Abstract
Blood malignancies can be cured with hematopoietic cell transplantation from HLA-matched unrelated donors, however, acute graft-versus-host disease (GVHD) affects up to 80% of patients and contributes to increased mortality. To test the hypothesis that undetected patient-donor differences for non-HLA genetic variation within the MHC could confer risks after HLA-matched transplantation, we conducted a discovery-validation study of 4205 transplants for 1120 MHC-region single nucleotide polymorphisms (SNPs). Two SNPs were identified as markers for disease-free survival and acute GVHD. Among patients with two or more HLA-matched unrelated donors identified on their search, SNP genotyping of patients and their potential donors demonstrated that most patients have a choice of SNP-matched donors. In conclusion, the success of HLA-matched unrelated donor hematopoietic cell transplantation depends on non-HLA MHC region genetic variation. Prospective SNP screening and matching provides an approach for lowering risks to patients.
INTRODUCTION
The MHC on chromosome 6p21.3 is the most gene-dense region of the human genome, encoding an estimated 300 loci of which 10–20% have immune-related function (1). The best characterized loci in the MHC are the classical HLA-A, -C, -B, -DR, -DQ and -DP genes that encode antigens which stimulate B and T cell responses (2). Furthermore, HLA-C (and some HLA-B and A) antigens participate in the innate immune response by serving as ligands for inhibitory natural killer immunoglobulin receptors (2). Over 100 diseases are associated with HLA antigens including diabetes, multiple sclerosis, rheumatoid arthritis and asthma (1). HLA antigens influence host susceptibility to infection including tuberculosis, malaria, and HIV-AIDS (1, 3, 4).
The classical HLA genes define the histocompatibility barrier by governing acceptance of transplanted tissues (5). In unrelated donor hematopoietic cell transplantation, HLA matching of the patient and donor is undertaken to lower the risks of graft rejection and graft-versus-host disease (GVHD). GVHD is the leading cause of early complications and death after unrelated donor hematopoietic cell transplantation, and arises when the newly engrafted donor immune system recognizes mismatched patient HLA antigens that are not present in the donor (6, 7). Even with HLA matching, life-threatening GVHD is a major cause of transplant failure (7, 8). We hypothesized that risks after HLA-matched unrelated donor hematopoietic cell transplantation might be conferred by undetected genetic variation within the MHC of patients and donors. We retrospectively surveyed a 4.9 megabase region of the MHC in patients and their HLA-matched unrelated donors to identify non-HLA genetic variants associated with GVHD and survival.
RESULTS
SNP minor allele frequencies and matching
The minor allele frequencies of SNP markers within the human MHC have been well characterized for several world populations (10). To determine whether patients and their HLA-matched unrelated transplant donors have similar MHC SNP allele frequencies as those previously described, minor allele frequencies were determined and found to exceed 5% for 1076 of the 1120 SNPs among Caucasian individuals in the discovery cohort (Fig. 1, Table 1, table S1), consistent with previous observations (10). The MHC is characterized by long-range positive linkage disequilibrium of genetic markers (11). Since the unrelated donors were originally selected for transplantation based on allele identity at each of the HLA-A, C, B, DRB1 and DQB1 loci, and the identification of putative functional variants using SNP markers could be signaled through patient-donor SNP matching or mismatching, the frequency of patient-donor SNP (mis)matching was determined for each transplant pair at each SNP position. The highest frequencies of patient-donor SNP matching were observed for SNPs residing 50 kilobases (kb) 5′ and 3′ of HLA-A, -C, -B, -DRB1 and -DQB1 loci, reflecting patient-donor HLA allele matching (Fig. 2A). In contrast, 17% of pairs were HLA-DPB1-matched and the average SNP match rate was 60% for SNPs residing 50 kb 5′ or 3′ of HLA-DPB1 (table S1).
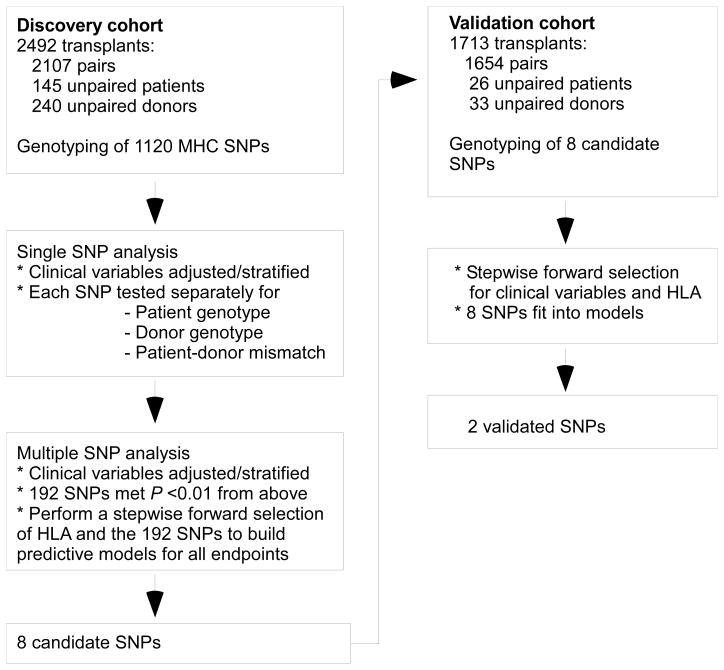
Fig. 1.
Study design. The study population consisted of a total of 4205 patients transplanted from HLA-A, -C, -B, -DRB1, -DQB1 allele-matched unrelated donors. The 2492 transplants in the discovery cohort included 2107 patient-donor pairs and 385 unpaired patients or donors. The 1713 transplants in the validation cohort included 1654 patient-donor pairs and 59 unpaired patients or donors. Transplants for whom only the patient or only the donor sample was available were informative for testing associations of patient SNP genotype or donor SNP genotype to clinical outcome, but not for analyses of patient-donor SNP mismatching. Samples in the discovery cohort were genotyped for 1120 SNPs residing within the MHC. Candidate SNPs were genotyped in the independent validation cohort.
Table 1.
Characteristics of the study population.
| Characteristic | Discovery cohort (n = 2492 transplants) | Validation cohort (n = 1713 transplants) |
|---|---|---|
| Age, yrs - n (%) | ||
| Patient | ||
| 0–19 | 408 (16%) | 267 (16%) |
| 20–39 | 896 (36%) | 472 (28%) |
| 40 and older | 1188 (48%) | 974 (57%) |
| Donor | ||
| 18–19 | 23 (<1%) | 40 (<1%) |
| 20–39 | 1588 (64%) | 1182 (69%) |
| 40 and older | 808 (32%) | 442 (26%) |
| Unknown | 73 (3%) | 49 (3%) |
| Year of transplantation - n (%) | ||
| 1986–1994 | 374 (15%) | 83 (5%) |
| 1995–1999 | 628 (25%) | 164 (10%) |
| 2000–2003 | 786 (32%) | 168 (10%) |
| 2004–2008 | 704 (28%) | 1298 (76%) |
| Patient-donor sex - n (%) | ||
| Male-male | 999 (40%) | 694 (41%) |
| Male-female | 594 (24%) | 464 (27%) |
| Female-male | 444 (18%) | 264 (15%) |
| Female-female | 451 (18%) | 286 (17%) |
| Unknown | 4 (<1%) | 5 (<1%) |
| Disease - n (%)/early, intermediate, late or advanced, other or unknown - n* | ||
| Acute myeloid leukemia | 878 (35%)/318, 247, 309, 4 | 749 (44%)/317, 177, 248, 7 |
| Acute lymphoblastic leukemia | 545 (22%)/180, 245, 119, 1 | 311 (18%)/114, 134, 60, 3 |
| Chronic myeloid leukemia | 808 (32%)/567, 207, 34, 0 | 294 (17%)/175, 94, 23, 2 |
| Myelodysplastic syndrome | 261 (10%)/55, 0, 92, 114 | 359 (21%)/64, 0, 148, 147 |
| Patient-donor serologic status for cytomegalovirus - n (%) | ||
| Negative-negative | 904 (36%) | 538 (31%) |
| Negative-positive | 349 (14%) | 197 (12%) |
| Positive-negative | 746 (30%) | 622 (36%) |
| Positive-positive | 439 (18%) | 274 (16%) |
| Unknown | 54 (2%) | 82 (5%) |
| Transplant type - n (%)† | ||
| Myeloablative | 2090 (84%) | 1249 (73%) |
| Reduced-intensity/non-myeloablative | 390 (16%) | 443 (26%) |
| Unknown | 12 (<1%) | 21 (1%) |
| Source of cells - n (%) | ||
| Bone marrow | 1672 (67%) | 742 (43%) |
| Peripheral blood stem cells | 820 (33%) | 971 (57%) |
| GVHD prophylaxis n (%) | ||
| Cyclosporine with or without other agents | 1371 (55%) | 507 (30%) |
| Tacrolimus with or without other agents | 842 (34%) | 860 (50%) |
| T-cell depletion | 216 (9%) | 122 (7%) |
| Other combinations | 56 (2%) | 216 (13%) |
| Missing | 7 (<1%) | 8 (<1%) |
| Karnofsky performance score (%) - n (%) | ||
| 10–80 | 543 (22%) | 498 (29%) |
| 90–100 | 1584 (64%) | 1017 (59%) |
| Missing | 365 (15%) | 198 (12%) |
| Patient-donor HLA-DPB1 - n (%) | ||
| Matched | 281 (11%) | 62 (4%) |
| GVH mismatch | 162 (7%) | 50 (3%) |
| HVG mismatch | 166 (7%) | 54 (3%) |
| Bidirectional mismatch | 956 (38%) | 263 (15%) |
| Missing | 927 (37%) | 1284 (75%) |
| Patient ethnicity | ||
| Hispanic | 26 (1%) | 76 (4) |
| Non-Hispanic‡ | ||
| Caucasian | 2325 (93%) | 1584 (92%) |
| African American | 53 (2%) | 10 (<1%) |
| Asian/Pacific Islander | 47 (2%) | 14 (<1%) |
| Native American | 3 (<1%) | 4 (<1%) |
| Other | 29 (1%) | 6 (<1%) |
| Unknown | 9 (<1%) | 19 (1%) |
| Donor ethnicity | ||
| Hispanic | 31 (1%) | 53 (3%) |
| Non-Hispanic‡ | ||
| Caucasian | 1983 (80%) | 1468 (86%) |
| African American | 43 (2%) | 11 (<1%) |
| Asian/Pacific Islander | 75 (3%) | 10 (<1%) |
| Native American | 19 (<1%) | 5 (<1%) |
| Other | 57 (2%) | 46 (3%) |
| Unknown | 284 (11%) | 120 (7%) |
Human leukocyte antigen (HLA); total body irradiation (TBI); graft-versus-host disease (GVHD).
Disease status prior to transplant is categorized as early (first complete remission [CR] of acute myeloid leukemia [AML] or acute lymphoblastic leukemia [ALL], first chronic phase [CP] of CML, refractory anemia [RA] or refractory anemia with ring sideroblasts [RARS] of myelodysplastic syndrome [MDS]); intermediate (second or higher CR of AML or ALL, second or higher CP or accelerated phase [AP] of CML); late or advanced (primary induction failure or first or higher relapse of AML or ALL, blast phase (or blast crisis) of CML, MDS RA with excess blasts or excess blasts in transformation); other (mainly unnamed MDS) or unknown.
Myeloablative conditioning regimens: cyclophosphamide (Cy) with total body irradiation (TBI) given as a single dose greater than 500 centigrey (cGY) or as fractionated TBI greater than 800 cGY total; CY/etoposide (VP16)/TBI; busulfan (BU)/CY; TBI 500 cGY or greater single dose; TBI 800 cGY or greater fractionated; melphalan greater than 150 mg/m2; BU greater than 9 mg/kg; BU/melphalan. Reduced intensity regimens: TBI less than 500 cGY single dose; TBI less than 800 cGY fractionated; melphalan 150 mg/m2 or less; BU 9 mg/kg or less; carmustine (BCNU)/VP16/cytarabine/melphalan (BEAM); CY/BCNU/VP16 (CBV); VP16/CY. Non-myeloblative regimens: TBI 200 cGY; fludarabine (FLU)/TBI 200 cGY; FLU/CY; FLU/cytosine arabinoside (ARA-C). For the discovery cohort, regimens contained TBI for 1524/2086 (73%), 22/219(10%), and 131/163 (80%) patients conditioned with ablative, reduced intensity, and non-myeloablative regimens, respectively. For the validation cohort, regimens contained TBI for 721/1249 (58%), 39/335 (12%), and 85/105 (81%) patients conditioned with ablative, reduced intensity, and non-myeloablative regimens, respectively.
Definitions follow the US Office of Management and Budget classification (9).
Fig. 2.
MHC region SNPs. (A) Map of the MHC encoding classical HLA-A, -C, -B, -DRB1, -DQB1 and -DPB1 genes and the frequency of patient-donor pairs who were matched at each of the 1120 SNPs genotyped in the discovery cohort (table S1) within each of the classically-defined MHC regions (1, 11). (B) Linkage plot of rs887464 in the class I region and rs2281389 in the class II region for Caucasian donors in the discovery cohort (table S2). Chromosome 6 drawing modified from the National Library of Medicine, the National Center for Biotechnology Information public website (12).
Impact of the number of SNP mismatches on clinical outcome
Risks associated with patient-donor mismatching for classical HLA antigens are additive with each additional HLA mismatch (6, 7). To determine whether risks after HLA-matched unrelated donor transplantation also depend on the number of mismatched SNPs, the association of the total number of mismatched SNPs to mortality was determined. Compared to pairs matched for 76–100% of the 1120 SNPs (n = 530), the hazard ratio (HR) of mortality for patients matched for 0–25% (n = 537), 26–50% (n = 504), and 51–75% (n = 516) of SNPs were 1.51 [95% conference interval (95% CI), 0.96–1.38; P = 0.12], 1.10 (95% CI, 0.91–1.32; P = 0.17), and 1.04 (95% CI, 0.87–1.25; P = 0.66), respectively (overall P = 0.36). These data suggest that clinical outcome does not depend on the total number of mismatched SNPs.
Impact of specific SNPs on clinical outcome
We hypothesized that risks after HLA-matched unrelated donor transplantation could be conferred through the SNP genotype of the patient, the SNP genotype of the donor, or through SNP mismatching between the patient and donor. Furthermore, the vector or direction of SNP mismatching between the patient and donor could be important if the causative gene tagged by the SNP functions in a way that is similar to classical HLA, that is, host-versus-graft (HVG) and graft-versus-host (GVH) recognition. When each of the 1120 SNPs was analyzed for association to clinical outcome in genotype and in vector-specific mismatch models, ten statistically significant associations (7 mismatch, 3 donor genotype) involving 8 SNPs were identified (Table 2). The SNP associations to clinical outcome were independent of HLA genotype, HLA-DPB1 mismatching, and clinical variables. When each of the 8 SNPs was tested in the validation cohort, rs887464 and rs2281389 remained statistically significantly associated with outcome (Fig. 3, Table 2).
Table 2.
Summary of SNP associations in the discovery and validation cohorts.*
| Discovery cohort | Validation cohort | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| Outcome | SNP | Gene/location | Model | Overall P value | Genotype or mismatch group | Number of events | HR (95% CI) | P value | Overall P value | Genotype or mismatch group | Number of events | HR (95% CI) | P value |
| Survival | rs887464 | † Centromeric to PSORS1C3 and POU5F1 | Mismatch | 1.22×10−4 | Matched | 1117/1960 | 1.00 | 0.13 | Matched | 827/1523 | 1.00 | ||
| HVG | 34/50 | 1.93 (1.32–2.81) | 6.26×10−4 | HVG | 20/37 | 1.23 (0.74–2.05) | 0.42 | ||||||
| ‡ GVH and Bi | 24/59 | 0.57 (0.37–0.89) | 1.42×10−2 | GVH and Bi | 30/67 | 0.69 (0.47–1.02) | 6.53×10−2 | ||||||
|
| |||||||||||||
| Disease-free survival | rs887464 | † Centromeric to PSORS1C3 and POU5F1 | Mismatch | 2.97×10−4 | Matched | 1166/1936 | 1.00 | 2.44×10−2 | Matched | 808/1444 | 1.00 | ||
| HVG | 34/46 | 1.86 (1.31–2.65) | 5.38×10−4 | HVG | 23/36 | 1.66 (1.04–2.66) | 3.36×10−2 | ||||||
| GVH and Bi | 26/59 | 0.67 (0.45–1.00) | 4.74×10−2 | GVH and Bi | 32/62 | 0.73 (0.49–1.07) | 0.11 | ||||||
|
| |||||||||||||
| Relapse | rs3094214¶ | CDSN/intron | Mismatch | 1.15×10−3 | Matched | 492/1838 | 1.00 | 0.14 | Matched | 374/1377 | 1.00 | ||
| HVG | 43/103 | 1.77 (1.28–2.45) | 5.25×10−4 | HVG | 23/91 | 0.92 (0.60–1.41) | 0.70 | ||||||
| GVH and Bi | 27/114 | 0.82 (0.55–1.22) | 0.33 | GVH and Bi | 33/91 | 1.42 (0.99–2.04) | 5.59×10−2 | ||||||
|
| |||||||||||||
| Grades II–IV acute GVHD | rs7774197 | TNXB/intron | Mismatch | 2.77×10−4 | Matched | 1022/1884 | 1.00 | 0.74 | Matched | 726/1463 | 1.00 | ||
| HVG | 54/78 | 1.75 (1.32–2.31) | 9.39×10−5 | HVG | 33/62 | 1.02 (0.72–1.46) | 0.90 | ||||||
| GVH and Bi | 30/67 | 0.85 (0.59–1.22) | 0.37 | GVH and Bi | 33/56 | 1.15 (0.81–1.64) | 0.44 | ||||||
| rs2859091 | HLA-DQA2/flanking 5′UTR | Donor genotype | 1.31×10−6 | TT | 489/819 | 1.00 | 0.74 | TT | 298/616 | 1.00 | |||
| AT | 462/917 | 0.72 (0.63–0.82) | 5.50×10−7 | AT | 376/725 | 1.06 (0.91–1.24) | 0.46 | ||||||
| AA | 155/293 | 0.75 (0.62–0.90) | 1.87×10−3 | AA | 118/240 | 1.01 (0.82–1.26) | 0.90 | ||||||
| rs2281389 | HLA-DPB1/flanking 3′UTR | Mismatch | 1.73×10−4 | Matched | 654/1273 | 1.00 | 5.37×10−3 | Matched | 459/973 | 1.00 | |||
| HVG | 200/353 | 1.19 (1.00–1.40) | 4.41×10−2 | HVG | 154/289 | 1.21 (1.00–1.46) | 4.95×10−2 | ||||||
| GVH | 217/344 | 1.43 (1.22–1.68) | 1.60×10−5 | GVH | 154/279 | 1.37 (1.14–1.65) | 8.65×10−4 | ||||||
| Bi | 35/59 | 1.29 (0.91–1.82) | 0.16 | Bi | 25/40 | 1.21 (0.80–1.82) | 0.37 | ||||||
|
| |||||||||||||
| Grades III–IV acute GVHD | rs3132486 | HLA-C/flanking 5′UTR | Donor genotype | 1.72×10−4 | AA | 174/620 | 1.00 | 0.83 | AA | 83/425 | 1.00 | ||
| AG | 215/1093 | 0.66 (0.54–0.81) | 5.35×10−5 | AG | 156/802 | 0.98 (0.75–1.29) | 0.90 | ||||||
| GG | 129/527 | 0.87 (0.69–1.10) | 0.25 | GG | 69/383 | 0.91 (0.65–1.27) | 0.57 | ||||||
| rs2859091 | HLA-DQA2/flanking 5′UTR | Donor genotype | 2.34×10−6 | TT | 252/897 | 1.00 | 0.36 | TT | 107/619 | 1.00 | |||
| AT | 195/1013 | 0.62 (0.51–0.75) | 7.01×10−7 | AT | 153/745 | 1.20 (0.94–1.54) | 0.15 | ||||||
| AA | 71/330 | 0.68 (0.52–0.89) | 5.53×10−3 | AA | 48/246 | 1.14 (0.81–1.61) | 0.45 | ||||||
|
| |||||||||||||
| Chronic GVHD | rs2523957 | HCG9/flanking 5′UTR | Mismatch | 4.27×10−5 | Matched | 931/1867 | 1.00 | 0.20 | Matched | 570/1426 | 1.00 | ||
| HVG | 21/57 | 0.64 (0.41–1.00) | 4.90×10−2 | HVG | 21/42 | 1.27 (0.82–1.98) | 0.29 | ||||||
| GVH and Bi | 36/53 | 1.99 (1.42–2.78) | 6.86×10−5 | GVH and Bi | 25/49 | 1.36 (0.90–2.05) | 0.14 | ||||||
| rs2071479 | HLA-DOB/intron | Mismatch | 1.07×10−4 | Matched | 930/1884 | 1.00 | 0.11 | Matched | 592/1460 | 1.00 | |||
| HVG | 25/48 | 0.96 (0.64–1.45) | 0.85 | HVG | 10/29 | 1.03 (0.54–1.94) | 0.94 | ||||||
| GVH | 33/45 | 2.19 (1.53–3.13) | 2.02×10−5 | GVH | 14/28 | 1.79 (1.04–3.06) | 3.42×10−2 | ||||||
For the discovery and validation cohorts, models adjusted or stratified for clinical variables influencing outcomes (Table 1). In the discovery cohort, HLA alleles (0, 1 or 2 copies) were associated with survival (HLA-B*52, C*08), disease-free survival (HLA-B*52), relapse (HLA-B*27, C*14, DRB1*15), transplant-related mortality (HLA-A*33, DQB1*02), grades II–IV acute GVHD (HLA-C*16), grades III–IV acute GVHD (HLA-B*13) and chronic GVHD (HLA-C*05). In the validation cohort, associations were observed for survival (HLA-B*52), disease-free survival (HLA-DRB1*08), transplant-related mortality (HLA-B*49), grades III–IV acute GVHD (HLA-B*40, B*52). The effects of these HLA alleles were adjusted in the models.
Rs887464 is located 244 bp centromeric to the PSORS1C3 locus and 7469 bp centromeric to the POU5F1 locus (12).
When the bidirectional (Bi) mismatch group contained 10 or fewer pairs (rs887464, rs3094214, rs7774197, and rs2523957), the pairs were combined with the GVH group.
GVH/bidirectional mismatching for rs3094214 was associated with transplant-related mortality in the validation cohort (HR 0.39; 95% CI, 0.21–0.73; P = 0.003), however, this SNP had no associations in the combined cohort. No SNP in the combined cohort was associated with relapse.
Fig. 3.

Probability of disease-free survival and grades II-IV acute GVHD (A) Probability of disease-free survival for patients according to patient-donor rs887464 match status: match (solid line), HVG mismatch (hatched line) or GVH/bidirectional mismatch (dotted line). (B) Probability of grades II-IV acute GVHD for patients according to patient-donor rs2281389 match status: matched (solid line), GVH mismatch (dotted line), HVG mismatch (hatched line), and bidirectional mismatch. Graft-versus-host (GVH); host-versus-graft (HVG).
SNP rs887464 resides 91 kb telomeric to HLA-C, hence formal evaluation of linkage disequilibrium with HLA-C alleles was performed. All patients and donors were HLA-C allele-matched, and 95% were rs887464-matched. Although the rs887464 G allele was positively correlated with HLA-C*02, *04, *05, *06, *08 and *14 in both patients and donors (P < 0.0001), none of these alleles were significantly associated with outcome (P > 0.01). HVG mismatching for rs887464 was associated with lower disease-free survival compared to matching (discovery cohort HR 1.86; 95% CI, 1.31–2.65; P = 0.0005; validation cohort HR 1.66; 95% CI, 1.04–2.66; P = 0.03; combined cohort HR 1.78; 95% CI, 1.32–2.38; P = 0.0001). HVG mismatching for rs887464 was statistically significantly associated with mortality in the discovery cohort, but not in the validation cohort (Table 2). However, in the combined discovery/validation cohorts, HVG mismatching for rs887464 was statistically significantly associated with increased risk of both transplant-related mortality (HR 1.96; 95% CI, 1.26–3.04; P = 0.003) and mortality (HR 1.98; 95% CI, 1.40–2.80; P = 0.0001). No SNP was associated with relapse in the combined cohort. Therefore, the clinical effect of rs887464 on disease-free survival was related to the significant negative effect on transplant-related mortality. Finally, 42% of HVG-mismatched pairs and 50% of GVH/bidirectionally-mismatched pairs had all three killer immunoglobulin receptor (KIR) ligands present (C1 and C2 defined by HLA-C, and Bw4 defined by HLA-B and HLA-A) (P = 0.33) (2). These results demonstrate that the rs887464 effects on clinical outcome cannot be attributed to the presence or absence of KIR ligands among these HLA-matched transplants.
To determine whether risk depends on the A or G allele of rs887464, two analyses were performed. First, HVG mismatches were evaluated for associations to survival, disease-free survival and transplant-related mortality for GG patients transplanted from AG donors relative to AA patients transplanted from AG donors. There were no statistically significant differences found, (HR 1.24, 95% CI 0.57–2.70, P = 0.06 for survival; HR 1.33, 95% CI 0.65–2.75, P = 0.044 for disease-free survival; HR 1.09, 95% CI 0.38–3.08, P = 0.88 for transplant-related mortality), although the number of pairs in each group was limited (23 AA-AG pairs and 27 GG-AG pairs). These results suggest that when transplant pairs are matched at this SNP position, the outcome for homozygous AA and homozygous GG pairs is similar. When patients and donors are mismatched at this SNP, however, the negative effects on disease-free survival of a homozygous donor for a heterozygous patient (HVG vector of mismatch) are evident. The second approach to evaluate potential association of rs887464A and G alleles to disease-free survival involved the analysis of 3405 rs887464-matched pairs in the combined discovery-validation cohort. Compared to AG patients transplanted from AG donors, the HR of disease-free survival for AA patients transplanted from AA donors was 1.06 (95% CI, 0.92–1.21; P = 0.42) and the HR of disease-free survival for GG patients transplanted from GG donors was 1.11 (95% CI, 0.99–1.24; P = 0.07). These results suggest that the effect of rs887464 on disease-free survival is likely defined by the presence of mismatching rather than patient or donor genotypes.
A survey of 59 SNPs residing 49 kb 5′ and 82 kb 3′ of rs887464 shows that rs887464 resides outside of a block of high linkage disequilibrium (r2 < 0.46) (Fig. 2B, table S2); no other SNP within 500 kb was associated with outcome. In the combined discovery-validation cohort, 344 pairs were matched at all 6 classical HLA loci (HLA-A, C, B, DRB1, DQB1, DPB1; “12/12” matched). Of these pairs, only 12 pairs were rs887464-mismatched and precluded analysis of the effects of mismatching for this SNP in the 12/12 matched setting. In summary, disease-free survival depends on mismatching for rs887464 and not to the effects of neighboring SNPs or to linked HLA-C alleles.
The second validated SNP, rs2281389, resides 2 kb centromeric to HLA-DPB1 in a 29 kb block (r2 > 0.80; Fig. 2B, table S2). World experience demonstrates that 80–85% of HLA-A, C, B, DRB1, DQB1-allele-matched patient-donor pairs are mismatched at HLA-DPB1 (7,16) and that DPB1 mismatching is a risk factor for acute GVHD (16). Consistent with previous observations, 17% of patient-donor pairs in the combined discovery-validation cohort were HLA-DPB1-matched, and of these transplants, 60% were rs2281389-matched. DPB1*03 and *04 alleles were positively associated with the rs2281389 A allele in both patients and donors (P < 0.0001). The presence of patient-donor HLA-DPB1 mismatching in the discovery cohort was associated with an increased risk of grades II IV acute GVHD (HR 1.2); after adjusting for HLA-DPB1 mismatching, GVH mismatching for rs2281389 conferred an increased risk of grades II-IV acute GVHD compared to matching (discovery cohort HR 1.43; 95% CI, 1.22–1.68; P = 0.00002; validation cohort HR 1.37; 95% CI, 1.14–1.65; P = 0.0009; combined cohort HR 1.34, 95% CI, 1.17–1.55; P = 0.00004). The increased risk of GVHD was not accompanied by a statistically significant reduction in relapse, although the absolute value of the hazard ratio was less than 1 (HR 0.84; 95% CI 0.65–1.09; P = 0.19). Only 10 of the 344 HLA-A, C, B, DRB1, DQB1, DPB1 allele-matched pairs in the combined discovery-validation cohort were rs2281389-mismatched and precluded an analysis of the effect of this SNP among HLA 12/12 matched pairs. Association of rs2281389 A and G alleles to the risk of grades II-IV acute GVHD was assessed in 2289 rs2281389-matched pairs. Compared to AA patients transplanted from AA donors, the HR of acute GVHD for GG patients transplanted from GG donors was 0.79 (95% CI, 0.19–3.36; P = 0.75). In summary, patient-donor rs2281389 mismatching rather than genotype is a risk factor for acute GVHD after HLA-matched unrelated donor transplantation.
The presence of certain high-frequency HLA haplotypes in the Japanese population is associated with transplant outcome and may be explained by the extreme SNP conservation of the MHC and lack of SNP mismatching (17). The predominantly Caucasian study population in the current analysis lacked sufficient numbers of HLA homozygous individuals to explore the potential association of specific extended HLA haplotypes to clinical outcome. Of the 3761 pairs in the combined discovery-validation cohorts, 60 (2%) were HLA-A, -C, - B, -DRB1, -DQB1-homozygous. Fifteen unique HLA haplotypes were represented of which HLA-A*01:01/C*07:01/B*08:01/DRB1*03:01/DQB1*02:01 was the most frequent (33/60). All pairs homozygous for this haplotype were rs887464-matched, but only 64% were rs2281389-matched.
Probability of identifying rs2281389-matched unrelated donors
The feasibility of integrating SNP genotyping and matching into prospective donor selection was evaluated by determining the probability of finding rs2281389-matched unrelated donors for 230 patients who had successfully identified two or more HLA-A, - C, -B, -DRB1, -DQB1-matched unrelated donors on their search at the Fred Hutchinson Cancer Research Center (FHCRC; Table 3). The number of HLA-matched donors studied per patient ranged from 2 to 9 donors. The likelihood of identifying rs2281389-matched donors increased with increasing numbers of donors tested per patient; furthermore, most patients had a choice of at least two SNP-matched HLA-matched donors. These results demonstrate that patients who successfully identify at least two HLA-A, -C, -B, -DRB1, -DQB1 allele-matched donors on their search, have rs2281389-matched donors from which to select the optimal donor for transplantation.
Table 3.
SNP rs2281389 matching for 230 patients with 2 or more HLA-A, -C, -B, -DRB1, - DQB1 allele-matched unrelated donors identified from the search prior to transplantation.
| Number of HLA-matched donors tested per patient | Number of patients | Number of patients with no rs2281389-matched donor (%) | Number of patients with only one rs2281389-matched donor (%) | Number of patients with more than one rs2281389-matched donor (%) |
|---|---|---|---|---|
| 2 | 131 | 24 (18 %) | 44 (34 %) | 63 (48%) |
| 3 | 39 | 5 (13 %) | 11 (28 %) | 23 (59 %) |
| 4 | 30 | 4 (13 %) | 7 (23 %) | 19 (63 %) |
| 5 | 9 | 1 (11%) | 2 (22 %) | 6 (67 %) |
| 6 | 17 | 1 (6 %) | 1 (6 %) | 15 (88 %) |
| 7 | 1 | 0 | 0 | 1 (100 %) |
| 8 | 2 | 0 | 0 | 2 (100 %) |
| 9 | 1 | 0 | 0 | 1 (100 %) |
DISCUSSION
HLA-matched unrelated donors and patients share the same HLA alleles, however they may differ for MHC variation that is linked to their HLA haplotypes (8). We hypothesized that the success of unrelated HCT might be influenced by undetected variation within the MHC, given the extreme density of immune-related genes in this region (1). We used a discovery-validation study design to identify non-HLA transplantation determinants and confirm putative variants in large homogeneous transplant populations. We found that HLA-matched patients and unrelated transplant donors can differ for SNPs across the 4.9 megabase MHC region, indicating that haplotype content can differ among individuals who share the same HLA alleles. Two SNPs were validated as robust markers for disease-free survival and acute GVHD risk. These results demonstrate that the transplant barrier is defined by not only by classical HLA genes, but also by non-HLA genetic variation within the MHC.
A hallmark of the human genome is the positive association of two or more genetic markers together (linkage disequilibrium) (13). The associated markers can be complex genes, simple bi-allelic SNPs, or other forms of variation (13). Nowhere in the human genome is linkage disequilibrium as strong and long-range as in the MHC, a phenomenon that hampers the identification of true causal variants in disease (1, 14, 15). To surmount these potential methodologic challenges, we studied only HLA-A, -C, -B, -DRB1, -DQB1-matched pairs (6, 7). After excluding the possibility that SNPs were markers for leukemia or myelodysplastic syndrome (Bonferroni P > 0.00004), we tested but did not find any effect on outcome of HLA-C alleles in positive linkage disequilibrium with rs887464. Associations between SNP genotype and outcome were elucidated in pairs matched for rs887464 and rs2281389; no association of rs887464 with disease-free survival, or of rs2281389 with risk of acute GVHD was evident. Finally, adjustment for HLA-DPB1 mismatching, a known risk factor for GVHD (16), yielded clear association of rs2281389 mismatching to GVHD risk. These results collectively demonstrate that undetected patient-donor mismatching for SNPs is an independent risk factor for transplant outcome after HLA-matched unrelated transplantation.
There are several limitations to the current study. In traditional disease-association mapping, the over (or under) representation of specific SNP genotypes in a disease cohort compared to healthy individuals signals the presence of potential causative genes (13, 19). In transplantation, not only could causative genes be defined by the genotype of SNPs carried by the patient or the donor, but if the causative gene functions in ways that are similar to the recognition of mismatched HLA antigens, then donor mismatching or patient mismatching for the SNP could be useful for mapping causative genes in transplantation. The vector of SNP incompatibility could serve as a useful tool for not only identifying the causative gene(s), but also could shed light on the mechanisms through which those genes provoke GVHD. However, if those genes are monomorphic then a vector analysis would not be enlightening. Fine mapping of the MHC near the SNPs of interest will help clarify the nature of the genetic variants responsible for risks after HLA-matched transplantation, and their potential mechanisms. Finally, certain world populations are distinguished by a few unique and highly conserved HLA haplotypes, resulting in a high frequency of HLA homozygosity with low SNP diversity (17). The presence (or absence) of such conserved haplotypes may themselves be associated with clinical outcome (17). The exceedingly low frequency (2%) of HLA homozygous patient-donor pairs in our predominately Caucasian study population precluded a definitive analysis of the clinical significance of specific HLA haplotypes. A larger transplant experience of HLA homozygous pairs will help to clarify the nature of risks associated with specific extended HLA haplotypes.
The association of patient-donor SNP mismatching with clinical outcome signals the presence of linked genetic variation that is biologically meaningful (13). Although the functional variants remain to be elucidated, the vector of the SNP mismatch is clinically relevant, suggesting genes or pathways that involve recognition of patient mismatching (rs2281389) or donor mismatching (rs887464). In this regard, it is intriguing that rs887464 is a putative expression quantitative locus (eQTL) for CCHCR1, TCF19, HLA-B, HLA-C, MICA and MICB genes within the MHC (18). Interestingly, SNP rs887464 is a risk marker for autoimmune diseases including type 1 diabetes (19, 20). The association of rs887464 mismatching to disease-free survival in the current study raises the possibility that the level of HLA class I gene expression is an important determinant of transplant outcome, as well as the potential role for the non-classical class I genes MICA and MICB as ligands for the innate immune system (2). Fine mapping of the regions 5′ and 3′ of rs887464 will clarify whether this SNP is a marker for a non-HLA genetic variant, or whether the SNP is effecting transplant outcome through differential expression of a target gene. If the latter is found, then linkage disequilibrium of rs887464 to the alleles of a target gene could help explain the association of rs887464 mismatching rather than genotype to outcome. If each of the two rs887464 alleles define haplotypes of polymorphic alleles of the target gene, then patient-donor mismatching at rs887464 could lead to clinical effects through allele-specific alterations in the expression of patient and/or donor alleles.
The association of rs2281389 mismatching with GVHD risk provides new information on the class II region. SNP rs2281389 maps 2 kb centromeric to the HLA-DPB1 3′ untranslated region and is a risk marker for pediatric asthma (21), rheumatoid arthritis (22), and Hodgkin lymphoma (23). In the current study, mismatching for rs2281389 and not rs9277535 was statistically significantly associated with acute GVHD, although rs9277535 showed similar trends (HR 1.37 for GVH mismatch). SNP rs9277535 is a putative trans-eQTL and micro RNA binding site in the HLA-DPB1 3′ untranslated region (18, 24). In this context, our observations suggest a possible role of HLA-DPB1 haplotype-linked expression in GVHD, where we surmise that rs2281389 alleles define haplotypes with DPB1 alleles; depending on the specific DPB1 mismatch combination in a given patient and donor, differential expression of the mismatched alleles could place a patient at increased or lowered risk of GVHD. Given that GVHD is a highly inflammatory state (25), it is not surprising that there may be pathways common to GVHD, autoimmunity, and infection.
Simple bi-allelic SNPs can be genotyped at high efficiency (13, 26). The results of the current analysis suggest that clinical outcome after HLA-matched unrelated donor transplantation can be improved through prospective donor evaluation of SNPs. The discovery-validation cohorts provide information on SNP match rates between the patient and the donor who was selected based on HLA match status but not on SNP criteria. Those data show that most HLA-matched patients and donors were matched for rs887464 whereas only 60% were matched for rs2281389, even among HLA homozygous pairs with common HLA haplotypes. Given a 60:40 match:mismatch rate for rs2281389, the results suggest that patients who prospectively identify three HLA-matched unrelated donors have a 90% chance that at least one donor will be rs2281389-matched. To demonstrate the feasibility of integrating SNP genotyping into donor selection, we analyzed patients who had identified two or more otherwise eligible HLA-matched donors on their search. The analysis of searchable donors provides information on rs2281389 match rates among a pool of individuals who share the same tissue type. The data demonstrate that SNP genotyping can be translated easily into donor selection. Furthermore, among patients with HLA-matched donors, the probability of having a choice of rs2281389-matched donors increases as the number of tested donors increases. Since patient-donor mismatching at HLA-DPB1 and rs2281389 each independently increase GVHD risk, future efforts to match donors for both genetic determinants are expected to lower risks for patients. With over 18 million unrelated donors registered worldwide (27), the potential to benefit future patients in need of a life-saving transplant is anticipated to be significant.
In conclusion, the transplant barrier is comprised of classical HLA loci as well as non-HLA variation within the gene-dense MHC region. Two new genetic markers are informative for disease-free survival and acute GVHD after HLA-matched unrelated donor transplantation. The identification of MHC resident transplantation determinants provides clinicians with tools to lower post-transplant risks through comprehensive donor matching, and to identify patients at highest risk for complications who might benefit from directed preventive measures that include optimization of GVHD prophylaxis. This study provides the foundation for future fine-mapping approaches to identify the specific nature of the genes and their mechanisms in health and disease.
MATERIALS AND METHODS
Study design and population
Discovery-validation cohort
We used a classic discovery-validation study design to identify putative non-HLA transplantation determinants and confirm the markers with strongest associations to clinical outcome (Fig. 1). Patients received a first allogeneic transplant from an HLA-A, -C, -B, -DRB1, -DQB1 allele-matched unrelated donor for the treatment of acute myeloid leukemia, acute lymphoblastic leukemia, chronic myeloid leukemia or myelodysplastic syndrome at the FHCRC (n = 858), or at one of 147 other centers in the Center for International Blood and Marrow Transplant Research network (n = 3347) (Table 1). The 4205 transplants were defined as a discovery (n = 2492) and a validation (n = 1713) cohort according to the chronological availability of research DNA samples (Fig. 1, Table 1). All subjects provided informed consent for participation in this study and protocols were approved by the institutional review boards of the FHCRC and the National Marrow Donor Program (NMDP).
Patients and their searchable donors
To determine the probability of identifying rs2281389-matched HLA-matched unrelated donors, we studied all consecutive patients referred to the FHCRC for initiation of an unrelated donor search between 2000–2008 for whom a minimum of two HLA-A, -C, -B, -DRB1, -DQB1-allele matched NMDP donors were identified on search and for whom sufficient waste DNA was available. All materials were approved by the institutional review boards of the FHCRC and the NMDP.
Genotyping
Patient and donor HLA-A, -C, -B, -DRB1, -DQB1 alleles were genotyped as previously described (6, 7). Samples in the discovery cohort (n = 4927) were genotyped for 1228 MHC region SNPs using the Illumina® FastTrack Genotyping Service (Illumina, Inc.), a robust platform for interrogating the MHC (28, 29). Quality control measures included 233 inter- and intra-experiment duplicate samples (0.997 concordance rate). Two SNPs did not meet Hardy-Weinberg equilibrium in Caucasian donors (rs2395031 and rs11756897, P < 0.0001); manual inspection of the scatter plots showed good genotype segregation and therefore these SNPs were included. A total of 65 samples (1.3%) failed genotyping and 30 samples (0.6%) failed duplicate controls, yielding 4599 samples and 1120 SNPs for analysis of transplant outcomes in the discovery cohort (0.977 genotyping rate) (Fig. 1). Eight SNPs that were associated with clinical outcome in the discovery cohort were genotyped in the 3391 samples in the validation cohort using TaqMan chemistry (Fig. 1, Table 2) (30). Fifteen inter-experiment duplicates had a 0.992 concordance rate; nine samples failed genotyping, yielding 3367 samples for analysis in the validation cohort (0.991 genotyping rate). For the analysis of patients and their searchable donors, DNA was genotyped for rs2281389 using TaqMan chemistry (30).
Statistical methods
The clinical endpoints were survival, disease-free survival, transplant-related mortality, relapse, grades II-IV and III-IV acute GVHD, and chronic GVHD. For each endpoint, the impact of single SNPs was evaluated in the discovery cohort, and the effects of multiple SNPs were tested in the discovery cohort, validation cohort as well as the combined discovery and validation cohort, using multivariate models that adjusted for factors that influenced clinical outcome (Table 2). An assessment of potential risk factors for each outcome was evaluated in multivariate analyses using Cox proportional hazards regression. First, the proportionality assumption was tested for each factor by adding a time-dependent covariate. When tests indicated differential effects over time (non-proportional hazards), the factors were adjusted through stratification. Second, a stepwise forward/backward model selection approach was used to identify other significant risk factors. Clinical factors which were significant at a 5% level were selected and retained in the final model. Different sets of models were built for the discovery and validation populations to control for demographic differences (Table 1).
In single SNP analysis, each SNP was tested separately in models by assessing the number of copies (zero, one or two) of minor alleles carried by each patient and donor, and SNP mismatching in patient-donor pairs (figs. S1-S7). Homozygous minor allele genotypes occurring in ten or fewer individuals were combined with the heterozygous genotype. SNP mismatching was defined according to the vector of mismatching: host-versus-graft (HVG; homozygous AA or GG patient with heterozygous AG donor); graft-versus-host (GVH; homozygous AA or GG donor with heterozygous AG patient), and bidirectional (AA patient with GG donor and vice versa). The association of patient genotype, donor genotype and patient-donor mismatching to each of the seven clinical endpoints was evaluated with adjustment for appropriate clinical prognostic factors as described above (Table 1). The two validated SNPs, rs887464 and rs2281389, reside in close proximity to HLA-C and DPB1, respectively. Accordingly, potential associations of HLA-C and DPB1 alleles to outcome were examined by grouping four-digit HLA-C and DPB1 alleles by their first two digits, and the association of zero, one and two allele copies to clinical endpoints was determined; alleles which were significant at a 5% level were selected and retained in the final model. To control for the family-wise error rate, the Bonferroni criterion was applied to adjust for the multiple testing. Donor rs2859091 genotype was associated with grades II-IV (adjusted P = 0.02) and III-IV (adjusted P = 0.005) acute GVHD, and patient-donor mismatching for rs2523957 was associated with chronic GVHD (P = 0.03). Permutation tests for each endpoint were conducted and yielded results consistent with the Bonferroni correction. No additional SNPs were detected beyond the single SNP analysis that used the Bonferroni adjustment.
A model selection procedure was conducted to test for potential associations from multiple SNPs. Since the total number of informative patient-donor pairs differed for each SNP (table S1), we used a two-step strategy. For each outcome, a set of SNPs in the single SNP analysis of patient genotype, donor genotype, or patient-donor mismatch was identified at a 0.01 significance level. A stepwise selection procedure was then performed on the set of SNPs for each endpoint at a 0.001 significance level, with inclusion of the clinical prognostic factors selected in the preliminary models
Genotypic associations between HLA alleles and SNPs were tested using chi-square (or Fisher’s exact for cells with five or fewer individuals). All analyses were performed using SAS version 9.2 (SAS Institute, Cary, NC). Hardy-Weinberg equilibrium, minor allele frequencies and linkage disequilibrium (r2) between SNPs were estimated using “Haploview” (31), PLINK version 1.07 (32), and R package “Genetics” version 1.3.4 (33). Plots were displayed with Matlab (MathWorks Inc., Natick, MA).
Supplementary Material
Fig. S1. Univariate P value plots for survival.
Fig. S2. Univariate P value plots for disease-free survival.
Fig. S3. Univariate P value plots for relapse.
Fig. S4. Univariate P value plots for transplant-related mortality.
Fig. S5. Univariate P value plots for grades II-IV acute GVHD.
Fig. S6. Univariate P value plots for grades III-IV acute GVHD.
Fig. S7. Univariate P value plots for chronic GVHD.
Table S1. SNP identifiers, minor allele frequencies in Caucasian patients and donors, and percent of patient-donor pairs matched for each SNP.
Table S2. r2 values for SNPs rs887464 and rs2281389 (Fig. 2B).
Acknowledgments
Funding: Supported by grants AI069197 (E.W.P., M.M., T.A.G., S.R.S., M.D.H., M.M.H., T.W.), CA100019 (E.W.P., M.M., T.A.G.), CA18029 (E.W.P., M.M., T.A.G.) and CA76518 (M.M.H.) from the National Institutes of Health; HHSH234200637015C (M.M.H.) from the Health Resources and Services Administration, and N00014-10-1-0204 and N00014-1-1-0339 (S.R.S., M.D.H., M.M.H.) from the Office of Naval Research.
We thank Mark Gatterman, Dawn Moran and Charlie Du for their outstanding technical assistance in SNPs genotyping; Dr. Shalini Pereira, Lisa Getzendaner and Brenda Nisperos for searchable donor samples, and Gary Schoch for database support. We are grateful to Dr. John Klein for his helpful comments, and Franco Mendolia for computational support.
Footnotes
Competing interests: The authors have no competing interests.
Data and materials: All reasonable requests for data will be fulfilled provided that material transfer agreements, data sharing agreements and institutional review board requirements are met between FHCRC and the requester.
Author contributions: E.W.P. developed the hypotheses and designed the study. E.W.P., M.M., S.R.S., M.D.H. and M.M.H. provided HLA and clinical data. M.M. managed DNA genotyping and data. T.W. performed the statistical analysis. T.A.G. provided statistical support. E.W.P., M.M., T.W., T.A.G. analyzed the data. All authors contributed to the preparation of the paper and approved the final manuscript.
References and Notes
- 1.Trowsdale J. The MHC, disease and selection. Immunol Lett. 2011;137:1–8. doi: 10.1016/j.imlet.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Norman PJ, Parham P. Complex interactions, the immunogenetics of human leukocyte antigen and killer cell immunoglobulin-like receptors. Semin Hematol. 2005;42:65–75. doi: 10.1053/j.seminhematol.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Hill AV. The immunogenetics of human infectious diseases. Annu Rev Immunol. 1998;16:593–617. doi: 10.1146/annurev.immunol.16.1.593. [DOI] [PubMed] [Google Scholar]
- 4.Carrington M, Nelson GW, Martin MP, Kissner T, Vlahov D, Goedert JJ, Kaslow R, Buchbinder S, Hoots K, O’Brien SJ. HLA and HIV-1, heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 5.Dausset J. Leuco-agglutinins. IV. Leuco-agglutinins and blood transfusion. Vox Sang. 1954;4:190–198. [Google Scholar]
- 6.Petersdorf EW, Hansen JA, Martin PJ, Woolfrey A, Malkki M, Gooley T, Storer B, Mickelson E, Smith A, Anasetti C. Major-histocompatibility-complex class I alleles and antigens in hematopoietic-cell transplantation. N Engl J Med. 2001;345:1794–1800. doi: 10.1056/NEJMoa011826. [DOI] [PubMed] [Google Scholar]
- 7.Lee SJ, Klein J, Haagenson M, Baxter-Lowe LA, Confer DL, Eapen M, Fernandez-Vina M, Flomenberg N, Horowitz M, Hurley CK, Noreen H, Oudshoorn M, Petersdorf E, Setterholm M, Spellman S, Weisdorf D, Williams TM, Anasetti C. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110:4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 8.Petersdorf E, Malkki M, Gooley TA, Martin PJ, Guo Z. MHC haplotype matching for unrelated hematopoietic cell transplantation. PLoS Medicine. 2007;4:e8. doi: 10.1371/journal.pmed.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker KS, Davies SM, Majhail NS, Hassebroek A, Klein JP, Ballen KK, Bigelow CL, Frangoul HA, Hardy CL, Bredeson C, Dehn J, Friedman D, Hahn T, Hale G, Lazarus HM, LeMaistre CF, Loberiza F, Maharaj D, McCarthy P, Setterholm M, Spellman S, Trigg M, Maziarz RT, Switzer G, Lee SJ, Rizzo JD. Race and socioeconomic status influence outcomes of unrelated donor hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2009;15:1543–1554. doi: 10.1016/j.bbmt.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC, Jr, Wright MW, Wain HM, Trowsdale J, Ziegler A, Beck S. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 12.National Library of Medicine, the National Center for Biotechnology Information public website. (as seen July 15, 2011, at http://www.ncbi.nlm.nih.gov/projects/mapview/maps.cgi?taxid=9606&chr=6)
- 13.Palmer LJ, Cardon LR. Shaking the tree, mapping complex disease genes with linkage disequilibrium. Lancet. 2005;366:1223–1234. doi: 10.1016/S0140-6736(05)67485-5. [DOI] [PubMed] [Google Scholar]
- 14.Yunis EJ, Larsen CE, Fernandez-Viña M, Awdeh ZL, Romero T, Hansen JA, Alper CA. Inheritable variable sizes of DNA stretches in the human MHC, conserved extended haplotypes and their fragments or blocks. Tissue Antigens. 2003;62:1–20. doi: 10.1034/j.1399-0039.2003.00098.x. [DOI] [PubMed] [Google Scholar]
- 15.Miretti MM, Walsh EC, Ke X, Delgado M, Griffiths M, Hunt S, Morrison J, Whittaker P, Lander ES, Cardon LR, Bentley DR, Rioux JD, Beck S, Deloukas P. A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:634–646. doi: 10.1086/429393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaw BE, Gooley TA, Malkki M, Madrigal JA, Begovich AB, Horowitz MM, Gratwohl A, Ringdén O, Marsh SG, Petersdorf EW. The importance of HLA-DPB1 in unrelated donor hematopoietic cell transplantation. Blood. 2007;110:4560–4566. doi: 10.1182/blood-2007-06-095265. [DOI] [PubMed] [Google Scholar]
- 17.Morishima S, Ogawa S, Matsubara A, Kawase T, Nannya Y, Kashiwase K, Satake M, Saji H, Inoko H, Kato S, Kodera Y, Sasazuki T, Morishima Y. Japan Marrow Donor Program, Impact of highly conserved HLA haplotype on acute graft-versus-host disease. Blood. 2010;115:4664–4670. doi: 10.1182/blood-2009-10-251157. [DOI] [PubMed] [Google Scholar]
- 18.SCAN. SNP and CNV Annotation Database. (as seen February 2, 2011, at http://www.scandb.org/newinterface/about.html)
- 19.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common disease and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sirota M, Schaub MA, Batzoglou S, Robinson WH, Butte AJ. Autoimmune disease classification by inverse association with SNP alleles. PLoS Genet. 2009;5:e1000792. doi: 10.1371/journal.pgen.1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noguchi E, Sakamoto H, Hirota T, Ochiai K, Imoto Y, Sakashita M, Kurosaka F, Akasawa A, Yoshihara S, Kanno N, Yamada Y, Shimojo N, Kohno Y, Suzuki Y, Kang MJ, Kwon JW, Hong SJ, Inoue K, Goto Y, Yamashita F, Asada T, Hirose H, Saito I, Fujieda S, Hizawa N, Sakamoto T, Masuko H, Nakamura Y, Nomura I, Tamari M, Arinami T, Yoshida T, Saito H, Matsumoto K. Genome-wide association study identifies HLA-DP as a susceptibility gene for pediatric asthma in Asian populations. PLoS Genet. 2011;7:e1002170. doi: 10.1371/journal.pgen.1002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plenge RM, Cotsapas C, Davies L, Price AL, de Bakker PI, Maller J, Pe’er I, Burtt NP, Blumenstiel B, DeFelice M, Parkin M, Barry R, Winslow W, Healy C, Graham RR, Neale BM, Izmailova E, Roubenoff R, Parker AN, Glass R, Karlson EW, Maher N, Hafler DA, Lee DM, Seldin MF, Remmers EF, Lee AT, Padyukov L, Alfredsson L, Coblyn J, Weinblatt ME, Gabriel SB, Purcell S, Klareskog L, Gregersen PK, Shadick NA, Daly MJ, Altshuler D. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39:1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moutsianas L, Enciso-Mora V, Ma YP, Leslie S, Dilthey A, Broderick P, Sherborne A, Cooke R, Ashworth A, Swerdlow AJ, McVean G, Houlston RS. Multiple Hodgkin lymphoma-associated loci within the HLA region at chromosome 6p21.3. Blood. 2011;118:670–674. doi: 10.1182/blood-2011-03-339630. [DOI] [PubMed] [Google Scholar]
- 24.TargetScan Human Database Release 6.0. (as seen November 15, 2011, at http://www.targetscan.org)
- 25.Ferrara JL, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet. 2009;373:1550–1561. doi: 10.1016/S0140-6736(09)60237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kruglyak L. The road to genome-wide association studies. Nat Rev Genet. 2008;9:314–318. doi: 10.1038/nrg2316. [DOI] [PubMed] [Google Scholar]
- 27.Foeken LM, Green A, Hurley CK, Marry E, Wiegand T, Oudshoorn M. The Donor Registries Working Group of the World Marrow Donor Association (WMDA), Monitoring the international use of unrelated donors for transplantation, the WMDA annual reports. Bone Marrow Transplant. 2010;45:811–818. doi: 10.1038/bmt.2010.9. [DOI] [PubMed] [Google Scholar]
- 28.Fan JB, Gunderson KL, Bibikov M, Yeakley JM, Chen J, Wickham Garcia E, Lebruska LL, Laurent M, Shen R, Barker D. Illumina universal bead arrays. Methods Enzymol. 2006;410:57–73. doi: 10.1016/S0076-6879(06)10003-8. [DOI] [PubMed] [Google Scholar]
- 29.Illumina MHC exon centric panel. (as seen December 1, 2005, at http://www.illumina.com/products/mhc_exon_centric_panel.ilmn)
- 30.Livak KJ, Marmaro J, Todd JA. Towards fully automated genome-wide polymorphism screening. Nat Genet. 1995;4:341–342. doi: 10.1038/ng0495-341. [DOI] [PubMed] [Google Scholar]
- 31.Barrett JC, Fry B, Maller J, Daly MJ. Haploview, analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 32.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK, a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R Package “Genetics” 1.3.4, G. Warnes with contributions from G. Gorjanc, F. Leisch and M. Man (2009). (as seen August 20, 2008, at http://cran.r-project.org/web/packages/genetics/).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Univariate P value plots for survival.
Fig. S2. Univariate P value plots for disease-free survival.
Fig. S3. Univariate P value plots for relapse.
Fig. S4. Univariate P value plots for transplant-related mortality.
Fig. S5. Univariate P value plots for grades II-IV acute GVHD.
Fig. S6. Univariate P value plots for grades III-IV acute GVHD.
Fig. S7. Univariate P value plots for chronic GVHD.
Table S1. SNP identifiers, minor allele frequencies in Caucasian patients and donors, and percent of patient-donor pairs matched for each SNP.
Table S2. r2 values for SNPs rs887464 and rs2281389 (Fig. 2B).