Abstract
Although cell-matrix adhesive interactions are known to regulate stem cell differentiation, the underlying mechanisms, in particular for direct three-dimensional (3D) encapsulation within hydrogels, are poorly understood. Here, we demonstrate that in covalently crosslinked hyaluronic acid (HA) hydrogels, the differentiation of human mesenchymal stem cells (hMSCs) is directed by the generation of degradation-mediated cellular-traction, independent of cell morphology or matrix mechanics. hMSCs within HA hydrogels of equivalent elastic moduli that either permit (restrict) cell-mediated degradation exhibited high (low) degrees of cell spreading and high (low) tractions, and favoured osteogenesis (adipogenesis). In addition, switching the permissive hydrogel to a restrictive state via delayed secondary crosslinking reduced further hydrogel degradation, suppressed traction, and caused a switch from osteogenesis to adipogenesis in the absence of changes to the extended cellular morphology. Also, inhibiting tension-mediated signalling in the permissive environment mirrored the effects of delayed secondary crosslinking, whereas upregulating tension induced osteogenesis even in the restrictive environment.
Adhesive interactions with extracellular matrix direct many aspects of cell behaviour, including viability1–3, morphogenesis4,5, and differentiation6,7. As such, it is important to understand interfacial interactions between stem cells and biomaterials towards their utility in therapeutic applications. For cells seeded on hydrogels, the modulus of the substrate can influence stem cell spreading, traction generation, and fate8–10, including in the absence of soluble differentiation factors11. Beyond the modulus, stem cell fate atop 2D substrates can also be directed via geometric constraints on cell adhesion or surface topography, both of which restrict cell spreading and tension generation12–15. Despite these advances, the influence of biophysical properties on stem cell fate when presented a 3D environment is not well understood. Heubsch et al.16 recently showed that within non-degradable, ionically crosslinked alginate hydrogels, encapsulated mesenchymal stem cell (MSC) differentiation is dictated by matrix stiffness irrespective of cell morphology since MSCs remained rounded independent of stiffness. Specifically, despite the lack of hydrogel degradation, the physically crosslinked alginate was adequately mobile to enable cellular re-organization of bound adhesive ligands, traction generation, and differentiation, with magnitudes and fate dependent on hydrogel crosslink density (i.e., matrix stiffness). However, many hydrogels behave quite differently from physically crosslinked alginate hydrogels on a molecular level. For example, covalently crosslinked hydrogels exhibit bonds that are stable rather than dynamic. Given the significant amount of work using covalently crosslinked hydrogels for stem cell encapsulation17–22, it is important to understand how differences in hydrogel structure and behavior modulate encapsulated stem cell fate. The work presented here reveals that fate is regulated by cell-generated tension that is enabled through cell-mediated degradation of the covalently crosslinked matrix, and emphasizes that the mechanisms by which stem cells respond to biophysical cues are highly dependent on the type of hydrogel used.
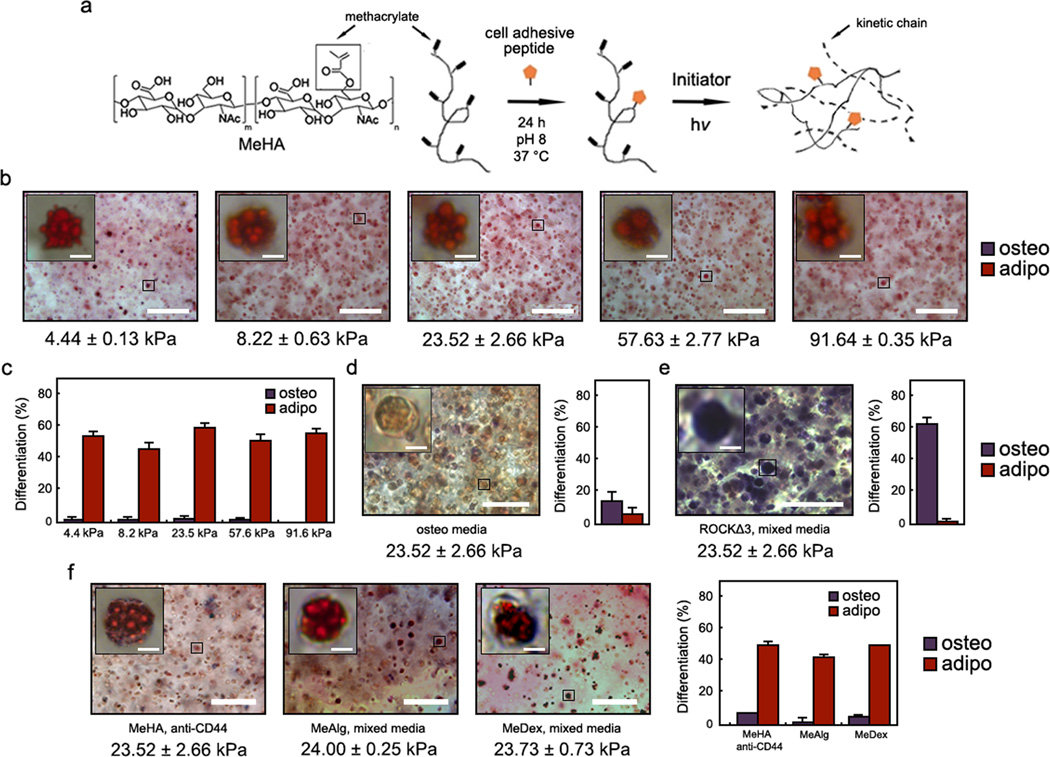
To assess the influence of crosslink density in covalently crosslinked hydrogels on encapsulated human MSC (hMSC) fate (cultured in a bipotential adipogenic/osteogenic media formulation), we replicated the experiment performed in physically crosslinked alginate gels with photopolymerized RGD-modified methacrylated hyaluronic acid (MeHA) hydrogels (Fig. 1a), where hydrogel moduli were tuned by MeHA macromer concentration23,24. In contrast to the crosslink density dependent response within alginate gels, hMSCs in MeHA gels of all moduli (~4 – 91 kPa) underwent almost exclusively adipogenesis relative to osteogenesis, as visualized by dual histological staining (Fig. 1b) and quantification (Fig. 1c) for alkaline phosphatase (ALP, osteogenesis) and neutral lipids (adipogenesis). Staining for nuclei confirmed that the lipid droplets corresponded to single cells rather than clusters (Supplementary Fig. S1). When hMSCs were blocked for CD44 interactions with HA via incubation with primary antibodies prior to encapsulation into MeHA hydrogels, or when untreated hMSCs were encapsulated within methacrylated alginate (MeAlg) or methacrylated dextran (MeDex) hydrogels that lack any HA moieties, the same trend in differentiation was observed across a similar range of mechanics (~4 – 95 kPa; Supplementary Fig. S2), including at an elastic modulus corresponding to osteogenesis in the physically crosslinked alginate system (~20 kPa; Fig. 1f). These findings suggest that the hydrogel structural cues resulting from covalent crosslinking, rather than direct interactions with the HA chemistry itself, mediate hMSC behavior and fate.
Figure 1.
hMSC matrix interactions and fate choice within photopolymerized MeHA hydrogels. a, Schematic of RGD conjugation to and photopolymerization of MeHA. b,d,e, Representative brightfield images of hMSCs within MeHA gels following b, 7 days mixed osteogenic/adipogenic media incubation, d, 7 days osteogenic media incubation, or e, mixed osteogenic/adipogenic media with encapsulated hMSCs transfected with constitutively active ROCK (ROCKΔ3). Oil red O (ORO) stains neutral lipids (adipogenesis) red and fast blue RR stains alkaline phosphatase (ALP, osteogenesis) purple. c–e, Percentage differentiation of hMSCs toward osteogenic or adipogenic lineages for these same groups. f, Representative brightfield images and percentage differentiation of hMSCs following 7 days mixed media incubation within MeHA hydrogels (with hMSCs incubated with primary anti-CD44 antibodies prior to encapsulation), MeAlg hydrogels, or MeDex hydrogels of elastic modulus corresponding to osteogenesis in the physically crosslinked alginate system (~20 kPa). For all mixed media groups, the percentage differentiation was significantly different between osteogenesis and adipogenesis (p < 0.001, t test). Error bars represent standard errors for the mean. Scale bars: b,d,e,f 100 µm, 5 µm (inset).
hMSCs transfected with a GFP-vinculin reporter and stained for actin showed limited focal adhesion formation and diffuse, unpolymerized actin that decreased in expression throughout the 7 d culture (Supplementary Fig. S3). In contrast, hMSCs seeded atop MeHA gels of similar elastic modulus (~25 kPa) and presenting the same RGD concentration exhibited punctate focal adhesions (Supplementary Fig. S4) and underwent primarily osteogenic differentiation9. 3D traction force microscopy (3D TFM)25 was used to monitor embedded bead displacements and revealed minimal deformation of the surrounding gels by encapsulated hMSCs in all formulations (Supplementary Fig. S5). Cell spreading was quantified as a dimensionless metric termed “circularity” (ranging from 0 to 1, with values near 0 representing highly spread cells and near 1 representing rounded cells) (Supplementary Fig. S6); high hMSC circularity (i.e., little spreading) was observed across the range of mechanics (Supplementary Fig. S5). hMSC encapsulation within 23.5 kPa (a modulus that led to osteogenesis in physically crosslinked alginate gels) MeHA gels was then repeated with hMSCs infected with lentivirus containing constitutively active Rho kinase (ROCKΔ3). The ROCKΔ3 hMSCs underwent a fate switch from adipogenesis to osteogenesis (Fig. 1e) despite remaining rounded, suggesting that the formation of load-bearing adhesions toward an osteogenic phenotype can only be rescued by manual activation of tension in a non-degradable, covalently crosslinked hydrogel system. In contrast, when MeHA gels containing non-transfected hMSCs were incubated in osteogenic media alone, little differentiation was observed (Fig. 1d). Taken together, these findings indicate, consistent with previous reports14,15, that ROCK-induced cytoskeletal tension is downstream of adhesive and soluble microenvironmental cues and is required for osteogenic versus adipogenic hMSC differentiation. Further, these results clearly illustrate differences in hMSC adhesion and differentiation based on the type (i.e., ionically crosslinked alginate versus covalently crosslinked MeHA) and dimensionality (i.e., 2D versus 3D) of the hydrogel used.
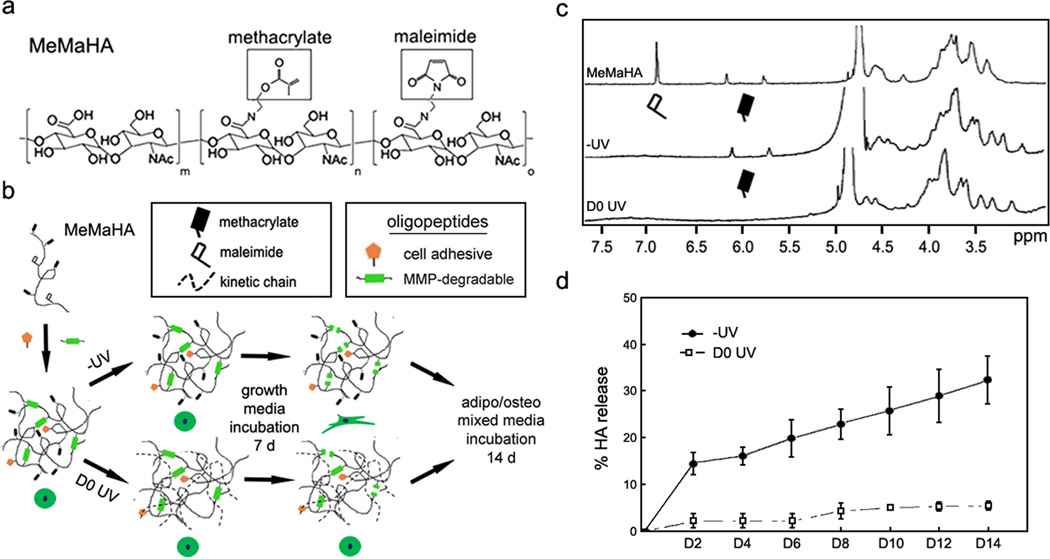
The above work indicates that crosslink density has little influence on stem cell fate in non-degradable covalently crosslinked systems, even when the network presents adhesive ligands; however, cell-mediated degradation can also be incorporated into these systems through the introduction of proteolytically cleavable crosslinks26–29. To accomplish this, HA was functionalized with both methacrylate and maleimide groups (MeMaHA; Fig. 2a; 14% & 10.5% modification, respectively) and subjected to a multi-step crosslinking protocol for cellular encapsulation (Fig. 2b). In the primary crosslinking step, a “−UV” hydrogel was formed using Michael-type reactions between MeMaHA maleimides and thiols on monofunctional cell adhesive oligopeptides and bifunctional MMP-degradable peptides (100% theoretical maleimide consumption). In a secondary step, −UV hydrogels were incubated with I2959 photoinitiator and exposed to light to initiate free radical photopolymerization of methacrylates (“D0 UV”), introducing kinetic chains that impede proteolytic degradation. 1H NMR analysis of −UV and D0 UV hydrogels (solubilized by incubation with hyaluronidases) demonstrated that primary and secondary crosslinking consumed all maleimides and methacrylates, respectively (Fig. 2c). When incubated with 20 nM exogenous MMP-2 for 14 d (Fig. 2d), −UV hydrogels exhibited rapid HA release consistent with proteolytic degradation, while D0 UV gels exhibited little HA release. The same trends were observed upon incubation of −UV and D0 UV gels with 10 nM MT1-MMP (pro and catalytic form) (Supplementary Fig. S7), illustrating that the oligopeptide sequence used in this study is susceptible to degradation by multiple proteases, and that secondary polymerization universally restricts proteolytic degradation. To investigate the influence of cell-mediated degradation on hMSC behavior, the MeMaHA macromer concentrations used were tuned to provide similar initial elastic moduli for formulations either “permissive” (−UV) or “inhibitory” (D0 UV) to cell-mediated degradation (2.5 wt% MeMaHa for −UV hydrogels: E = 4.30 ± 0.11 kPa, 1.5 wt% MeMaHA for D0 UV hydrogels: E = 4.49 ± 0.18 kPa).
Figure 2.
MeMaHA sequential crosslinking schematic, characterization & proteolytic degradation kinetics. a, MeMaHA chemical structure (m = .755, n = .14, o = .105). b, Schematic of sequential crosslinking of MeMaHA using a primary addition and (nominally) secondary radical polymerization to create “−UV” and “D0 UV” hydrogels, respectively. c, 1H NMR spectra (D2O) showing uncrosslinked MeMaHA polymer, −UV and D0 UV hydrogels, respectively. d, Degradation kinetics of −UV and D0 UV hydrogels in the presence of 20 nM MMP-2. For all timepoints % HA release was greater from −UV relative to D0 UV gels (p < 0.01, t test). Error bars represent standard errors for the mean.
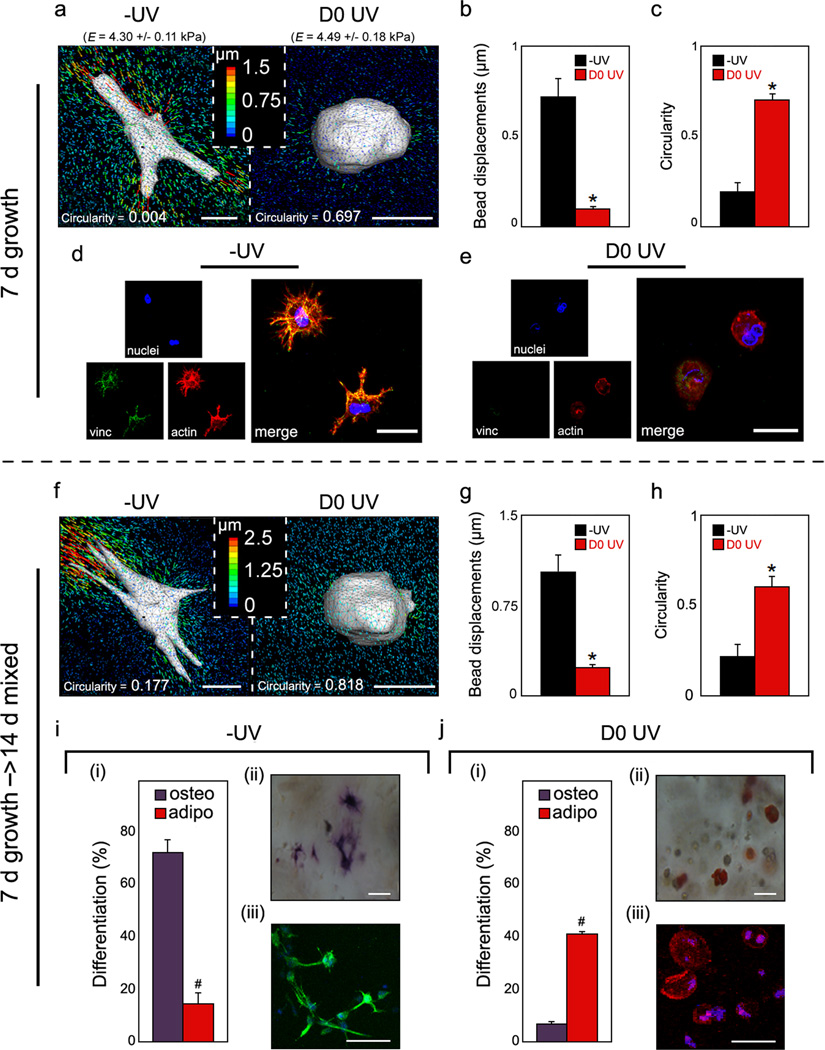
To assess the importance of local degradability on cell behaviour, hMSCs were uniformly encapsulated by re-suspension in the polymer solution immediately prior to addition of the crosslinker peptide (Supplementary Fig. S8). At day 7 of growth media incubation, hMSCs spread within −UV hydrogels and deformed the surrounding matrix to a greater extent than rounded hMSCs within D0 UV gels (Fig. 3a). Bead displacements (~7-fold greater in −UV versus D0 UV) and cellular circularity (~3.5-fold greater in D0 UV versus −UV) were found to be significantly different between the gels (Fig. 3b–c). Rheological testing confirmed that bead encapsulation did not alter MeMaHA gelation or the final elastic modulus (Supplementary Fig. S9). Staining for actin demonstrated a robust network of stress fibers within cells in −UV gels, with vinculin concentrated at the tips of extended processes (Fig. 3d). In contrast, only diffuse, depolymerized actin and no organized vinculin were observed within cells in D0 UV gels (Fig. 3e), suggesting that secondary crosslinking restricted hMSC matrix adhesion and cytoskeletal organization as observed in MeHA gels. To confirm that this switch was mediated by a change in degradation cues and not an adverse effect of light exposure on the hMSCs, cell viability and total DNA content were evaluated in the −UV and D0 UV conditions, as well as in gels exposed to light in the absence of photoinitiator (UV + light), after 7 days growth media incubation (Supplementary Fig. S10). Similarly high viability was observed in all conditions; additionally, total DNA content, as well as hMSC circularity, were similar between the −UV and −UV + light groups, indicating that light exposure did not adversely affect the encapsulated hMSCs. MALDI analysis confirmed that light exposure did not damage the RGD-containing cell adhesive oligopeptide (Supplementary Fig. S11). Further, the concentration of RGD peptide was quantitatively determined to be equivalent between −UV and D0 UV gels (Supplementary Fig. S12). Encapsulation of hMSCs into MeMaHA gels without RGD peptide resulted in rounded cells exhibiting lower viability and minimal tractions (Supplementary Fig. S13), indicating that the hMSC spreading and traction responses were mediated through integrin-RGD binding. Although this specific ligand may not be necessary for the observed results, adhesion appears to be needed for hydrogel degradation and traction generation. Finally, gene expression and biochemical staining for ALP and lipids indicated that encapsulated hMSCs remained undifferentiated after the growth media incubation period (Supplementary Fig. S14).
Figure 3.
MeMaHA hydrogel structure-dependent hMSC matrix interactions & fate choice. a,f, Representative 3D TFM images of hMSCs, b,g, average drift-corrected bead displacements within 15 µm of the cell surface (*p < 0.001, t test), and c,h, average circularity of hMSCs within −UV or D0 UV cells (*p < 0.001, t test), following a–c, 7 days incubation in growth media or f–h, an additional 14 days incubation in mixed osteogenic/adipogenic media. d–e, Representative staining for hMSC vinculin (green), actin (red), and nuclei (blue) in d −UV and e D0 UV gels. i–j, hMSC differentiation following 14 d mixed media incubation. i(i),j(i), percentage differentiation of hMSCs toward osteogenic or adipogenic lineages in i(i) −UV or j(i) D0 UV hydrogels (#p < 0.005, t test). i(ii–iii)–j(ii–iii), Representative bright field and fluorescent images of hMSCs; i(ii),j(ii), staining for ALP (osteogenesis) and lipid droplets (adipogenesis), or i(iii),j(iii), immunocytochemistry for osteocalcin (OC, osteogenesis) and fatty acid binding protein (FABP, adipogenesis) in i(ii–iii), −UV or j(ii–iii), D0 UV hydrogels, respectively. Error bars represent standard errors for the mean. Scale bars: a,f, 10 µm; d,e, 15 µm; i(ii),j(ii), 25 µm; i(iii),j(iii), 20 µm.
Upon switching the media to a bipotential adipo/osteo media for 14 days following the 7 day growth media incubation (i.e., for day 7 – 21 of culture), the same population trends in cell spreading and traction generation were observed (Fig. 3f–h). With respect to differentiation, hMSCs within −UV and D0 UV gels underwent primarily osteogenesis and adipogenesis, respectively, based on dual staining for ALP and lipids (Fig. 3i(ii), 3j(ii)). Lineage commitment was quantified by counting cells stained for each marker and dividing by total nuclei; osteogenesis was significantly greater in −UV (72.4% ± 10.0%) versus D0 UV (14.9% ± 8.1%) gels, while adipogenesis was significantly greater in D0 UV (41.1% ± 1.5%) versus −UV (5.5% ± 0.3%) gels. The population differentiation trends were confirmed at day 21 via immunocytochemistry for osteocalcin (OC, osteogenesis) and fatty acid binding protein (FABP, adipogenesis) (Fig. 3i(iii), 3j(iii)) and via gene expression (Supplementary Fig. S14). Please note that there is upregulation for both adipogenic and osteogenic genes in all systems after culture in bipotential media; yet, this only translates to protein staining with the appropriate environmental signals. These same differentiation trends were observed when hMSCs were encapsulated at the seeding density corresponding to 3D TFM studies (60,000 cells mL−1) (Supplementary Fig. S15), confirming that cellular traction generation via matrix adhesion, rather than cell density and associated cell-cell interactions, directed cell fate. Additionally, the images acquired for TFM showed uniform embedded bead distribution immediately surrounding the cells; this suggests that hMSC-mediated degradation in this system was localized to the site of initial encapsulation, and that cell motility and migration was not a significant factor. Thus, within hydrogels of the same initial modulus, osteogenesis was favoured in systems where cells were able to spread and pull on the surrounding matrix and adipogenesis was favoured in systems where cells remained rounded and were unable to displace the surrounding matrix. It is also interesting to note that these moduli are much lower than those that supported hMSC spreading and osteogenesis in 2D8,9,30,31, again highlighting dimensionality as a biophysical cue that impacts stem cell behaviour.
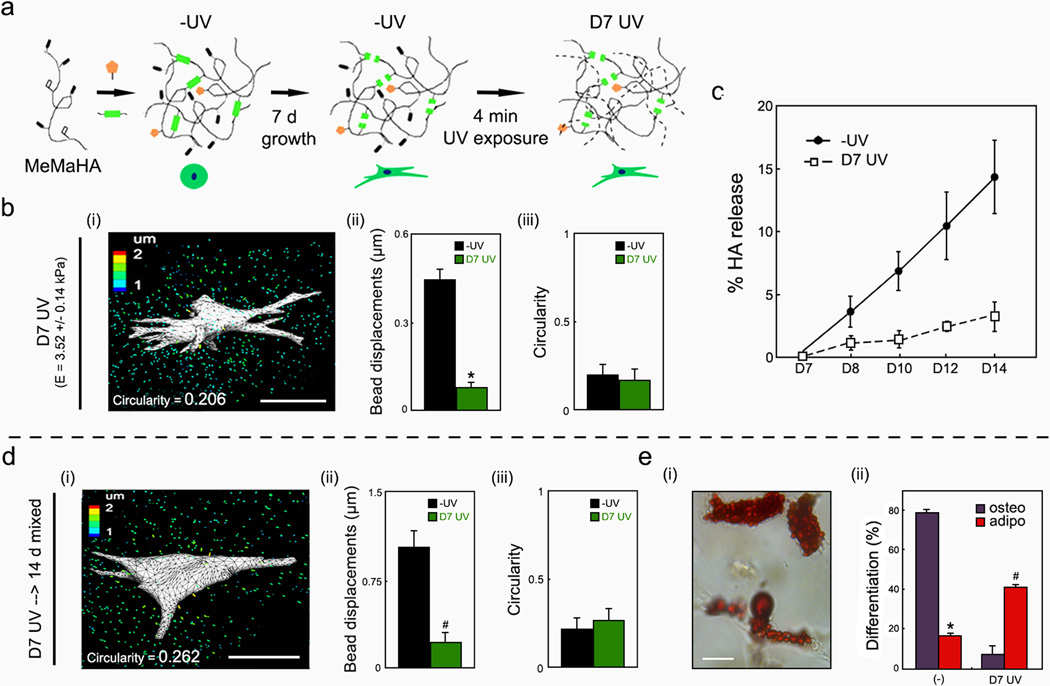
To decouple the influence of spreading and hydrogel degradation, the MeMaHA system was again used, but the secondary crosslinking was applied at day 7 (“D7 UV”), after the hMSCs were allowed to spread. This delayed crosslinking resulted in a significant increase in the elastic modulus and a significant decrease in bead displacements compared to the same hydrogel formulation without UV exposure, as well as prevented further degradation of the network (Supplementary Fig. S16). Specifically, acellular D7 UV hydrogels exhibited minimal degradation when incubated with 20 nM MMP-2 compared to −UV gels (Fig. 4c), indicating that delayed crosslinking impedes further proteolytic degradation in a manner similar to D0 UV exposure. 3D TFM analysis revealed significantly reduced deformations within D7 UV gels 24 hours after exposure relative to control gels of the same hydrogel formulation without UV exposure at the same time; however, these gels have different moduli (Supplementary Fig. S16). Bead displacements were also reduced when compared to −UV gels (Fig. 4b(i–ii)) with similar moduli (2.5 wt% MeMaHa for −UV hydrogels: E = 4.30 ± 0.11 kPa, 1.5 wt% MeMaHA for D7 UV hydrogels: E = 3.52 ± 0.14 kPa following crosslinking) and no differences in cell morphology (Fig. 4b(iii)). Thus, the introduction of non-degradable crosslinks after cell spreading prevented further deformation of the matrix. It was also confirmed that the exposure of hMSCs to UV light itself (i.e., in the absence of photoinitiator) did not affect cellular tractions (Supplementary Fig. 17). Following incubation of the −UV and D7 UV groups with similar moduli in mixed inductive media for an additional 14 days, there was no change in cell traction or morphology (Fig. 4d(i–iii)); however, adipogenesis was significantly increased relative to osteogenesis in the D7 UV hydrogels, resulting in morphologically spread cells with extensive lipid droplet formation (Fig. 4e). Additionally, hMSCs within the D7 UV gels exhibited minimal cytoskeletal organization, in contrast to robust cytoskeletal organization within −UV gels (Supplementary Fig. S18). Thus, these findings indicate that the ability of a cell to degrade and interact with the hydrogel during the differentiation stage dictates cellular interactions with the gel and fate decisions, regardless of whether the hMSC is spread.
Figure 4.
Delayed secondary crosslinking re-directs hMSC matrix interactions & fate choice without altering cell shape. a, Schematic of delayed UV exposure following 7 d growth media incubation. b(i),d(i), Representative TFM images, b(ii),d(ii), hydrogel deformations (*p < 0.001, t test), and b(iii),d(iii) circularity of hMSCs within MeMaHA hydrogels following b, D7 UV exposure or d, D7 UV exposure and an additional 14 d mixed media incubation. c, HA release from D7 UV versus −UV hydrogels (normalized to total HA content) in the presence of 20 nM MMP-2. e(i), Representative brightfield image of a D7 UV hydrogel, with encapsulated hMSCs stained for ALP (osteogenesis) and lipid droplet (adipogenesis), and e(ii), relative frequency of lineage commitment within D7 UV hydrogels following 14 d mixed media incubation (*p < 0.001, #p < 0.005, t test). Error bars represent standard errors for the mean. Scale bars: b,d, 25 µm; e(i),10 µm.
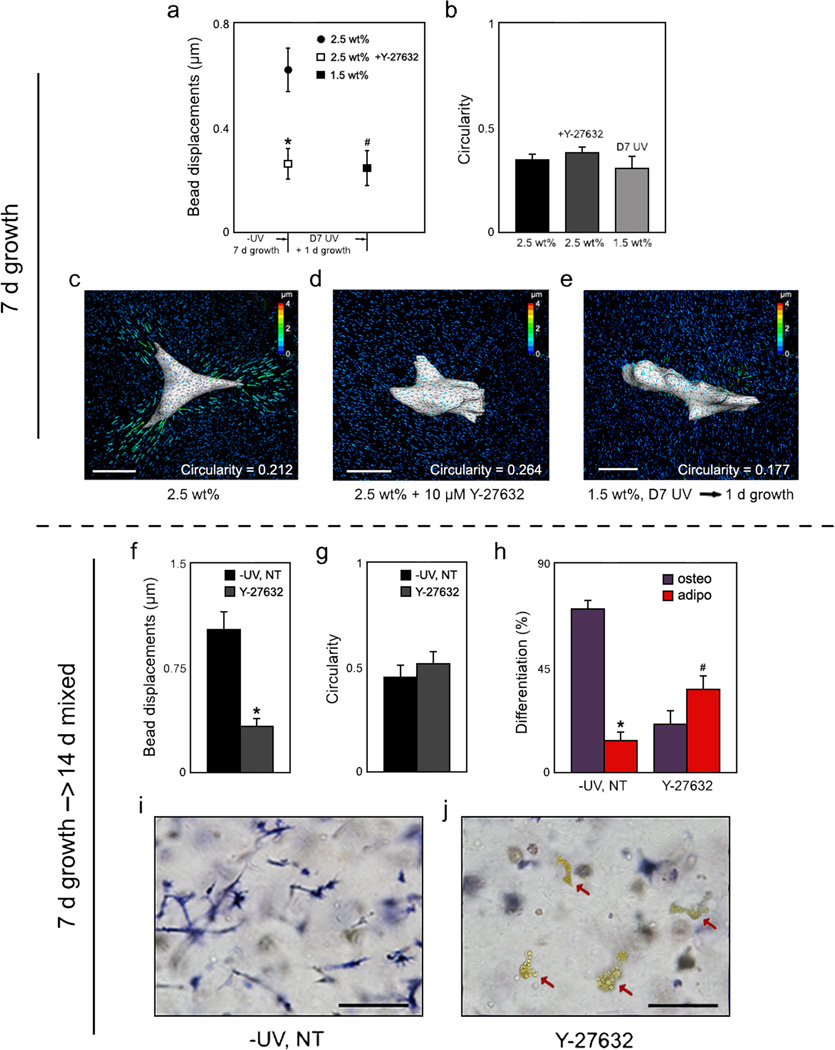
To further understand the mechanism by which delayed secondary crosslinking abrogates hMSC tractions and re-directs fate, −UV gels were treated with the Y-27632, an inhibitor of ROCK, the RhoA effector that induces non-muscle myosin-mediated contractility. Y-27632 (10 µM) was administered daily during either the 7 day growth media (day 0 – 7) or 14 day mixed media incubation (day 7 – 21) periods to prevent the assembly of a robust actin cytoskeleton (Fig. 5). When treated with 10 µM Y-27632 daily during the 7 day growth media incubation, hMSCs deformed the matrix to a much lesser extent (Fig. 5a), yet displayed similar spreading (Fig. 5b) to hMSCs without the pharmacological inhibitor treatment. When hMSCs in −UV gels were treated with Y-27632 only during the differentiation phase (day 7–21), a decrease in gel deformation (Fig. 5f) and no change in cell morphology (Fig. 5g) was again observed when compared to untreated hMSCs. However, a fate switch from primarily osteogenesis to adipogenesis was also observed via biochemical staining for ALP (Fig. 5i–j) and quantification (Fig. 5h), similar to that observed when non-degradable crosslinks were introduced with D7 UV exposure. Also similar to D7 UV gels, lipid-filled hMSCs within the −UV Y-27632 treated gels exhibited minimal cytoskeletal organization relative to hMSCs within untreated gels (Supplementary Fig. S18). To rule out off target effects of the pharmacologic inhibitor, hMSCs in 2.5 wt% −UV gels also were treated with an inhibitor of non-muscle myosin II, blebbistatin during the differentiation phase (Supplementary Fig. S19). Blebbistatin (50 µM) treatment also significantly reduced cellular tractions and osteogenesis, and increased adipogenesis, though to a less pronounced extent relative to Y-27632 treatment. In addition to its effects on myosin, Y-27632 inhibition of ROCK has been shown also to destabilize F-actin via inhibition of LIM kinase and Cofilin, further amplifying its effects32,33. Additionally, a recent report implicated ROCK-induced cytoskeletal tension as a necessary component in hMSC osteogenesis14,34. In contrast, blebbistatin acts specifically to inhibit non-muscle myosin-II35,36; though this is a primary myosin isoform upregulated in hMSC osteogenesis, recent reports have implicated that multiple myosin isoforms may also be active during osteogenesis, including even smooth muscle myosin37. Taken together, these findings suggest that the introduction of non-degradable crosslinks mediates a switch in hMSC behavior and fate by blocking traction generation in a manner similar to direct pharmacological inhibition of myosin activity.
Figure 5.
hMSC matrix interactions & lineage commitment upon pharmacologically induced changes in cytoskeletal tension. a, Cell-induced bead displacements (*p < 0.001, #p < 0.005 relative to 2.5 wt%, t test) and b, circularity analysis of hMSCs within 2.5 wt% −UV MeMaHA gels following 7 d growth media incubation with or without daily 10 µM Y-27632, or within 1.5 wt% D7 UV MeMaHA gels plus one additional day growth media incubation. c–d, Representative TFM images of hMSCs within 2.5 wt%, −UV MeMaHA hydrogels following 7 d growth media incubation either c, without or d, with daily 10 µM Y-27632. e, Representative TFM image of an hMSC within a 1.5 wt%, D7 UV hydrogel. f, Cell-induced bead displacements (*p < 0.001 relative to 2.5 wt%, t test), g, circularity analysis, and h, percentage differentiation fate of hMSCs toward osteogenic or adipogenic lineages within 2.5 wt% −UV MeMaHA gels following an additional 14 d mixed media incubation. i–j, Representative bright field images of these same groups stained for ALP (osteogenesis); lipid-containing cells (red arrows) appear yellow. Error bars represent standard errors for the mean. Scale bars: c–e, 25 µm; i–j, 50 µm.
Collectively, the present work provides new insights on the role of traction generation in hMSC fate choice in 3D hydrogels. Within covalently crosslinked hydrogels in particular, traction is dependent on hMSCs being able to degrade their surroundings and assemble focal adhesions and cytoskeletal structures. Unlike cell behaviour atop 2D substrates, these results highlight the importance of degradability as a parameter separate from previously described effects of substrate crosslinking or cell morphology. Additionally, the work stresses the importance of understanding stem cell interactions with each hydrogel type (e.g., covalently versus ionically crosslinked), whose degradability and molecular structure may drive divergent outcomes and ultimately impact the successful design of hydrogels in stem cell-based therapies.
Methods
For quantification of hMSC differentiation fate and circularity, n ≥ 45 cells per condition were analysed. All other experiments were performed in quadruplicate (n=4). For further methods please refer to the Supplementary Information.
Encapsulation of hMSCs within HA hydrogels
Previously described methods were used to synthesize MeHA30 (~92% methacrylation) from sodium hyaluronate (Lifecore), MeAlg (~70% modification) from sodium alginate (Sigma), and MeDex (~50% modification) from dextran (Sigma). MeMaHA with ~14% and ~10.5% methacrylate and maleimide modification, respectively, was synthesized via the coupling of the tetrabutylammonium salt of NaHA (HA-TBA) with 2-amino methacrylate hydrochloride (Sigma) and 2-amino maleimide trifluoroacetate salt (Sigma) (Supplementary Fig. S20). The chemical structures and 1H NMR spectra of MeHA, MeAlg, MeDex and MeMaHA are provided in Supplementary Fig. S21, Supplementary Fig. S22, Supplementary Fig. S23, and Supplementary Fig. S24, respectively. The integrin binding peptide GCGYRGDSPG (Genscript; italics indicates cell adhesive domain) was conjugated to MeHA, MeAlg, and MeDex (754 µM, matching that used in the described physically crosslinked alginate studies), and to MeMaHA (1 mM) via 30 min reaction in pH 8.0 PBS at 25 °C prior to crosslinking. Passage 3 hMSCs (Lonza) were encapsulated either into MeMaHA (1 million hMSCs ml−1) hydrogels using Michael addition reactions between MeMaHA maleimides and the MMP degradable peptide GCRDVPMS↓MRGGDRCG (Genscript; down arrow indicates cleavage site by MMP-2), or into MeHA, MeAlg, or MeDex (15 million hMSCs ml−1) using photo-initiated free radical polymerization (Exfo Omnicure S1000 lamp with a 320–390 nm filter, exposure of 10 mW cm−2 for 5 min) in the presence of 0.05 wt% Irgacure 2959 (I2959; Ciba), a photoinitiator chosen for its aqueous solubility and good cytocompatibility38. For CD44 blocking studies, hMSCs were incubated with anti-CD44 (3/1000, mouse mAb CD44, Abcam) in a buffer (2 mM EDTA and 2% FBS in PBS) for 45 min on ice, washed twice in buffer, and resuspended in growth media prior to encapsulation. All gels were transferred to FBS-supplemented MEM-α (Invitrogen). MeMaHA hydrogels were secondarily photopolymerized at day 0 (“D0 UV”) or day 7 (“D7 UV”) by incubating with I2959 and exposing to UV light as described above. The elastic modulus of the hydrogels was measured via parallel plate compression testing at 10% ramped strain min−1 as previously reported27. For differentiation studies, following 7 days of incubation in growth media, hydrogels were transferred to a 1:1 mixture of adipogenic:osteogenic media (R&D Systems), with media changes every 3 days. For ROCK inhibition studies, selected −UV gels were treated with 10 µM Y-27632 (Sigma) daily during either the growth media (day 1–7) or mixed media (day 7–21) incubation periods.
Assessment of hMSC matrix interactions, differentiation, viability, and proliferation
To evaluate the extent of matrix adhesions, hMSCs transfected with lentiviral vinculin conjugated with green fluorescent protein were encapsulated into MeHA and MeMaHA hydrogels. After growth and/or mixed media incubation, the gels were fixed in 4% formalin and stained for actin using rhodamine-phalloidin (Invitrogen). For lineage analysis following 7 d (MeHA gels) or 14 d (MeMaHA gels) mixed media incubation, encapsulated hMSCs were stained for biochemical markers ALP (Fast Blue) and neutral lipid droplets (Oil Red O) as previously reported9,27. The differentiation trends were confirmed via immunocytochemistry for OC and FABP, also as previously reported27. Percentage differentiation toward each lineage was quantified by counting the number of positively stained cells and dividing by total nuclei. hMSC viability following 7 d growth media incubation in either −UV, D0 UV, or −UV + light (in the absence of photoinitiator) hydrogels was assessed using a live/dead staining kit (Molecular Probes) and reporting (# viable cells)/(# total cells). To evaluate proliferation, total dsDNA content was determined using the PicoGreen assay as previously reported39.
3-dimensional TFM analysis
Round cover slides were functionalized with methacrylate groups as previously reported30. Immediately prior to crosslinking, MeHA and MeMaHA solutions with 60,000 hMSCs ml−1 were pipetted between the slide and a sterilized PDMS mold to immobilize the gel to the slide. Two types of fluorescent beads (0.2 µm diameter, non-functionalized yellow-green (Polysciences) and Suncoast yellow (Bangs Labs)) were co-encapsulated at ~2 × 1010 beads ml−1 each. Encapsulated cells were imaged using an inverted microscope (Olympus IX71) equipped with a spinning disk confocal scan head (Yokogawa Electric) and live-cell incubator (Pathology Devices). A 151 × 151 × 200 µm volume was imaged around each cell prior to and 45 min following cell lysis with 0.5% SDS (Bio-Rad). 2D rendering of the cell surface using image processing software (Amira) and fluorescent bead displacement tracking using a feature-vector based algorithm (Matlab) were then performed as previously reported40. Further details can be found in the Supplementary Information.
Cell circularity analysis
A dimensionless term describing the roundness of encapsulated hMSCs was developed (Supplementary Fig. S6). The distance of each cell surface node to the center of a box inscribing the cell was calculated and normalized to the maximum distance over all nodes. Circularity was then calculated as the standard deviation of these normalized values multiplied by a scaling constant. As a result, the circularity range is from 0, representing the maximally non-circular cell across all studies, to 1, corresponding to a perfect sphere. Further quantitative details and examples can be found in Supplementary Fig. S6.
Supplementary Material
Acknowledgements
This work was supported by funding from a Fellowship in Science and Engineering from the David and Lucile Packard Foundation (JAB), a CAREER award (JAB) and Graduate Research Fellowship (SK) from the National Science Foundation, and grant GM74048 from the National Institutes of Health (CSC). The authors would like to thank Ross Marklein and Colin Choi for helpful discussions and experimental assistance.
Footnotes
Author contributions. S.K. and J.A.B. conceived the ideas and designed the experiments. S.K., M.G., W.R.L., and D.M.C. conducted the experiments and analyzed the data. S.K., W.R.L., C.S.C., and J.A.B. interpreted the data and wrote the manuscript.
References
- 1.Nuttelman CR, Tripodi MC, Anseth KS. Synthetic hydrogel niches that promote hMSC viability. Matrix Biol. 2005;24:208–218. doi: 10.1016/j.matbio.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Ruoslahti E, Reed JC. Anchorage dependence, integrins, and apoptosis. Cell. 1994;77:477–478. doi: 10.1016/0092-8674(94)90209-7. [DOI] [PubMed] [Google Scholar]
- 3.VandeVondele S, Vörös J, Hubbell JA. RGD-grafted poly-L-lysine-graft-(polyethylene glycol) copolymers block non-specific protein adsorption while promoting cell adhesion. Biotechnol Bioeng. 2003;82:784–790. doi: 10.1002/bit.10625. [DOI] [PubMed] [Google Scholar]
- 4.Docheva D, Popov C, Mutschler W, Schieker M. Human mesenchymal stem cells in contact with their environment: surface characteristics and the integrin system. J Cell Mol Med. 2007;11:21–38. doi: 10.1111/j.1582-4934.2007.00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meredith JE, Jr, Fazeli B, Schwartz MA. The extracellular matrix as a cell survival factor. Mol Biol Cell. 1993;4:953–961. doi: 10.1091/mbc.4.9.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guilak F, et al. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reilly GC, Engler AJ. Intrinsic extracellular matrix properties regulate stem cell differentiation. J Biomech. 2010;43:55–62. doi: 10.1016/j.jbiomech.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 9.Guvendiren M, Burdick JA. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat Commun. 2012;792 doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- 10.Fu J, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert PM, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guvendiren M, Burdick JA. The control of stem cell morphology and differentiation by hydrogel surface wrinkles. Biomaterials. 2010;31:6511–6518. doi: 10.1016/j.biomaterials.2010.05.037. [DOI] [PubMed] [Google Scholar]
- 13.Kilian KA, Bugarija B, Lahn BT, Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc Natl Acad Sci U S A. 2010;107:4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 15.Ruiz SA, Chen CS. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells. 2008;26:2921–2927. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huebsch N, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benoit DS, Schwartz MP, Durney AR, Anseth KS. Small functional groups for controlled differentiation of hydrogel-encapsulated human mesenchymal stem cells. Nat Mater. 2008;7:816–823. doi: 10.1038/nmat2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira LS, et al. Bioactive hydrogel scaffolds for controllable vascular differentiation of human embryonic stem cells. Biomaterials. 2007;28:2706–2717. doi: 10.1016/j.biomaterials.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ifkovits JL, Burdick JA. Photopolymerizable and degradable biomaterials for tissue engineering applications. Tissue Eng. 2007;13:2369–2385. doi: 10.1089/ten.2007.0093. [DOI] [PubMed] [Google Scholar]
- 20.Mann BK, Gobin AS, Tsai AT, Schmedlen RH, West JL. Smooth muscle cell growth in photopolymerized hydrogels with cell adhesive and proteolytically degradable domains: synthetic ECM analogs for tissue engineering. Biomaterials. 2001;22:3045–3051. doi: 10.1016/s0142-9612(01)00051-5. [DOI] [PubMed] [Google Scholar]
- 21.Nicodemus GD, Bryant SJ. Cell encapsulation in biodegradable hydrogels for tissue engineering applications. Tissue Eng Part B Rev. 2008;14:149–165. doi: 10.1089/ten.teb.2007.0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khetan S, Katz JS, Burdick JA. Sequential crosslinking to control cellular spreading in 3-dimensional hydrogels. Soft Matter. 2009;5:1601–1606. [Google Scholar]
- 23.Burdick JA, Chung C, Jia X, Randolph MA, Langer R. Controlled degradation and mechanical behavior of photopolymerized hyaluronic acid networks. Biomacromolecules. 2005;6:386–391. doi: 10.1021/bm049508a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erickson IE, et al. Macromer density influences mesenchymal stem cell chondrogenesis and maturation in photocrosslinked hyaluronic acid hydrogels. Osteoarthritis Cartilage. 2009;17:1639–1648. doi: 10.1016/j.joca.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legant WR, et al. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat Methods. 2010;7:969–971. doi: 10.1038/nmeth.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahn MS, Miller JS, West JL. Three-dimensional biochemical and biomechanical patterning of hydrogels for guiding cell behavior. Adv Mater. 2006;18:2679–2684. [Google Scholar]
- 27.Khetan S, Burdick JA. Patterning network structure to spatially control cellular remodeling and stem cell fate within 3-dimensional hydrogels. Biomaterials. 2010;31:8228–8234. doi: 10.1016/j.biomaterials.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 28.West JL, Hubbell JA. Polymeric Biomaterials with Degradation Sites for Proteases Involved in Cell Migration. Macromolecules. 1999;32:241–244. [Google Scholar]
- 29.Miller JS, et al. Bioactive hydrogels made from step-growth derived PEG-peptide macromers. Biomaterials. 2010;31:3736–3743. doi: 10.1016/j.biomaterials.2010.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marklein RA, Burdick JA. Spatially controlled hydrogel mechanics to modulate stem cell interactions. Soft Matter. 2010;6:136–143. [Google Scholar]
- 31.Hudson JE, et al. A synthetic elastomer based on acrylated polypropylene glycol triol with tunable modulus for tissue engineering applications. Biomaterials. 2010;31:7937–7947. doi: 10.1016/j.biomaterials.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Maekawa M, et al. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science. 1999;285:895–898. doi: 10.1126/science.285.5429.895. [DOI] [PubMed] [Google Scholar]
- 33.Vardouli L, Moustakas A, Stournaras C. LIM-kinase 2 and cofilin phosphorylation mediate actin cytoskeleton reorganization induced by transforming growth factor-beta. J Biol Chem. 2005;280:11448–11457. doi: 10.1074/jbc.M402651200. [DOI] [PubMed] [Google Scholar]
- 34.Shih YR, Tseng KF, Lai HY, Lin CH, Lee OK. Matrix stiffness regulation of integrin-mediated mechanotransduction during osteogenic differentiation of human mesenchymal stem cells. J Bone Miner Res. 2011;26:730–738. doi: 10.1002/jbmr.278. [DOI] [PubMed] [Google Scholar]
- 35.Duxbury MS, Ashley SW, Whang EE. Inhibition of pancreatic adenocarcinoma cellular invasiveness by blebbistatin: a novel myosin II inhibitor. Biochem Biophys Res Commun. 2004;313:992–997. doi: 10.1016/j.bbrc.2003.12.031. [DOI] [PubMed] [Google Scholar]
- 36.Even-Ram S, et al. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat Cell Biol. 2007;9:299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- 37.Bennett KP, et al. Preoteomics reveals multiple routes to the osteogenic phenotype in mesenchymal stem cells. BMC Genomics. 2007;8:380. doi: 10.1186/1471-2164-8-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams CG, Malik AN, Kim TK, Manson PN, Elisseeff JH. Variable cytocompatibility of six cell lines with photoinitiators used for polymerizing hydrogels and cell encapsulation. Biomaterials. 2005;26:1211–1218. doi: 10.1016/j.biomaterials.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 39.Singer VL, Jones LJ, Yue ST, Haugland RP. Characterization of PicoGreen reagent and development of a fluorescence-based solution assay for double-stranded DNA quantitation. Anal Biochem. 1997;249:228–238. doi: 10.1006/abio.1997.2177. [DOI] [PubMed] [Google Scholar]
- 40.Gao Y, Kilfoil ML. Accurate detection and complete tracking of large populations of features in three dimensions. Opt Express. 2009;17:4685–4704. doi: 10.1364/oe.17.004685. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.