/blood-2006-08-043232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Roodman GD. Role of the bone marrow microenvironment in multiple myeloma. J Bone Miner Res. 2002;17(11):1921–5. doi: 10.1359/jbmr.2002.17.11.1921. [DOI] [PubMed] [Google Scholar]
- 120.Anderson KC, Dalton WS. Synopsis of a research roundtable presented on cell signaling in myeloma: regulation of growth and apoptosis--opportunities for new drug discovery. Mol Cancer Ther. 2002;1(14):1361–5. [PubMed] [Google Scholar]
- 121.Zhan F, Hardin J, Kordsmeier B, et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99(5):1745–57. doi: 10.1182/blood.v99.5.1745. [DOI] [PubMed] [Google Scholar]
- 122.Tarte K, Zhan F, De Vos J, et al. Gene expression profiling of plasma cells and plasmablasts: toward a better understanding of the late stages of B-cell differentiation. Blood. 2003;102(2):592–600. doi: 10.1182/blood-2002-10-3161. [DOI] [PubMed] [Google Scholar]
- 123.Derksen PW, Keehnen RM, Evers LM, et al. Cell surface proteoglycan syndecan-1 mediates hepatocyte growth factor binding and promotes Met signaling in multiple myeloma. Blood. 2002;99(4):1405–10. doi: 10.1182/blood.v99.4.1405. [DOI] [PubMed] [Google Scholar]
- 124.Seidel C, Borset M, Hjertner O, et al. High levels of soluble syndecan-1 in myeloma-derived bone marrow: modulation of hepatocyte growth factor activity. Blood. 2000;96(9):3139–46. [PubMed] [Google Scholar]
- 125.Standal T, Seidel C, Hjertner O, et al. Osteoprotegerin is bound, internalized, and degraded by multiple myeloma cells. Blood. 2002;100(8):3002–7. doi: 10.1182/blood-2002-04-1190. [DOI] [PubMed] [Google Scholar]
- 126.Lacey DL, Timms E, Tan HL, et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93(2):165–76. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 127.Zhan F, Huang Y, Colla S, et al. The molecular classification of multiple myeloma. Blood. 2006;108(6):2020–8. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yang Y, MacLeod V, Dai Y, et al. The syndecan-1 heparan sulfate proteoglycan is a viable target for myeloma therapy. Blood. 2007 doi: 10.1182/blood-2007-04-082495. prepublished online May 29, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Naggi A, Casu B, Perez M, et al. Modulation of the heparanase-inhibiting activity of heparin through selective desulfation, graded N-acetylation, and glycol splitting. J Biol Chem. 2005;280(13):12103–13. doi: 10.1074/jbc.M414217200. [DOI] [PubMed] [Google Scholar]