Abstract
Previously, we cloned and characterized an insect (Sf9) cell cDNA encoding a class II α-mannosidase with amino acid sequence and biochemical similarities to mammalian Golgi α-mannosidase II. Since then, it has been demonstrated that other mammalian class II α-mannosidases can participate in N-glycan processing. Thus, the present study was performed to evaluate the catalytic properties of the Sf9 class II α-mannosidase and to more clearly determine its relationship to mammalian Golgi α-mannosidase II. The results showed that the Sf9 enzyme is cobalt-dependent and can hydrolyze Man5GlcNAc2 to Man3GlcNAc2, but it cannot hydrolyze GlcNAcMan5GlcNAc2. These data establish that the Sf9 enzyme is distinct from Golgi α-mannosidase II. This enzyme is not a lysosomal α-mannosidase because it is not active at acidic pH and it is localized in the Golgi apparatus. In fact, its sensitivity to swainsonine distinguishes the Sf9 enzyme from all other known mammalian class II α-mannosidases that can hydrolyze Man5GlcNAc2. Based on these properties, we designated this enzyme Sf9 α-mannosidase III and concluded that it probably provides an alternate N-glycan processing pathway in Sf9 cells.
Glycoproteins constitute a large and diverse group of macromolecules with central roles in many biological processes. Among them are the Asn-linked glycoproteins, which consist of a polypeptide backbone and an amide-linked oligosaccharide side chain or N-glycan. The N-glycan precursor, Glc3Man9GlcNAc2, is initially transferred to newly synthesized proteins in the lumen of the endoplasmic reticulum (ER)1 and is converted to various mature forms by processing enzymes distributed along the secretory pathway. Processing begins with cleavage of the three glucose residues by ER α-glucosidases, which produce Man9GlcNAc2, generally known as a “high mannose” N-glycan. In higher eukaryotes, high mannose N-glycans may be further processed by ER and Golgi α-mannosidases, which cleave up to six mannose residues. Cleavage of these mannose residues is required before N-glycans can be converted to “hybrid” or “complex” N-glycans by various glycosyltransferases (1).
Three types of α-mannosidases are involved in N-glycan processing: endo-α-mannosidase, class I α-mannosidases, and class II α-mannosidases (2–5). Endo-α-mannosidase cleaves the reducing terminus of the mannose residue to which the glucose residues are attached if a glycoprotein containing Glc(1–3)Man9GlcNAc2 exits the ER. Class I α-mannosidases cleave the four α1,2-linked mannose residues, converting Man9GlcNAc2 to Man5GlcNAc2 in the ER and Golgi complex. Historically, the only class II α-mannosidase thought to be involved in N-glycan processing was Golgi α-mannosidase II. This enzyme requires the prior action of N-acetylglucosaminyltransferase I and cleaves the terminal α1,3- and α1,6-linked mannose residues from GlcNAcMan5GlcNAc2 producing GlcNAcMan3GlcNAc2. Golgi α-mannosidase II was considered to be essential for the conversion of N-glycan precursors to complex structures. However, when the mouse Golgi α-mannosidase II gene was inactivated, most cell types still produced complex N-glycans (6). This observation revealed that mice have at least one additional processing class II α-mannosidase, which compensated for the absence of Golgi α-mannosidase II. Furthermore, although null mouse cell extracts could not hydrolyze GlcNAcMan5GlcNAc, they could convert Man5GlcNAc to Man3GlcNAc. This suggested that N-glycan processing was mediated by an α-mannosidase activity, termed α-mannosidase III, which produced a novel Man3GlcNAc2 intermediate in these cells. However, the enzyme(s) responsible for this activity were not purified, nor was the gene encoding this enzyme identified.
In a previous study, we cloned and characterized a cDNA encoding a class II α-mannosidase from the lepidopteran insect cell line, Sf9 (7). The deduced amino acid sequence of this enzyme is similar to those of various mammalian Golgi α-mannosidases II. Like the latter enzymes, the Sf9 α-mannosidase is an integral membrane glycoprotein with type II topology. It can hydrolyze p-nitrophenyl α-D-mannopyranoside (pNP-α-Man), and it is inhibited by swainsonine. In the present study, we further examined the catalytic properties of this Sf9 class II α-mannosidase to more clearly determine its relationship to mammalian Golgi α-mannosidase II. The results showed that this enzyme is actually distinct from Golgi α-mannosidase II as its activity is stimulated by cobalt and it can hydrolyze various substrates containing terminal mannose residues but not GlcNAcMan5GlcNAc2. In view of these properties, we have designated this enzyme Sf9 α-mannosidase III (SfManIII), and we propose that it functions in an alternate N-glycan processing pathway in Sf9 cells.
EXPERIMENTAL PROCEDURES
Cells
Sf9 cells are derived from IPLB-Sf21-AE, a cell line originally isolated from Spodoptera frugiperda (fall armyworm) ovaries (8). Sf9 cells were maintained in suspension between 0.3 and 3.0 × 106 cells/ml in TNM-FH medium (9) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (HyClone, Logan, UT) and 0.1% (w/v) pluronic F68 (BASF Wynandotte Corp., Parsippany, NJ; Ref. 10).
Isolation of a Recombinant Baculovirus Expressing a Glutathione S-transferase (GST)-tagged, Secreted Form of SfManIII
A cDNA fragment encoding the SfManIII ectodomain (amino acids 35–1130) was subcloned into pAcSecG2T (PharMingen, San Diego, CA). The resulting plasmid, pAcGST-SfManIII, encoded a hybrid protein consisting of a cleavable signal peptide, the GST protein, and the SfManIII ectodomain under the transcriptional control of a baculovirus polyhedrin promoter. This plasmid was used to produce a recombinant baculovirus, AcGST-SfManIII, which expresses the GST-tagged, secreted form of SfManIII during the very late phase of infection. AcGST-SfManIII was produced by homologous recombination (9, 11) with Bsu36I-digested BacPAK6 viral DNA (12), plaque-purified twice, amplified in Sf9 cells, and titered by plaque assay (9).
Expression and Purification of GST-SfManIII
Sf9 cells were seeded at a density of 12 million cells/75-cm2 flask and infected at a multiplicity of infection of 5 plaque-forming units/cell with either AcGST-SfManIII or wild type baculovirus. The cells were cultured for 18 h in TNM-FH medium containing 10% (v/v) heat-inactivated fetal bovine serum, then the medium was removed, and the cells were washed and fed with Grace’s medium (13) containing 0.5% (v/v) heat-inactivated fetal bovine serum. At 36 h post infection, the medium was harvested and clarified by centrifugation for 5 min at 1,000 × g. The supernatant was harvested, and virus particles were removed by centrifugation for 30 min at 20,000 × g. This second supernatant was adjusted to 0.5% (v/v) Triton X-100 and applied at room temperature to a column containing 400 μl of immobilized glutathione-agarose (Pierce) equilibrated with buffer A (100 mM NaMES, pH 6.3, 0.5% Triton X-100). The flow-through was passed back over the column, then the column was washed with 10 ml of buffer A, and the packing was removed, resuspended in a final volume of 1 ml of buffer A, and used for enzyme assays.
For SDS-polyacrylamide gel electrophoresis (14) analyses, samples of the starting material, second flow-through, and wash were mixed with an equal volume of 2× protein sample buffer. A sample of the bound material was obtained by treating glutathione-agarose beads with an equal volume of 2× protein sample buffer. All samples were heated at 70 °C for 15 min and clarified by centrifugation at 13,000 × g for 5 min prior to the analysis. Proteins were visualized by either Coomassie Blue staining or immunoblotting. For immunoblots, proteins were electrophoretically transferred from the polyacrylamide gel to an Immobilon™-P membrane (Millipore Corp., Bedford, MA; Ref. 15). The membrane was then probed with rabbit anti-GST (Sigma) and alkaline phosphatase-conjugated goat anti-rabbit IgG (Promega, Inc., Madison, WI), and antibody binding was detected by a color reaction (16).
Mannosidase Activity Assays with pNP-α-Man
Hydrolysis of pNP-α-Man was assayed in a 37 °C incubator-shaker using 5 μl of GST-SfManIII in a final volume of 200 μl of buffer containing 50 mM NaMES, 0.25% (v/v) Triton X-100, and 5 mM pNP-α-Man. The enzyme was assayed in triplicate in each experiment and the results were presented as averages with standard deviations. To examine the effects of metal ions, SfManIII activity assays were done at pH 6.3 in the presence of 1 mM EDTA or various concentrations of CoCl2, MnCl2, ZnCl2, or NiCl2. For these assays, the enzyme was preincubated with EDTA or the metal ions for 2 h at 37 °C prior to the addition of pNP-α-Man. To determine the influence of pH, assays were done at various pH values in the presence of 1 mM CoCl2. To measure the effect of swainsonine, the assays were done at pH 6.3 in the presence of 1 mM CoCl2 and various concentrations of swainsonine (Genzyme Corp., Cambridge, MA). All assays were terminated by adding 800 μl of a buffer containing 133 mM glycine, 67 mM NaCl, and 83 mM Na2CO3 (pH 10.4). After removing the beads by centrifugation for 3 min at 13,000 × g, the absorbance of the supernatant was measured at 405 nm using a Beckman model DU® 7400 spectrophotometer (Beckman Instruments).
Mannosidase Activity Assays with Glycan Substrates
Hydrolysis of various pyridylamine (PA)-tagged glycans was assayed in a 37 °C incubator-shaker using 200 μl of GST-SfManIII in a final volume of 207 μl of buffer A supplemented with 1 mM CoCl2 and 100 pmol of substrate. After various incubation times, the beads were removed by centrifugation for 3 min at 13,000 × g, and the supernatants were boiled for 3 min. The reaction products were then resolved on a Hypersil APS-2 NH2 HPLC column as previously described (17).
Isolation of a Recombinant Baculovirus Expressing a GFP-tagged Form of SfManIII
A cDNA fragment encoding full-length SfManIII was subcloned into the immediate early baculovirus transfer plasmid, pAcP+E1TV3 (18). The sequence encoding enhanced green fluorescent protein, a mutant form of GFP with enhanced fluorescence at 507 nm (CLONTECH, Palo Alto, CA), was then inserted immediately downstream, in-frame with the SfManIII coding sequence. The resulting plasmid, pAcP+IE1-SfManIII-GFP, encoded a hybrid protein consisting of the full-length SfManIII protein with GFP fused to its C terminus under the transcriptional control of a baculovirus immediate early promoter. This plasmid was used to produce a recombinant baculovirus, AcP+IE1-SfManIII-GFP, which expresses the enhanced green fluorescent protein-tagged SfManIII protein during the immediate early phase of infection. AcP+IE1-SfManIII-GFP was produced by homologous recombination (9, 11) with Bsu36I-digested BacPAK6 viral DNA (12), plaque-purified twice, amplified in Sf9 cells, and titered by plaque assay (9).
Confocal Microscopy
Sf9 cells were seeded at a density of 300,000 cells/chamber into LAB-TEK® two-chamber slides (Nalge Nunc International Corp., Napeville, IL), infected with 3 plaque-forming units/cell of AcP+IE1-SfManIII-GFP, and cultured for 18 h in TNM-FH medium containing 10% (v/v) heat-inactivated fetal bovine serum. The cells were then incubated with either Golgi- or lysosome-specific dyes, rinsed with phosphate-buffered saline, and examined with a Leica TSD-4D CSLM confocal microscope (Leica, Heidelberg, Germany). To stain the Golgi, the cells were treated for 40–120 min at 27 °C with Grace’s medium containing 0.5% (v/v) heat-inactivated fetal bovine serum and 250 nM BODIPY®TR ceramide (Molecular Probes Inc., Eugene, OR). To stain the lysosomes, the cells were treated for 4 min at room temperature with Grace’s medium containing 0.5% (v/v) heat-inactivated fetal bovine serum and 25 nM LysoTracker™ Red DND-99 (Molecular Probes).
RESULTS
Expression and Purification of GST-SfManIII
AcGST-SfManIII is a recombinant baculovirus that encodes a GST-tagged, secreted form of SfManIII. Sf9 cells were infected with either this recombinant virus or a wild type control, and the extracellular media were harvested and analyzed by immunoblotting with rabbit anti-GST as described under “Experimental Procedures.” The results revealed that there were two specifically immunoreactive proteins in the extracellular growth medium from the AcGST-SfManIII-infected cells (Fig. 1A, lane 1). Neither was detected in the medium from wild type virus-infected cells (data not shown). The major protein had an apparent molecular mass of ~150 kDa suggesting that it is intact GST-SfManIII, which has a calculated molecular mass of 157.5 kDa. The other, relatively minor protein had an apparent molecular mass of ~130 kDa and is probably a degradation product. Another, weakly immunoreactive protein of ~68 kDa was also detected, but its presence in the negative controls (data not shown) indicated that it was not specific. Coomassie Blue staining revealed that the growth medium contained at least three proteins, including the 68-kDa protein, which were present in vast excess relative to the 150- and 130-kDa immunoreactive proteins (Fig. 1B, lane 1).
FIG. 1. Affinity purification of GST-SfManIII.
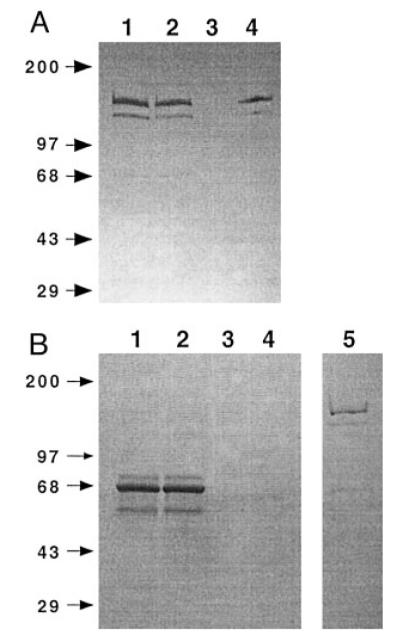
Extracellular medium from Sf9 cells infected for 36 h with AcGST-SfManIII was used for affinity purification as described under “Experimental Procedures.” Equivalent samples of medium (lanes 1), flow-through (lanes 2), wash (lanes 3), and bound material (lanes 4) were analyzed by SDS-polyacrylamide gel electrophoresis followed by immunoblotting with polyclonal anti-GST (panel A) or Coomassie Blue staining (panel B). A 50-fold excess of the Coomassie Blue-stained GST-SfManIII preparation is shown in panel B, lane 5. The positions of protein markers are indicated by their sizes (kDa) on the left.
Affinity chromatography on an immobilized glutathione column was used to isolate the extracellular immunoreactive proteins as described under “Experimental Procedures.” The affinity column retained the specifically immunoreactive proteins detected in the growth medium (compare Fig. 1A, lanes 1 and 4), but not the nonspecifically immunoreactive and non-immunoreactive proteins detected by Coomassie Blue staining (compare Fig. 1B, lanes 1 and 4). About half of the α-mannosidase activity in the medium was retained by the column as determined by pNP-α-Man assays (data not shown). Multiple attempts to elute active GST-SfManIII from the column with reduced glutathione were unsuccessful, but the column-bound enzyme had high levels of pNP-α-Man activity (data not shown). Therefore, the immobilized glutathione-agarose beads were removed from the column, resuspended in buffer A, and used as the source of GST-SfManIII for this study. The bound GST-SfManIII retained its activity for at least 1 week at 4 °C but was inactivated by freezing.
The Coomassie Blue-stained gel profile of an excess amount of the material extracted from the immobilized glutathione-agarose beads is shown in lane 5 of Fig. 1B. This analysis revealed that the SfManIII preparations consisted of a major protein with an apparent molecular mass of ~150 kDa, a minor protein with an apparent molecular mass of ~130 kDa, and no other detectable proteins. Presumably, the two proteins stained by Coomassie Blue corresponded to those detected by immunoblotting.
Biochemical Characterization of GST-SfManIII
A series of enzyme assays was performed to characterize the biochemical properties of GST-SfManIII. One set of assays was designed to evaluate the influence of various metal ions on GST-SfManIII activity (Table I). The addition of 1 mM EDTA reduced hydrolysis of pNP-α-Man to less than 10% of control levels indicating that GST-SfManIII has a divalent cation requirement. Indeed, GST-SfManIII activity was greatly enhanced by the addition of CoCl2 and, to a lesser extent, MnCl2. NiCl2 had little effect, and GST-SfManIII activity was inhibited by ZnCl2. The effect of calcium was not examined because TNM-FH and Grace’s media contain high concentrations of CaCl2.
TABLE I.
Influence of metal ions on GST-SFMANIII activity pNP-a-Man assays were performed in the presence of the indicated supplements, as described under “Experimental Procedures.” Activity levels are expressed as percentages of the level measured in control assays containing no supplements
| Supplement | 1 mM | 0.1 mM | 0.01 mM |
|---|---|---|---|
| % | % | % | |
| None | 100 | ||
| EDTA | 8.9 | ||
| CoCl2 | 2340.7 | 3019.1 | 858 |
| MnCl2 | 605.8 | 148.6 | 98 |
| ZnCl2 | 8.3 | 21.8 | 44 |
| NiCl2 | 116.7 | 119.5 | 108 |
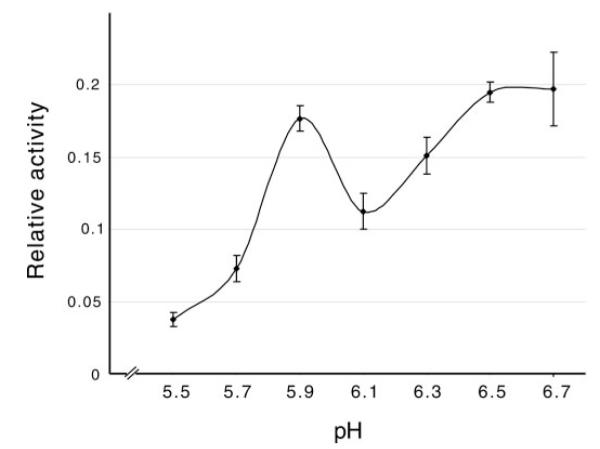
Another set of assays was performed to examine the influence of pH on GST-SfManIII activity against pNP-α-Man in the presence of 1 mM CoCl2 (Fig. 2). GST-SfManIII activity was negligible at pH 5.5, increased at pH 5.7 and 5.9, decreased at pH 6.1, and finally rose to a plateau at pH 6.5–6.7. These data showed that GST-SfManIII had significantly more activity around neutral pH than at acidic pH, which is consistent with the idea that it is a processing, not a lysosomal, enzyme. The reason for the reproducible decline in activity observed at pH 6.1 is unknown.
FIG. 2. Influence of pH on GST-SfManIII activity.
GST-SfManIII activity against pNP-α-Man was measured at various pH values as described under “Experimental Procedures.” The error bars in this and subsequent figures indicate the standard deviations obtained with triplicate samples.
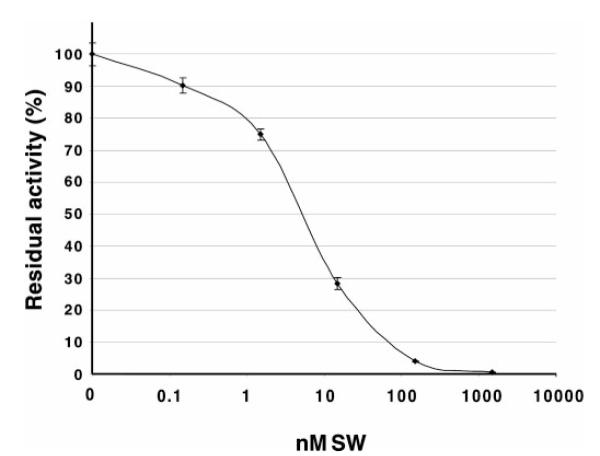
Finally, a set of pNP-α-Man assays was performed to determine the effect of swainsonine on GST-SfManIII activity (Fig. 3). GST-SfManIII was clearly sensitive to swainsonine in assays performed at pH 6.3 in the presence of 1 mM CoCl2. The IC50 was ~10 nM, and 1.5 μM swainsonine reduced GST-SfManIII activity to less than 0.5% of control values.
FIG. 3. Influence of swainsonine on GST-SfManIII activity.
GST-SfManIII activity against pNP-α-Man was measured in the presence of various concentrations of swainsonine (SW) as described under “Experimental Procedures.”
Thus, GST-SfManIII shares some biochemical properties with Golgi α-mannosidase II including its neutral pH optimum and swainsonine sensitivity, but its dependence on cobalt clearly distinguishes this enzyme from Golgi α-mannosidase II.
Substrate Specificity of GST-SfManIII
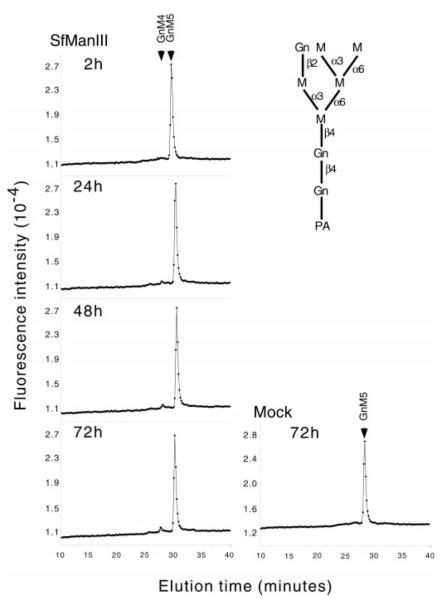
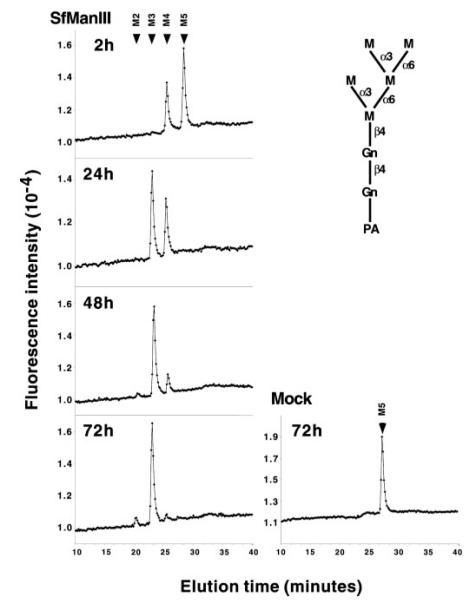
Whereas it was well established that Golgi α-mannosidase II hydrolyzes GlcNAcMan5GlcNAc2 to GlcNAcMan3GlcNAc2 (2), the natural substrate specificity of SfManIII had not been determined. Therefore, experiments were undertaken to examine the action of GST-SfManIII on various glycan substrates. GST-SfManIII had little effect on GlcNAcMan5GlcNAc2-PA even after extended incubation times (Fig. 4). Thus, GST-SfManIII failed to hydrolyze the natural substrate of Golgi α-mannosidase II. On the other hand, GST-SfManIII effectively hydrolyzed Man5GlcNAc2-PA through a Man4GlcNAc2-PA intermediate to Man3GlcNAc2-PA (Fig. 5). This activity was not due to contamination with endogenous α-mannosidases as there was no detectable hydrolysis when Man5GlcNAc2-PA was incubated for 72 h with a mock affinity-purified enzyme preparation from wild type baculovirus-infected Sf9 cells (Fig. 5, Mock). Thus, GST-SfManIII can hydrolyze the proposed substrate for α-mannosidase III, the enzyme thought to mediate the alternate N-glycan processing pathway in mice (6).
FIG. 4. Action of GST-SfManIII on GlcNAcMan5GlcNAc2.
GlcNAcMan5GlcNAc2-PA (top right) was incubated for the indicated times with GST-SfManIII, and the oligosaccharide products were analyzed by HPLC (left-hand panels) as described under “Experimental Procedures.” The panel on the bottom right shows the results of a control reaction in which GlcNAcMan5GlcNAc2-PA was incubated for 72 h with a mock-purified enzyme preparation made from cells infected with wild type baculovirus. The arrows mark the positions of glycan standards. GnM5 and GnM4 refer to GlcNAcMan5GlcNAc2-PA and GlcNAcMan4GlcNAc2-PA, respectively.
FIG. 5. Action of GST-SfManIII on Man5GlcNAc2.
Man5GlcNAc2-PA (top right) was incubated for the indicated times with GST-SfManIII, and the oligosaccharide products were analyzed by HPLC (left-hand panels) as described under “Experimental Procedures.” The panel on the bottom right shows the results of a control reaction in which Man5GlcNAc2-PA was incubated for 72 h with a mock-purified enzyme preparation made from cells infected with wild type baculovirus. The arrows mark the positions of glycan standards. M5 to M2 refer to Man(5–2)GlcNAc2-PA.
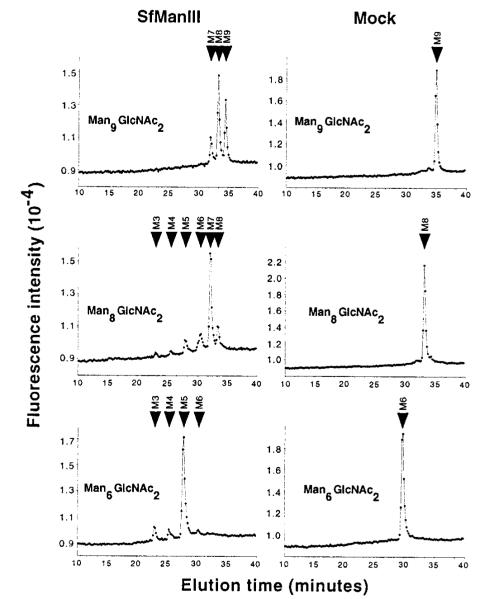
Considering its unusual substrate specificity, we also evaluated the ability of GST-SfManIII to hydrolyze other glycans, which had terminal α1,2-linked mannose residues (Fig. 6). GST-SfManIII was incubated for 72 h with Man9GlcNAc2-PA, Man8GlcNAc2-PA (mixed isomers) or Man6GlcNAc2-PA (isomer C with one α1,2-linked mannose residue on the middle arm). Over half of the Man9GlcNAc2-PA was consumed producing Man8GlcNAc2-PA and a small amount of Man7GlcNAc2-PA. Similarly, most of the Man8GlcNAc2-PA was consumed producing Man7GlcNAc2-PA and small amounts of Man6GlcNAc2-PA and Man5GlcNAc2-PA. Finally, Man6GlcNAc2-PA was completely consumed producing mainly Man5GlcNAc2-PA and small amounts of Man4GlcNAc2-PA and Man3GlcNAc2-PA. None of these activities were due to contaminating endogenous α-mannosidases as there was no detectable hydrolysis of any of these substrates when they were incubated for 72 h with a mock affinity-purified enzyme preparation from wild type baculovirus-infected Sf9 cells (Fig. 6, Mock column). These results demonstrated that GST-SfManIII can hydrolyze a variety of glycan substrates containing terminal α1,2-linked mannose residues in addition to Man5GlcNAc2-PA. Interestingly, SfManIII produced much less Man7GlcNAc2-PA from the Man9GlcNAc2-PA substrate than from the Man8GlcNAc2-PA (mixed isomer) substrate. The reason why Man9GlcNAc2-PA was not completely converted to Man7GlcNAc2-PA is unclear. It is possible that this simply reflects the relatively slow conversion of Man9GlcNAc2-PA to Man8GlcNAc2-PA (Fig. 6, top left panel). Alternatively, it is possible that this reflects the composition of the Man8GlcNAc2-PA (mixed isomer) substrate, which consisted of ~75% isomer A, 20% isomer B, and 5% isomer C (data not shown). This mixture might be a better substrate for SfManIII because isomers A and B are better substrates for SfManIII. In contrast, SfManIII digestion of Man9GlcNAc2-PA could yield a totally different mixture of Man8GlcNAc2-PA isomers, which is relatively resistant to further digestion by SfManIII. These possibilities were not examined in this study.
FIG. 6. Action of GST-SfManIII on Man(9–6)GlcNAc2.
Man9GlcNAc2-PA, Man8GlcNAc2-PA (mixed isomers), or Man6GlcNAc2-PA (isomer C) were incubated for 72 h with GST-SfManIII, and the oligosaccharide products were analyzed by HPLC (left-hand panels) as described under “Experimental Procedures.” The right-hand panels show the results of control reactions in which each glycan substrate was incubated for 72 h with a mock-purified enzyme preparation made from cells infected with wild type baculovirus. The arrows mark the positions of glycan standards. M9–M3 refer to Man(9–3)GlcNAc2-PA.
All of the assays of GST-SfManIII activity against glycan substrates were performed in the presence of 1 mM CoCl2. Therefore, additional assays were performed to specifically evaluate the cobalt requirement for Man5GlcNAc2-PA hydrolysis by GST-SfManIII. Hydrolysis was greatly reduced when CoCl2 was not added to the assays and was completely abolished in the presence of 5 mM EDTA (data not shown) indicating that cobalt is required for the hydrolysis of Man5GlcNAc2-PA by GST-SfManIII. The effect of swainsonine on Man5GlcNAc2-PA hydrolysis by GST-SfManIII was also evaluated, and the results were similar to those obtained in the pNP-α-Man assays (data not shown) indicating that hydrolysis of Man5GlcNAc2-PA by GST-SfManIII is sensitive to swainsonine, as well.
Intracellular Distribution of SfManIII-GFP
All of the properties of SfManIII were consistent with the interpretation that this enzyme plays a role in N-glycan processing in Sf9 cells. If SfManIII has this role in vivo, it should reside in the Golgi compartment. To examine its intracellular distribution, confocal microscopy was used to examine live Sf9 cells infected with a baculovirus vector encoding a full-length, GFP-tagged version of SfManIII. This baculovirus expression vector expresses SfManIII-GFP under the transcriptional control of a baculovirus promoter that is active immediately after infection and provides relatively low expression levels (18). This promoter was used to circumvent the potential problem of aberrant localization of SfManIII-GFP, which might happen if it had been overexpressed. Sf9 cells were infected with this baculovirus for 18 h, and then labeled with the red fluorescent dyes BODIPY®TR ceramide to stain the Golgi (Fig. 7, top panels) or LysoTracker™ Red DND-99 to stain the lysosomes (Fig. 7, bottom panels). Confocal microscopy revealed that the SfManIII-GFP was localized in punctate fluorescent structures dispersed throughout the cytoplasm but excluded from the nuclei(Fig. 7, left panels). This staining pattern overlapped significantly with the pattern obtained with the Golgi dye (Fig. 7, top middle panel) but not with the pattern obtained with the lysosomal dye (Fig. 7, bottom middle panel). It is noteworthy that a previous study established the dispersed nature of the Golgi elements in Sf9 cells (19). Thus, the confocal microscopy results described above indicate that SfManIII is localized in the Golgi compartment of Sf9 cells.
FIG. 7. Intracellular distribution of GST-SfManIII.
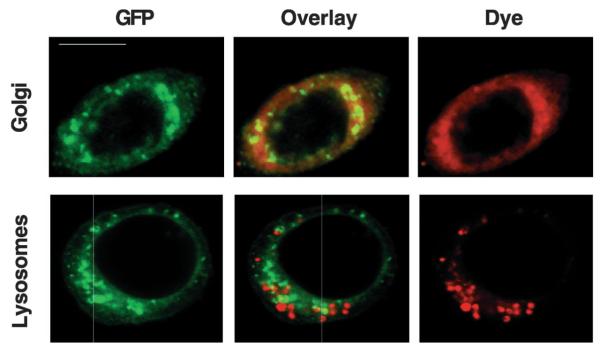
Sf9 cells were infected for 18 h with AcP(+)IE1-SfManIII-GFP, then labeled with red-fluorescent dyes specific for the Golgi (top panels) or lysosomes (bottom panels), and unfixed cells were examined by confocal microscopy. The panels on the left show the pattern of fluorescence obtained with SfManIII-GFP, whereas those on the right show the pattern obtained with the dyes. The panels in the middle show overlays of the panels on either side. Bar, 10 μ m.
DISCUSSION
This study demonstrated that Sf9 cells encode a class II α-mannosidase that is distinct from Golgi α-mannosidase II and has a unique set of properties. Based on these properties, and in accordance with the nomenclature used by Chui et al.(6), we named this enzyme SfManIII. Unlike Golgi α-mannosidase II, SfManIII is activated by cobalt, it can hydrolyze Man(9–5)GlcNAc2, and it can convert Man5GlcNAc2 to Man3GlcNAc2 but cannot hydrolyze GlcNAcMan5GlcNAc2. On the other hand, SfManIII has the same neutral pH optimum and intracellular distribution as Golgi α-mannosidase II. These properties, together with the fact that Man3GlcNAc2 can be elongated by N-acetylglucosaminyltransferase I (20, 21) and further processed to complex structures, suggest that SfManIII functions in N-glycan processing in Sf9 cells. However, the current study did not establish whether or not SfManIII actually plays this role in vivo.
The early steps of N-glycan processing, including mannose trimming, appear to be similar or identical in Sf9 and mammalian cells (22, 23). A cDNA encoding a class I α-mannosidase has been cloned from Sf9 cells, and its product has been extensively characterized (19, 24, 25). The results of these studies have shown that SfManI is virtually identical in structure, function, and intracellular distribution to the mammalian α-mannosidases IA and IB. In addition, Sf9 cells have a class II α-mannosidase that acts on GlcNAcMan5GlcNAc2 without a metal ion requirement and appears to be functionally identical to mammalian Golgi α-mannosidase II (26). In contrast, whereas SfManIII has the biochemical properties of Class II mannosidases, a mammalian ortholog with similar catalytic characteristics has not yet been purified or cloned (5).
Considering the similarities between the other processing α-mannosidases of Sf9 and mammalian cells, there is probably a mammalian ortholog of SfManIII as well. One likely candidate is the cobalt-activated α-mannosidase activity(ies) that convert Man5GlcNAc to Man3GlcNAc in null mice lacking Golgi α-mannosidase II (6). This activity, termed α-mannosidase III, compensates for the absence of Golgi α-mannosidase II in these mice by providing an alternate pathway for N-glycan processing. However, it is currently impossible to unequivocally decipher the relationship between this activity and SfManIII because the mouse enzyme(s) that provide this activity have not yet been identified. In fact, several enzymes probably contribute to the α-mannosidase III activity detected in mouse cell lysates including both swainsonine-resistant enzymes that are not involved in N-glycan processing, as well as one or more swainsonine-sensitive, processing enzymes.
One mammalian enzyme that could account for the Man5GlcNAc-hydrolyzing activity in the null mouse cell lysates is an α-mannosidase first characterized in BHK cells (27) and later purified from rat liver (28, 29). This BHK/rat liver enzyme is cobalt-activated, relatively resistant to swainsonine, and can convert Man5GlcNAc2 to Man3GlcNAc2 but cannot hydrolyze pNP-α-Man. This enzyme does not appear to be a processing enzyme because there is no complex N-glycan biosynthesis in BHK cells treated with swainsonine (30) indicating that all of the processing class II α-mannosidases in these cells are swainsonine sensitive. But, like the null mice discussed above, mutant BHK cells lacking detectable Golgi α-mannosidase II activity can still synthesize some complex N-glycans (31). Thus, these cells must have another, swainsonine sensitive α-mannosidase III activity that provides the alternate N-glycan processing pathway.
A second mammalian enzyme that could account for the α-mannosidase III activity is an isozyme of Golgi α-mannosidase II that has been termed α-mannosidase IIx (32). This enzyme shares sequence similarity with both the human and insect class II α-mannosidases, but its N-glycan substrate specificity has not yet been characterized.
There also is another group of mammalian enzymes, generally classified as ER/cytosolic α-mannosidases, which could account for the Man5GlcNAc-hydrolyzing activity detected in the null mouse cell lysates. These enzymes are activated by cobalt, are relatively resistant to swainsonine, and can convert Man5GlcNAc to Man3GlcNAc. However, they act efficiently only on glycans with a single GlcNAc residue on their reducing end and appear to function in N-glycan catabolism rather than processing (33–36).
SfManIII is clearly distinct from both the swainsonine resistant BHK/rat liver enzyme and the ER/cytosolic α-mannosidases. SfManIII is sensitive to swainsonine, can hydrolyze pNP-α-Man, and can efficiently hydrolyze glycans with chitobiose cores. Considering these properties and the fact that Sf9 cells have another α-mannosidase, which is functionally equivalent to mammalian Golgi α-mannosidase II (26), SfManIII is probably equivalent to the enzyme providing the alternate pathway in the null mice (6) and mutant BHK cells (31). However, an unequivocal test of this hypothesis awaits purification of the mammalian enzyme(s) responsible for the α-mannosidase III activity.
Acknowledgments
We thank Carla Weinkauf and Brian Francis for technical assistance during the preliminary phases of this study.
Footnotes
This work was supported by National Institutes of Health Grants GM49734 (to D. L. J.) and GM47533 and RR05351 (to K. W. M.).
The abbreviations used are: ER, endoplasmic reticulum; pNP-α-Man, p-nitrophenyl α-D-mannopyranoside; SfManIII, Spodoptera frugiperda α-mannosidase III; GST, glutathione S-transferase; NaMES, sodium 2-[morpholino]ethanesulfonic acid; PA, pyridylamine; HPLC, high pressure liquid chromatography; BHK, baby hamster kidney.
REFERENCES
- 1.Kornfeld R, Kornfeld S. Ann. Rev. Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 2.Moremen KW, Trimble RG, Herscovics A. Glycobiology. 1994;4:113–125. doi: 10.1093/glycob/4.2.113. [DOI] [PubMed] [Google Scholar]
- 3.Daniel PF, Winchester B, Warren CD. Glycobiology. 1994;4:551–566. doi: 10.1093/glycob/4.5.551. [DOI] [PubMed] [Google Scholar]
- 4.Herscovics A. Biochim. Biophys. Acta. 1999;1473:96–107. doi: 10.1016/s0304-4165(99)00171-3. [DOI] [PubMed] [Google Scholar]
- 5.Moremen KW. In: Oligosaccharides in Chemistry and Biology: A Comprehensive Handbook. Ernst B, Hart G, Sinay P, editors. Vol. 2. John Wiley and Sons, Inc.; New York: 2000. pp. 81–117. [Google Scholar]
- 6.Chui D, Oh-Eda M, Liao YF, Panneerselvam K, Lal A, Marek KW, Freeze HH, Moremen KW, Fukuda MN, Marth JD. Cell. 1997;90:157–167. doi: 10.1016/s0092-8674(00)80322-0. [DOI] [PubMed] [Google Scholar]
- 7.Jarvis DL, Bohlmeyer DA, Liao YF, Lomax KK, Merkle RK, Weinkauf C, Moremen KW. Glycobiology. 1997;7:113–127. doi: 10.1093/glycob/7.1.113. [DOI] [PubMed] [Google Scholar]
- 8.Vaughn JL, Goodwin RH, Thompkins GJ, McCawley P. In Vitro. 1977;13:213–217. doi: 10.1007/BF02615077. [DOI] [PubMed] [Google Scholar]
- 9.Summers MD, Smith GE. Tex. Agric. Exp. Stn. Bull. 1987:1555. [Google Scholar]
- 10.Murhammer DW, Goochee CF. Bio/Technology. 1988;6:1411–1418. [Google Scholar]
- 11.O’Reilly DR, Miller LK, Luckow VA. Baculovirus Expression Vectors. W. H. Freeman and Company; New York: 1992. [Google Scholar]
- 12.Kitts PA, Possee RD. BioTechniques. 1993;14:810–817. [PubMed] [Google Scholar]
- 13.Grace TD. Nature. 1962;195:788–789. doi: 10.1038/195788a0. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Towbin H, Staehelin T, Gordon J. Proc. Natl. Acad. Sci. U. S. A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blake MS, Johnston KH, Russell-Jones GJ, Gotschlich EC. Anal. Biochem. 1984;136:175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez DS, Karaveg K, Vandersall-Nairn AS, Lal A, Moremen KW. J. Biol. Chem. 1999;274:21375–21386. doi: 10.1074/jbc.274.30.21375. [DOI] [PubMed] [Google Scholar]
- 18.Jarvis DL, Weinkauf C, Guarino LA. Protein. Expression Purif. 1996;8:191–203. doi: 10.1006/prep.1996.0092. [DOI] [PubMed] [Google Scholar]
- 19.Kawar Z, Jarvis DL. Insect Biochem. Mol. Biol. 2001;31:289–297. doi: 10.1016/s0965-1748(00)00121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schachter H. Biochem. Cell Biol. 1986;64:163–181. doi: 10.1139/o86-026. [DOI] [PubMed] [Google Scholar]
- 21.Altmann F, Kornfeld G, Dalik T, Staudacher E, Glossl J. Glycobiology. 1993;3:619–625. doi: 10.1093/glycob/3.6.619. [DOI] [PubMed] [Google Scholar]
- 22.Marz L, Altmann F, Staudacher E, Kubelka V. In: Glycoproteins. Montreuil J, Vliegenthart JFG, Schachter H, editors. Vol. 29a. Elsevier Science Publishers B.V.; Amsterdam: 1995. pp. 543–563. [Google Scholar]
- 23.Jarvis DL, Kawar ZS, Hollister JR. Curr. Opin. Biotechnol. 1998;9:528–533. doi: 10.1016/s0958-1669(98)80041-4. [DOI] [PubMed] [Google Scholar]
- 24.Kawar Z, Herscovics A, Jarvis DL. Glycobiology. 1997;7:433–443. doi: 10.1093/glycob/7.3.433. [DOI] [PubMed] [Google Scholar]
- 25.Kawar Z, Romero PA, Herscovics A, Jarvis DL. Glycobiology. 2000;10:347–355. doi: 10.1093/glycob/10.4.347. [DOI] [PubMed] [Google Scholar]
- 26.Ren J, Castellino FJ, Bretthauer RK. Biochem. J. 1997;324:951–956. doi: 10.1042/bj3240951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monis E, Bonay P, Hughes RC. Eur. J. Biochem. 1987;168:287–294. doi: 10.1111/j.1432-1033.1987.tb13419.x. [DOI] [PubMed] [Google Scholar]
- 28.Bonay P, Hughes RC. Eur. J. Biochem. 1991;197:229–238. doi: 10.1111/j.1432-1033.1991.tb15903.x. [DOI] [PubMed] [Google Scholar]
- 29.Bonay P, Roth J, Hughes RC. Eur. J. Biochem. 1992;205:399–407. doi: 10.1111/j.1432-1033.1992.tb16793.x. [DOI] [PubMed] [Google Scholar]
- 30.Foddy L, Feeney J, Hughes RC. Biochem. J. 1986;233:697–706. doi: 10.1042/bj2330697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes RC, Feeney J. Eur. J. Biochem. 1986;158:227–237. doi: 10.1111/j.1432-1033.1986.tb09742.x. [DOI] [PubMed] [Google Scholar]
- 32.Misago M, Liao YF, Kudo S, Eto S, Mattei MG, Moremen KW, Fukuda MN. Proc. Natl. Acad. Sci. U. S. A. 1995;92:11766–11770. doi: 10.1073/pnas.92.25.11766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bischoff J, Moremen K, Lodish HF. J. Biol. Chem. 1990;265:17110–17117. [PubMed] [Google Scholar]
- 34.Weng S, Spiro RG. Arch. Biochem. Biophys. 1996;325:113–123. doi: 10.1006/abbi.1996.0014. [DOI] [PubMed] [Google Scholar]
- 35.Grard T, Herman V, Saint-Pol A, Kmiecik D, Labiau O, Mir AM, Alonso C, Verbert A, Cacan R, Michalski JC. Biochem. J. 1996;316:787–792. doi: 10.1042/bj3160787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamashiro K, Itoh H, Yamagishi M, Natsuka S, Mega T, Hase S. J. Biochem., (Tokyo) 1997;122:1174–1181. doi: 10.1093/oxfordjournals.jbchem.a021878. [DOI] [PubMed] [Google Scholar]