Abstract
The cardiac muscle comprises dynamically interacting components that use allosteric/cooperative mechanisms to yield unique heart-specific properties. An essential protein in this allosteric/cooperative mechanism is cardiac muscle troponin T (cTnT), the central region (CR) and the T2 region of which differ significantly from those of fast skeletal muscle troponin T (fsTnT). To understand the biological significance of such sequence heterogeneity, we replaced the T1 or T2 domain of rat cTnT (RcT1 or RcT2) with its counterpart from rat fsTnT (RfsT1or RfsT2) to generate RfsT1-RcT2 and RcT1-RfsT2 recombinant proteins. In addition to contractile function measurements, dynamic features of RfsT1-RcT2 and RcT1-RfsT2 reconstituted rat cardiac muscle fibers were captured by fitting the recruitment-distortion model to force response of small amplitude (0.5%) muscle length changes. RfsT1-RcT2 fibers showed a ~40% decrease in tension and ~44% decrease in ATPase activity, but RcT1-RfsT2 fibers were unaffected. The magnitude of length-mediated increase in crossbridge recruitment (E0) decreased by ~33% and the speed of crossbridge recruitment (b) increased by ~100% in RfsT1-RcT2 fibers. Our data suggest the following: (1) the CR of cTnT modulates crossbridge recruitment dynamics; (2) the N-terminal end region of cTnT has a synergistic effect on the ability of CR to modulate crossbridge recruitment dynamics; (3) the T2 region is important for tuning the Ca2+ regulation of cardiac thin filaments. The combined effects of CR-Tm interactions and the modulating effect of the N-terminal end of cTnT on CR-Tm interactions may lead to the emergence of a unique property that tunes contractile dynamics to heart rates.
Keywords: Allosteric interaction, regulatory proteins, troponin T, thin filament activation, contractile dynamics, cardiac troponin T
INTRODUCTION
Allosteric/cooperative interactions among sarcomeric contractile proteins are central to striated muscle contraction. An essential protein in this allosteric cascade mechanism is Troponin T (TnT), which is important for full cooperative regulation of muscle contraction. In the absence of Ca2+, TnT inhibits the binding of myosin heads to actin and the ATPase activity, via its allosteric interactions with troponin C (TnC), troponin I (TnI), and tropomyosin (Tm) 1. In the presence of Ca2+ , TnT removes the inhibition of actin-myosin interactions by interacting strongly with TnC-TnI-Tm, thereby potentiating ATPase activity 2; 3. However, the allosteric/cooperative mechanisms regulated by TnT, which permits Ca2+ induced changes in TnC to be transduced to the actin-myosin interface, are not well understood in the cardiac muscle.
Based on extensive structural and biochemical solution studies on tryptic peptides of fast skeletal TnT (fsTnT), cardiac TnT (cTnT) may be roughly divided into two distinct domains, the N-terminus (1–193 residues of rat cTnT; T1) and the C-Terminus (194–289 residues; T2). The T1 region of cTnT may be further separated into two parts: (1) the N-Terminal end region (1–76 residues), which is not known to bind to any other thin filament protein 2; 4; and (2) the central region (77–193 residues; CR), which is known to strongly interact with Tm 5; 6. A cursory look at the amino acid sequences of cTnT and fsTnT across various species (mouse, rat, rabbit, porcine, bovine, and human) show substantial sequence heterogeneity not only in their N-Terminal end regions, but also in the CRs and T2 regions 7. Such sequence heterogeneity in different domains between cTnT and fsTnT may be important for tissue-specific regulation of thin filament activation. This notion is supported by our previous study, which showed that an exchange of rat cTnT (RcTnT) for rat fsTnT (RfsTnT) significantly altered maximal activation and crossbridge (XB) recruitment dynamics in rat cardiac muscle fibers 8. However, the previous study could not resolve as to which specific domain of cTnT was responsible for its effect on the cardiac contractile dynamics.
When the cTnT from different species are aligned, there is a high degree of similarity (~92–99%) in the CR, suggesting an evolutionarily conserved heart-specific functional role. This is further substantiated by the finding that the CRs of cTnT and fsTnT from different species share only a 66–69 % sequence similarity. Previous biochemical solution studies on the CR of cTnT have suggested the following: (1) it promotes the binding of Tm to actin 5; (2) it aids in the assembly of Tm on the actin filament by promoting Tm-Tm polymerization 9; (3) it is important for the folding stability and flexibility of TnT 10; 11; and (4) it regulates the myosin binding to actin and cooperative activation of the thin filament 12; 13; 14. The T2 region of cTnT also differs significantly from its counterpart in fsTnT (~51–57% similarity). Because the T2 region of cTnT interacts with TnC, TnI, and Tm in a Ca2+-dependent manner 15, sequence variation in the T2 region is also expected to confer tissue-specific function. Not surprisingly, cardiomyopathy-related substitutions and deletions in the T2 region of cTnT have been shown to alter its interaction with TnI/TnC, affecting both the inhibition and Ca2+-mediated activation of the actin-myosin ATPase activity 16; 17; 18; 19.
Because of the strategic location of cTnT in the cardiac thin filament, sequence variations in cTnT are expected to affect the following contractile mechanisms: (1) the extent of thin filament activation due to Ca2+ and strong XBs 20; 21; (2) the length-mediated activation of thin filaments 22; and (3) the tuning of the XB recruitment dynamics 8. All of the above mentioned contractile mechanisms are known to be different in the cardiac muscle, giving substantial credence to the idea that the sequence variations in the CR and T2 region of cTnT are essential for the contractile function of the heart. This is further highlighted by the observation that most of the known cardiomyopathy-related mutations are found both in the CR of cTnT (63%) and the T2 region of cTnT (32%). Therefore, the main objective of this study was to understand how sequence variations in the CR and T2 region of cTnT modulate and tune Ca2+-, XB-, and length-mediated activation of thin filaments in the cardiac muscle. We hypothesized that the cardiac-specific CR of cTnT plays a key role in “modulating” the XB recruitment dynamics, while the T2 region of cTnT aids in “tuning” the Ca2+-mediated regulation of cardiac thin filaments.
It is now widely appreciated that dynamic relationships, rather than static aspects of force-pCa and force-length relationships, dominate in conditions under which cardiac muscle functions. In vitro solution studies lack structural constraints imposed not only by the highly organized structure of the myofilament, but also by various allosteric/cooperative interactions that exist within this complex network of proteins. Therefore, it is important to study the properties of sarcomeric functions under conditions that retain most of the self-organized, interconnected, and interacting systems essential for the unique behavior of the heart. Therefore, we made selective alterations within the cTnT by substituting the T1 and T2 domains of RcTnT with those of RfsTnT to generate two recombinant rat chimeric TnT proteins: (1) RfsT1-RcT2 chimera, in which the T1 fragment of RcTnT (RcT1) was replaced with the T1 of RfsTnT (RfsT1) and (2) RcT1-RfsT2 chimera, in which T2 fragment of RcTnT (RcT2) was replaced with the T2 of RfsTnT (RfsT2). These substitutions allowed us to conserve other regions of cTnT, so that the functional differences could be attributed to the modified segments only. Contractile and dynamic measurements were made in RcTnT, RfsT1-RcT2 and RcT1-RfsT2 reconstituted muscle fibers.
Cooperative effects contribute to XB recruitment dynamics, but not to XB distortion dynamics 23; 24. Therefore, distinguishing between the effects on XB recruitment dynamics and the effects on XB distortion dynamics − as a consequence of a molecular change in the cTnT structure − helps to identify the mechanism of action of cTnT in regulating the unique functional features of contractile dynamics. To better determine the effect of specific alterations in cTnT, we used an interpretive mathematical model 25. We discuss these data in terms of an effect of the CR and T2 region on the XB recruitment dynamics and Ca2+-mediated activation of cardiac thin filaments.
RESULTS
Rationale for generating TnT chimeras
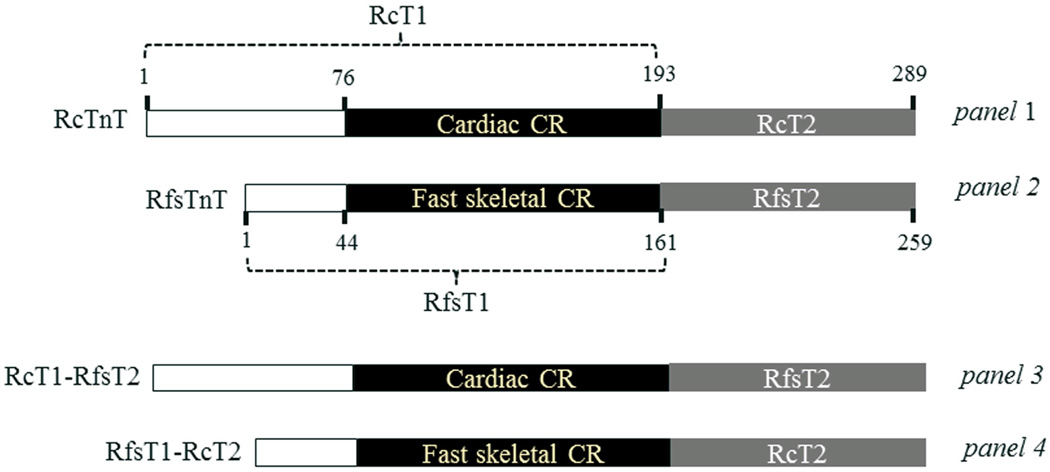
A sequence alignment of RcT1 and RfsT1 using LALIGN program 26 reveals an identity of only 56.2%, while a sequence comparison of RcT2 and RfsT2 reveals an identity of 54.5% (Fig. 1; panels 1 and 2). In order to understand the functional significance of the protein sequences in the cardiac-specific CR (residues 77–193) and T2 region (residues 194–289) of cTnT, we generated two rat TnT chimeras: one in which RcT2 was replaced by RfsT2 (RcT1-RfsT2; Fig. 1, panel 3) and the other in which RcT1 was replaced by RfsT1 (RfsT1-RcT2; Fig. 1; panel 4). Previous studies have demonstrated that the N-terminal 1–76 residues of RcTnT behave similar to 1–45 residues of rabbit fsTnT with regard to their effect on the thin filament activation 4; 27. Thus, by replacing the functionally corresponding N-Terminal end portion of RfsTnT into RcTnT, we were able to associate observed functional differences to either a sequence variation in the CR or in the T2 region of cTnT.
Figure 1. A schematic representation of the N-terminal end region, central region (CR), and T2 region of the recombinant TnT proteins.
A comparison of the N-terminal regions, CRs, and T2 regions between RcTnT (panel 1) and RfsTnT (panel 2). To investigate the physiological roles of CR (residues 77–193) and T2 region (residues 194–289) of RcTnT, two recombinant chimeric TnT proteins − RcT1-RfsT2 and RfsT1-RcT2 − were generated (panels 3 and 4, respectively). The T2 region of RcTnT was replaced by RfsT2 in RcT1-RfsT2 chimera, whereas, the T1 region of RcTnT was replaced by RfsT1 in RfsT1-RcT2 chimera. The CR and T2 regions of both proteins are shown in black and gray, respectively; the unshaded portions represent the N-terminal end regions.
Effect on the overall secondary structure of TnT chimeras
To examine whether the replacement of RcT1, RcT2 or RcTnT with its respective RfsTnT analog affected the overall secondary structure of the protein, we measured the far-ultraviolet (UV) circular dichroism (CD) spectral features of RcTnT, RcT1-RfsT2, RfsT1-RcT2, and RfsTnT proteins (Fig. 2). CD spectral data showed no significant differences in the α-helical content, β-sheet, and random coil between the four recombinant TnT proteins. Each protein contained approximately 40% α-helix, 15% β-sheet, and 45% random coil.
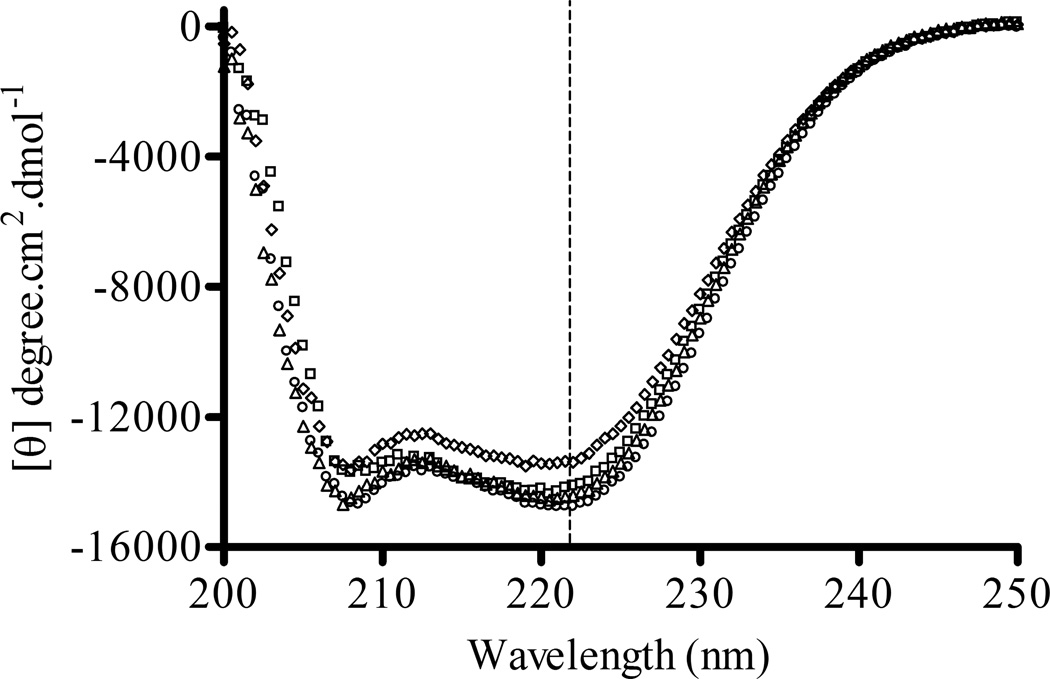
Figure 2. Comparison of far-UV circular dichroism spectral features of RcTnT, RcT1-RfsT2, RfsT1-RcT2, and RfsTnT.
Mean residue ellipticity (MRE) values were plotted against wavelength to illustrate the spectral features of the recombinant TnT proteins. The K2D algorithm from DICHROWEB 46; 47; 48, an online server for CD spectral analysis, was used to analyze the far-UV CD spectra to estimate the secondary structural content of the proteins. (○) represent recombinant RcTnT, (□) represent RcT1-RfsT2, (△) represent RfsT1-RcT2, and (◊) represent RfsTnT. Data for RfsTnT reconstituted fibers from our previous study 8 are used for comparisons in this study. Dotted line represents 222 nm, where the MRE was used to estimate the percentage α-helical content. Each recombinant TnT protein contained approximately 40% of α-helical content.
SDS-PAGE and Western blot analysis of detergent-skinned rat cardiac myofibers reconstituted with TnT chimeras
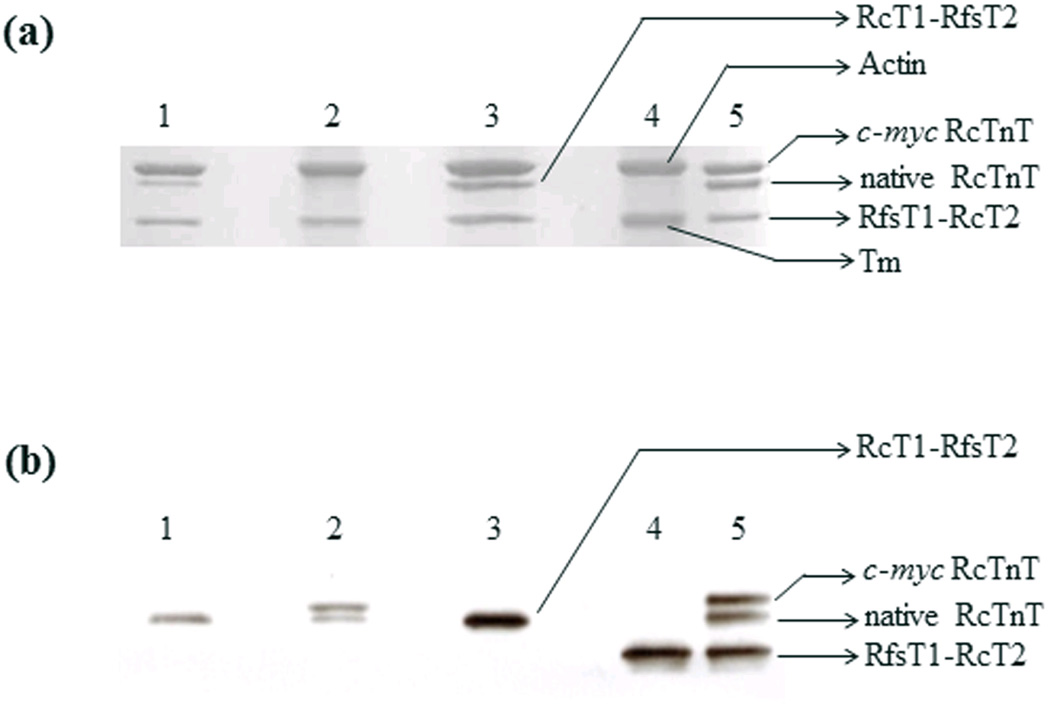
Protein preparations from fibers reconstituted with various recombinant proteins were dissolved in 2% SDS solution, solubilized in the gel-loading buffer, and separated on 10% SDS-gels as described in Methods. We used the c-myc-tagged cTnT to assess the level of incorporation in control fibers (i.e., RcTnT fibers). Under these conditions, an optimum separation of actin, tropomyosin, and TnT can be readily appreciated (Fig. 3a). However, the c-myc tagged RcTnT co-migrated with actin (lane 2) and RfsT1-RcT2 comigrated with Tm (lane 4); thus hindering our ability to estimate the amount of protein incorporated. Therefore, we used the Western blot analysis to assess the level of incorporation of recombinant TnT proteins in reconstituted muscle fiber preparations. Densitometric analysis of the band profiles from the Western blot demonstrated that the extent of incorporation of the c-myc-tagged RcTnT was 69% in RcTnT fibers (Fig. 3b, lane 2), while that of RfsT1-RcT2 chimera was ~100% in RfsT1-RcT2 fibers (Fig. 3b, lane 4). Because the molecular weight of RcT1-RfsT2 is similar to that of RcTnT, it comigrated with RcTnT (Fig. 3b, lane 3), making it difficult to exactly quantify the extent of RcT1-RfsT2 incorporation in cardiac myofibers. However, the following reason gives us confidence in stating that the level of incorporation is at least 69% or greater. In the RcT1-RfsT2 reconstituted fibers, two contractile parameters were altered (significant decrease in Ca2+ sensitivity (Fig. 5a) and a significant decrease in the magnitude of XB recruitment dynamics (Fig. 6d)); whereas all other contractile parameters (Ca2+-activated maximal tension (Fig. 4a), ATPase activity (Fig. 4b), number of strongly-bound XBs as assessed by the magnitude of infinite frequency stiffness (E∞; Fig 6b), rate constant of XB recruitment (b; Fig. 7d), and ktr (Fig. 7b)) were similar to the RcTnT reconstituted fibers. Thus, the functional effects observed between various reconstituted fiber groups in this study may be solely attributed to the specific structural alteration introduced in the TnT, but not to the differences in the efficiency of mutant TnT incorporation between groups.
Figure 3. SDS-PAGE and Western blot analysis of detergent-skinned papillary muscle fibers reconstituted with RcTnT, RcT1-RfsT2 or RfsT1-RcT2.
(a) 2% SDS solution was used to digest the reconstituted fibers, as described previously 27. Equal quantities of digested protein samples (10 µg per sample) were run on 10% SDS-PAGE to achieve molecular weight-based separation of various sarcomeric proteins. The gel was then stained with Coomassie brilliant blue (R-250, Bio-rad Laboratories, Hercules, CA) to visualize the protein content. (b) Western blot. TnT was transferred from 10% SDS-gel to a PVDF membrane, and TnT in each preparation was probed using an anti-TnT primary antibody and an anti-mouse secondary antibody. Densitometric analysis of the TnT band profiles from the Western blot was carried using the ImageJ software (National Institutes of Health). Lanes 1–5 in (panels A and B) represent the following: lane 1, untreated cardiac fibers; lane 2, fibers reconstituted with wild-type c-myc tagged RcTnT; lane 3, fibers reconstituted with RcT1-RfsT2 chimera; lane 4, fibers reconstituted with RfsT1-RcT2 chimera and; lane 5, purified recombinant proteins. Tm stands for tropomyosin.
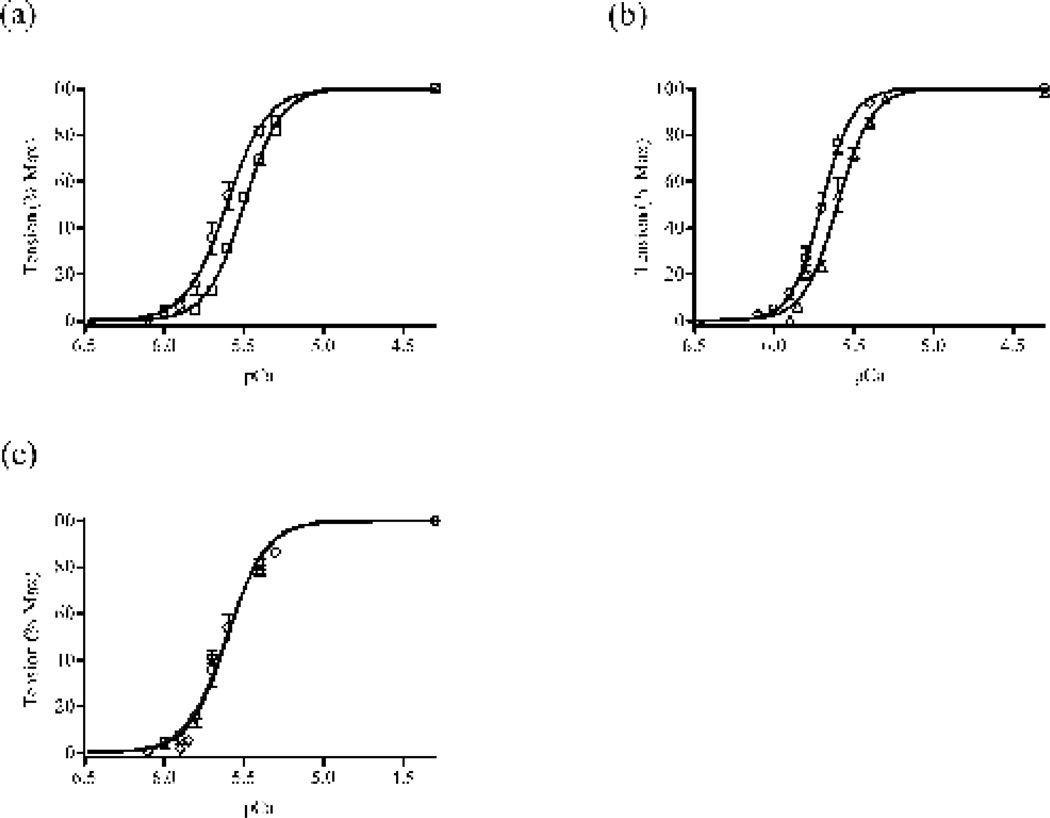
Figure 5. Normalized pCa-tension relationships in rat cardiac muscle fibers reconstituted with RcTnT, RcT1-RfsT2, RfsT1-RcT2 or RfsTnT.
(a) Normalized pCa-tension relationships in RcT1-RfsT2 (b) Normalized pCa-tension relationships in RfsT1-RcT2 fibers and (c) Normalized pCa-tension relationships in RfsTnT fibers. Isometric steady-state tension elicited by the fiber was measured in various pCa solutions and these measurements were normalized with their respective steady-state values in pCa 4.3 solution 49; 50. Normalized tension values were then plotted against pCa to construct pCa-tension relationships. Data for RfsTnT reconstituted fibers from our previous study 8 are used for comparisons in this study. Myofilament Ca2+ sensitivity (pCa50) and cooperativity (nH) were derived from the nonlinear fits of Hill’s equation to the normalized pCa-tension relationships (see Table 1). Curves presented here are Hill fits to the pCa-tension relationships. (○) represent data from RcTnT fibers, (□) represent data from RcT1-RfsT2 fibers, (Δ) represent data from RfsT1-RcT2 fibers, and (◊) represent data from RfsTnT. Differences in the tension measurements between RcTnT and RfsTnT reconstituted fibers were very small at each pCa level, causing a substantial overlap in their pCa-Tension curves (see panel c). Number of determinations is ≥10 for each group. Data are reported as mean ± SE. Standard error bars are smaller than symbols in some cases.
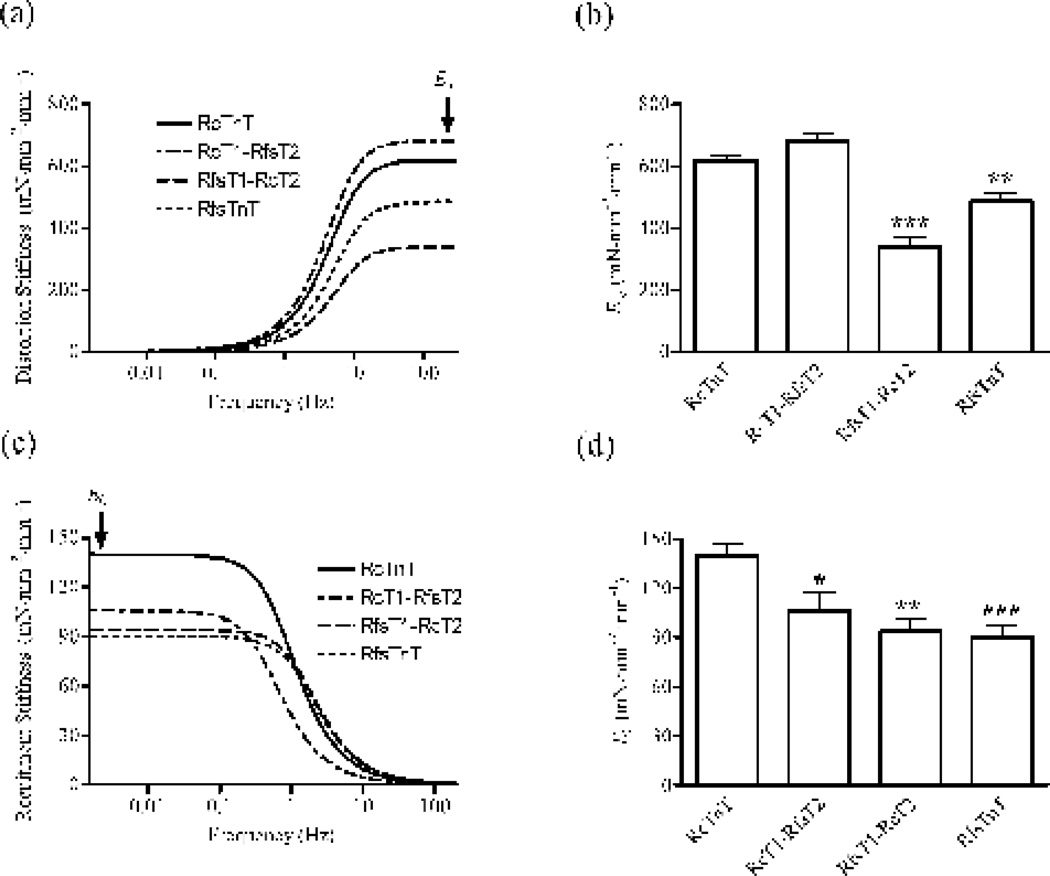
Figure 6. Muscle fiber stiffness parameters, E∞ and E0, in rat cardiac muscle fibers reconstituted with RcTnT, RcT1-RfsT2, RfsT1-RcT2 or RfsTnT.
We estimated E∞ and E0 by fitting the RD model to small changes in force elicited by the fiber in response to small changes in muscle length 25. Data for RfsTnT reconstituted fibers from our previous study 8 are used for comparisons in this study. (a) A plot of model-predicted distortion component of the stiffness as a function of frequency. The magnitude of the distortion stiffness at infinite frequency is termed E∞ (as illustrated by the arrow in panel a). (b) Comparison of E∞ in fibers reconstituted with RcTnT, RcT1-RfsT2, Rfs1-RcT2, or RfsTnT. (c) A plot of model-predicted recruitment component of the stiffness as a function of frequency. The magnitude of the recruitment stiffness at zero frequency is termed E0 (as illustrated by the arrow in panel c). (d) Comparison of E0 in fibers reconstituted with RcTnT, RcT1-RfsT2, Rfs1-RcT2, or RfsTnT. Statistical differences between fiber groups were analyzed by one-way ANOVA with the data from RcTnT reconstituted fibers as controls. Number of determinations was ≥10 for each group. Asterisks represents a statistically significant result compared to RcTnT reconstituted fibers (*P < 0.05; **P < 0.01; *** P < 0.001). Data are reported as mean ± SE.
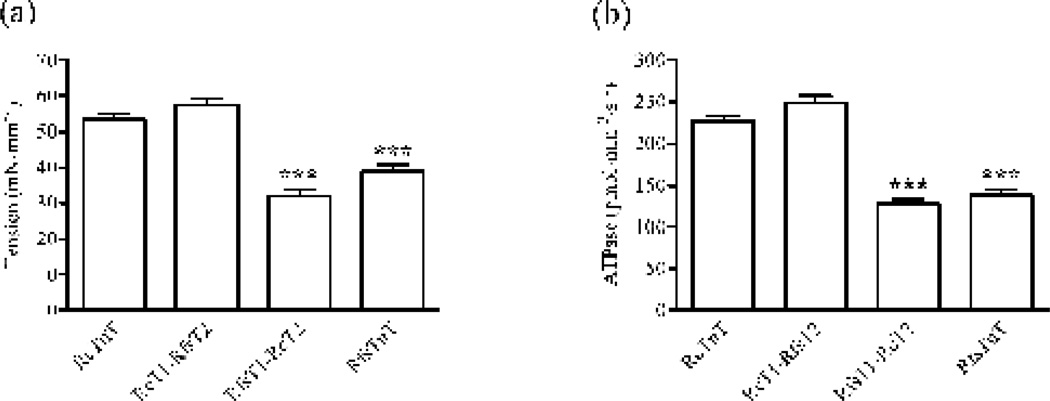
Figure 4. Ca2+-activated maximal tension and ATPase activity in rat cardiac muscle fibers reconstituted with RcTnT, RcT1-RfsT2, RfsT1-RcT2 or RfsTnT.
Ca2+-activated maximal tension and ATPase activity in reconstituted fibers were simultaneously measured in pCa 4.3 49; 50. (a) Effect of TnT chimeras on Ca2+-activated maximal tension. To determine the maximal tension, reconstituted fibers were bathed in pCa 4.3 solution until they reached a steady-state in force. The isometric steady-state force was then converted to tension by expressing it as force per cross-sectional area. (b) Effect of TnT chimeras on Ca2+-activated maximal ATPase activity. ATPase activity was measured using a coupled enzymatic assay, as described previously 49; 50; 54. Data for RfsTnT reconstituted fibers from our previous study 8 are used for comparisons in this study. Statistical differences between fiber groups were analyzed by one-way ANOVA, with the data from RcTnT fibers as controls. Number of determinations was ≥10 for each group. Asterisks represents a statistically significant result compared to cTnT reconstituted fibers (***P < 0.001). Data are reported as mean ± SE.
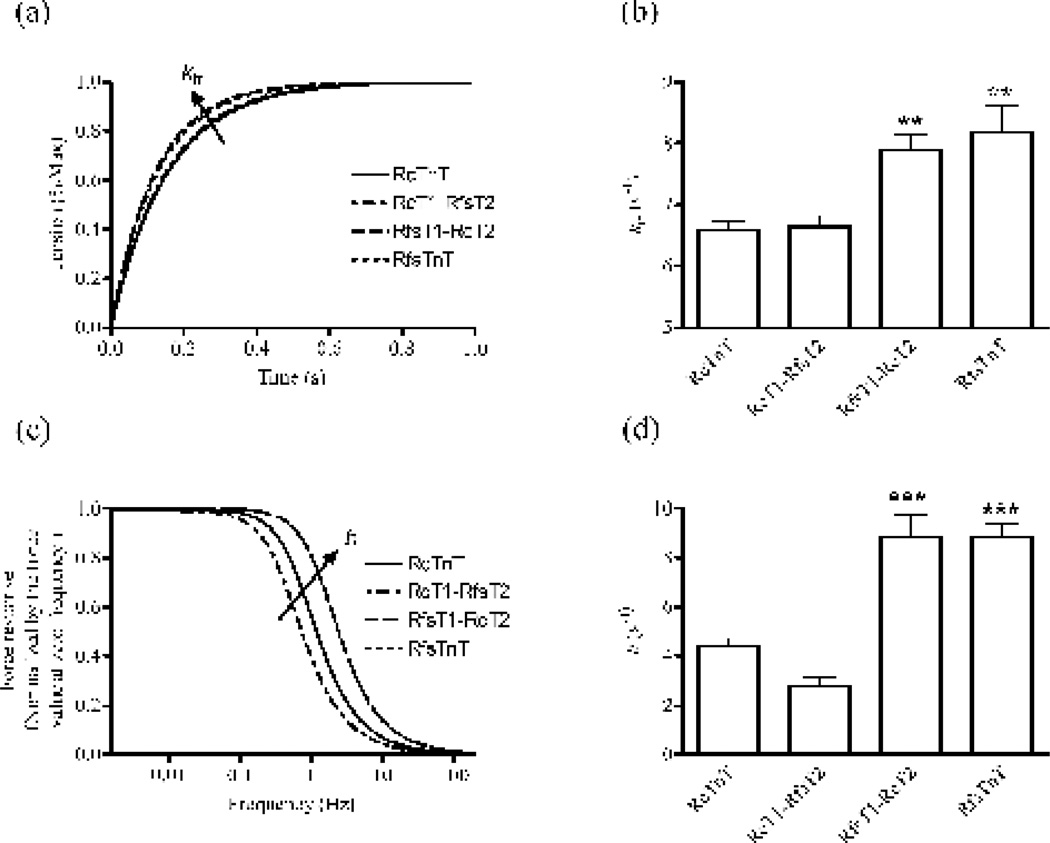
Figure 7. The rate constant of tension redevelopment, ktr, and the rate constant of lengthmediated XB recruitment, b, in rat cardiac muscle fibers reconstituted with RcTnT, RcT1-RfsT2, RfsT1-RcT2 or RfsTnT.
(a) Time course of averaged force responses to large-release restretch transient in maximally-activated fibers reconstituted with RcTnT, RcT1-RfsT2, RfsT1-RcT2, or RfsTnT. Data for RfsTnT reconstituted fibers from our previous study 8 are used for comparisons in this study. Force values were normalized by their respective new steady-state values following perturbation. (b) Effects of TnT chimeras on ktr. We estimated ktr by fitting a mono-exponential function to the rise of tension in response to a large-release restretch transient 53. (c) Contributions of the RD model-predicted recruitment component of the force response as a function of frequency in fibers reconstituted with RcTnT, RcT1-RfsT2, RfsT1-RcT2, or RfsTnT. Forces were normalized with their respective values at zero frequency. (d) Effects of TnT chimeras on b. The parameter b was estimated by fitting the RD model to small changes in force around the steady state in response to chirp length perturbations 25. Statistical differences between fiber groups were analyzed by one-way ANOVA with the data from RcTnT fibers as controls. Because the estimates of ER and b are similar between RfsT1-RcT2 and RfsTnT reconstituted fibers, there is a substantial overlap in their force responses as shown in the panel c. Number of determinations was ≥10 for each group. Asterisks represents a statistically significant result compared to RcTnT reconstituted fibers (**P < 0.01; ***P < 0.001). Data are reported as mean ± SE.
Effect of TnT chimeras on Ca2+-activated maximal tension and ATPase activity
The effect of chimeric TnT proteins on Ca2+-mediated activation of thin filaments was first assessed by measuring Ca2+-activated maximal tension (Fig. 4a) and ATPase activity (Fig. 4b). Replacement of RcT2 with RfsT2, as in RcT1-RfsT2 fibers, resulted in no significant change in either Ca2+ - activated maximal tension or ATPase activity. In contrast, substitution of the RcT1 region with RfsT1 region, as in RfsT1-RcT2 fibers, resulted in a dramatic decrease in both maximal tension (~40%; P < 0.001) and ATPase activity (~44%; P < 0.001). Similar decreases in maximal tension (~29%; P < 0.001) and ATPase activity (39%; P < 0.001) were also observed in RfsTnT fibers, demonstrating that the attenuation of maximal Ca2+ activation was only observed in fibers that lacked the RcT1 region − for example, RfsT1-RcT2 and RfsTnT reconstituted fibers.
Effect of TnT chimeras on pCa-tension relationships
Figures 5a–c show the comparisons of pCa-tension relationships between RcTnT, RcT1-RfsT2, RfsT1-RcT2, and RfsTnT fibers. The Hill’s equation was fitted to the pCa-tension relationships to estimate myofilament Ca2+ sensitivity (pCa50) and cooperativity (nH). The corresponding estimates of pCa50 and nH of tension in various reconstituted fiber groups are provided in Table 1. When compared to the estimates in RcTnT fibers, both RcT1-RfsT2 and RfsT1-RcT2 fibers displayed a significant decrease in pCa50 (Table 1), as indicated by the rightward shifts in the pCa-tension relationships (Fig. 5a and b, respectively). The corresponding decrease in the pCa50 (ΔpCa50) in RcT1-RfsT2 fibers was 0.12 (P < 0.01), while this was 0.07 (P < 0.05) in RfsT1-RcT2 fibers. However, myofilament Ca2+ sensitivity was not affected in RfsTnT reconstituted fibers (Fig. 5c and Table 1). The un-normalized tension in RfsT1-RcT2 fibers decreased uniformly by 40% in the pCa range 4.3–5.3, while this attenuation in tension was greater than 40% in the pCa range 5.3–6.0 (data not shown). Thus, when normalized, the pCa-Tension in RfsT1-RcT2 fibers showed only a small but significant rightward shift of 0.07 pCa units, as compared with that of RcTnT fibers.
Table 1. Myofilament Ca2+ sensitivity (pCa50) and cooperativity (nH) in detergent-skinned rat cardiac fibers reconstituted with RcTnT, RcT1-RfsT2, RfsT1-RcT2 or RfsTnT.
The Hill’s equation was fitted to the normalized pCa-tension and pCa-ATPase relationships using a nonlinear least squares regression procedure to derive pCa50 and nH. Hill parameters, pCa50 and nH, were determined separately from each muscle fiber experiment and the values were averaged.
| RcTnT | RcT1-RfsT2 | RfsT1-RcT2 | RfsTnTψ | |
|---|---|---|---|---|
| pCa-Tension relationships | ||||
| pCa50 | 5.62 ± 0.03 | 5.50 ± 0.01 ** | 5.55 ± 0.01 * | 5.60 ± 0.01 |
| nH | 4.38 ± 0.23 | 3.79± 0.17 | 3.46 ± 0.1 * | 4.25 ± 0. 06 |
| pCa-ATPase relationships | ||||
| pCa50 | 5.68 ± 0.03 | 5.57 ± 0.01 *** | 5.60 ± 0.01 ** | 5.70 ± 0.01 |
| nH | 5.25 ± 0.25 | 4.63 ± 0.22 | 4.22 ± 0.17 ** | 5.41 ± 0.10 |
Data for RfsTnT reconstituted fibers from our previous study 8 are used for comparisons in this study. Statistical differences between groups were analyzed by one-way ANOVA, with the data from RcTnT reconstituted fibers as controls. Number of determinations was ≥10 for each group.
Asterisks represents a statistically significant result compared to RcTnT fibers (*P < 0.05;
P < 0.01;
P < 0.01). Values are presented as mean ± SE.
The estimates of nH in RcT1-RfsT2 and RfsTnT fibers were not significantly different from those of RcTnT fibers; however, RfsT1-RcT2 fibers demonstrated a slight but significant decrease in nH (~21%; P < 0.05; Table 1). These observations suggested that selective substitution of just RcT1 with RfsT1, as in RfsT1-RcT2 fibers, had a significant impact on myofilament cooperativity; whereas, the replacement of RcT2 or RcTnT with its RfsTnT equivalent had no effect on myofilament cooperativity. Similar effects in the pCa-ATPase relationships of RcT1-RfsT2, RfsT1-RcT2, and RfsTnT fibers were also observed (pCa-ATPase curves not shown). The Hill’s equation was fitted to the pCa-ATPase relationships to estimate pCa50 and nH (Table 1). The effects observed in pCa50 and nH of ATPase in RcT1-RfsT2, RfsT1- RcT2, and RfsTnT fibers were similar to those of pCa-tension relationships (Table 1).
Effects of TnT chimeras on muscle fiber stiffness parameters, E∞ and E0
To examine whether the effects of Ca2+-activated maximal tension observed in RcT1-RfsT2, RfsT1-RcT2, and RfsTnT reconstituted fibers could be correlated to the changes in the maximum number of strongly-bound XBs, we measured force responses in RcTnT, RcT1-RfsT2, RfsT1-RcT2, and RfsTnT fibers to small amplitude (±0.5%) muscle length (ML) changes (see Methods). The recruitment-distortion (RD) model was fitted to these force responses with the imposed length changes as input, to separate the contributions of low-frequency recruitment and high-frequency distortion components of the force responses 25. Figure 6a shows the averaged distortion component of the stiffness response plotted as a function of frequency in fibers reconstituted with RcTnT, RcT1-RfsT2, RfsT1-RcT2 or RfsTnT. At high frequencies, the stiffness response is mostly dominated by the distortion of strongly-bound XBs because the high speed of the length change prevents recruitment of new XBs 25. The magnitude of this stiffness value at the infinite frequency (illustrated by an arrow in Fig. 6a) is termed infinite frequency stiffness, E∞. We have previously shown that E∞ is an approximate measure of strongly-bound XBs, as it represents the magnitude of stiffness increase due to the rapid distortion of the elastic elements within the strongly-bound XBs 25. Thus, a comparison of E∞ estimates between various reconstituted fiber groups may provide clue as to whether there are any changes in the number of strongly-bound XBs. As expected, Fig. 6b shows that our estimates of E∞ in RcT1-RfsT2 fibers were not significantly different from those of RcTnT fibers, while the estimates of E∞ were dramatically lower by 45% (P < 0.001) and 22% (P < 0.01) in RfsT1-RcT2 and RfsTnT fibers, respectively. Thus, a significant decrease in tension associated with a decrease in the number of strongly-bound XBs in both RfsT1-RcT2 and RfsTnT fibers confirms that the presence of cT1 is important for maximal activation in cardiac thin filaments.
Figure 6c shows the comparison of the averaged recruitment component of the stiffness response plotted as a function of frequency in RcTnT, RcT1-RfsT2, RfsT1-RcT2, and RfsTnT reconstituted fibers. E0 − as illustrated by the arrow in Fig. 6c − is termed the zero frequency stiffness; it represents the slope of the static force-length relationship. In other words, E0 is a measure of an increase in the number of strong XBs corresponding to an increase in the muscle length. A comparison of E0 estimates between various reconstituted fiber groups is shown in Fig. 6d. Interestingly, when compared to RcTnT reconstituted fibers, estimates of E0 in RcT1-RfsT2, RfsT1-RcT2, and RfsTnT fibers were significantly lower by 24% (P < 0.05), 33% (P < 0.01), and 35.7% (P < 0.001), respectively. Therefore, our data demonstrates that the substitution of RcT1, RcT2, or RcTnT by its respective RfsTnT analog affected the lengthmediated increase in the number of strongly-bound XBs.
Effect of TnT chimeras on tension cost and the rate constant for XB distortion dynamics (c)
In order to assess whether the chimeric cTnT proteins affected the XB detachment kinetics, we measured tension cost and c. Tension cost, which is the slope of the tension-ATPase relationship, was evaluated by simultaneous measurements of tension and ATPase activity. As outlined in Table 2, no significant differences were observed in tension cost values between RcTnT, RcT1-RfsT2, RfsT1-RcT2, and RfsTnT fibers. Previous studies have shown that tension cost is an indicator of the XB detachment rate 28 and that it is strongly correlated to XB distortion dynamics, c 25. Estimates of c in RcT1-RfsT2, RfsT1-RcT2 or RfsTnT fibers were also not significantly different from those of RcTnT fibers (Table 2). Similar effects observed in both c and tension cost values suggested that the substitution of RcT1, RcT2, or RcTnT with its RfsTnT equivalent had no effect on the rate of XB detachment.
Table 2. Tension cost and XB distortion dynamic, c, in rat cardiac muscle fibers reconstituted with RcTnT, RcT1-RfsT2, RfsT1-RcT2 or RfsTnT.
Ca2+-activated maximal tension and ATPase activity were simultaneously measured in pCa 4.3 solution. Tension cost was estimated as the slope of ATPase-tension relationship 8; 49; 50. c was determined by fitting the RD model to small changes in force elicited by the fiber around the steady-state to small changes in muscle length 25.
| RcTnT | RcT1-RfsT2 | RfsT1-RcT2 | RfsTnTψ | |
|---|---|---|---|---|
| Tension Cost (pmol mN−1·mm−1·s−1) | 4.58 ± 0.16 | 4.42 ± 0.20 | 4.30 ± 0.19 | 4.44 ± 0.15 |
| XB distortion dynamic, c (s−1) | 42.48 ± 2.05 | 38.4 ± 2.07 | 48.90 ± 3.19 | 48.67 ± 1.97 |
Data for RfsTnT reconstituted fibers from our previous study 8 are used for comparisons in this study. Statistical differences between fiber groups were analyzed by one-way ANOVA with the data from RcTnT fibers as controls. No significant differences were observed in tension cost and c between RcTnT, RcT1-RfsT2, RfsT1-RcT2, and RfsTnT fibers. Number of determinations was ≥10 for each group. Values are presented as mean ± SE.
Effects of TnT chimeras on the rate of tension redevelopment (ktr), XB recruitment dynamic (b), and the frequency of minimum stiffness (fmin)
ktr was estimated using a large release-restretch protocol and b was estimated by fitting the RD model to the force responses from fibers in response to length perturbations (see Methods). Figure 7a shows the averaged force responses in various groups normalized by their new steady-state values. It is evident that both RfsT1-RcT2 and RfsTnT fibers attain the steady-state with faster ktr, whereas, RcT1-RfsT2 reconstituted fibers demonstrated no effect. Pooled estimates of ktr in RfsT1-RcT2 and RfsTnT fibers showed a significant increase by 20% (P < 0.001), and 24% (P < 0.01), respectively (Fig. 7b). However, no effect was observed in ktr estimates of RcT1-RfsT2 fibers (Fig. 7b). Figure 7c shows a plot of model-predicted recruitment component of the force response as a function of frequency of length perturbation. Forces were normalized by their respective values at zerofrequency. Similar to the effects observed in ktr, both RfsT1-RcT2 and RfsTnT fibers exhibited a dramatic increase by 2-fold in the speed of XB recruitment (Fig. 7d), as illustrated by the significant rightward shift in the force response (Fig. 7c). Previously, we have demonstrated that ktr is correlated to b 8; 25 and that both ktr and b are valuable indicators of XB turnover rate 25. Therefore, similar effects of ktr and b suggest that the absence of RcT1, as in RfsT1-RcT2 and RfsTnT reconstituted fibers, augments XB turnover rate.
The transition of dominance from recruitment to distortion occurs at a frequency of minimum stiffness (fmin) in the heart muscle, and this is an important feature that tunes the dynamics of muscle contraction to the heart rate 8. To determine whether cT1 or cT2 is responsible for this effect on fmin, complex stiffness was calculated using the RD model parameter estimates in various reconstituted fiber groups. Estimates of fmin values in RcTnT, RcT1-RfsT2, RfsT1-RcT2, and RfsTnT were 0.72 ± 0.03, 0.62 ± 0.07, 1.08 ± 0.06, and 1.05 ± 0.02, respectively. Significant lower values of fmin observed in both RcTnT and RcT1-RfsT2 reconstituted fibers (P < 0.01), as opposed to RfsT1-RcT2 and RfsTnT fibers, suggests that cT1 plays a role in tuning the contractile dynamics of the heart muscle.
Binding of TnT chimeras to immobilized α-Tm
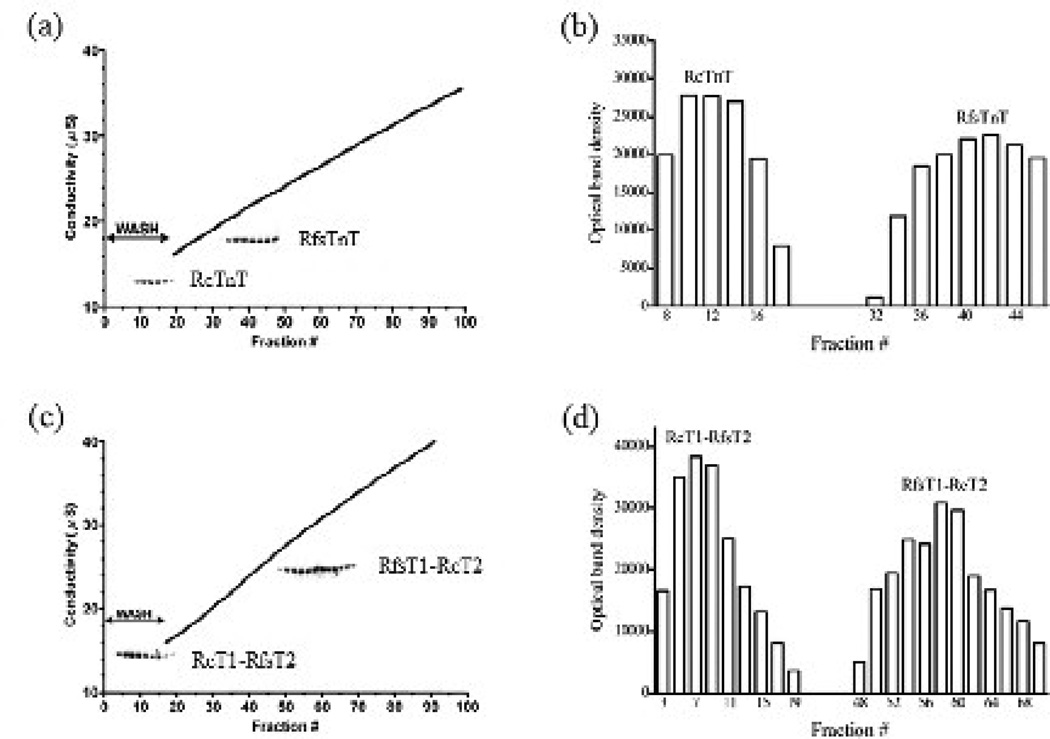
In order to understand how the T1 and T2 regions of cTnT and fsTnT contribute to the overall Tm affinity, the binding of RcTnT, RfsTnT, RcT1-RfsT2, and RfsT1-RcT2 to α-Tm was evaluated. Figure 8a shows a typical trace of fraction number versus conductivity of the eluate containing RcTnT or RfsTnT, overlaid with the SDS acrylamide gel bands corresponding to respective TnT in the eluted fractions. Figure 8b shows the optical band densities of gel bands corresponding to RcTnT and RfsTnT in the eluted fractions as a function of fraction number. Since RcTnT was eluted during the wash phase of the elution profile (fractions 8–20), its affinity for the immobilized α-Tm was relatively weak (Fig. 8a–b). In contrast, when applied to the α-Tm column, RfsTnT was eluted at higher conductivities of 20.1–24.0 µS (fractions 32–46), suggesting that its affinity for the α-Tm was comparatively stronger than that of RcTnT (Fig. 8a–b). The T2 region of TnT has also been implicated in Tm binding 29; 30; 31. To assess the impact of this region on Tm interaction, the binding of the TnT chimeras to α-Tm column was also evaluated. Figure 8c shows a typical trace of fraction number versus conductivity of the eluate containing RcT1-RfsT2 and RfsT1-RcT2 chimeras, while Fig. 8d shows the optical band densities of gel bands corresponding to these TnT chimeras in the eluted fractions as a function of fraction number. The RcT1-RfsT2 chimera, like RcTnT, eluted during the column wash phase (fractions 1–21) and, therefore, had relatively weaker binding affinity for α-Tm (Fig. 8c–d). Akin to RfsTnT, the RfsT1-RcT2 chimera showed stronger affinity for α-Tm since fractions containing RfsT1-RcT2 were only eluted at conductivities of 27.7–32.5 µS (fractions 48–70; Fig. 8c–d). These observations suggest that the affinity of RfsT1 for Tm is stronger than that of RcT1.
Figure 8. Binding affinity of RcTnT, RcT1-RfsT2, RfsT1-RcT2 or RfsTnT for Tm.
(a) Binding affinity of RcTnT and RfsTnT for α-Tm affinity column. (b) Optical band densities (in arbitrary units) of the gel bands corresponding to RcTnT and RfsTnT in the eluent as a function of fraction number. (c) Binding affinity of RcT1-RfsT2 and RfsT1-RcT2 for α-Tm affinity column. (d) Optical band densities (in arbitrary units) of the gel bands corresponding to RcT1-RfsT2 and RfsT1-RcT2 in the eluent as a function of fraction number. Each recombinant TnT protein was individually loaded onto the immobilized α-Tm affinity column and was eluted using a 150–700 mM NaCl gradient as described in Methods. SDS-PAGE analysis and conductivity measurement of fractions eluted from the α-Tm column showed the relative binding strength of each recombinant TnT protein to α-Tm. The inlaid coomassie-stained SDS gel bands in panels, (a) and (b), correspond to the fractions containing the respective TnT protein. The densitometric analysis of the gel bands corresponding to the TnT in the eluted fractions was carried out using the ImageJ software available from the World-Wide Web serves of NIH.
DISCUSSION
The biological significance of the protein sequence heterogeneity in the CR and T2 region of cTnT is unknown. Novel finding from our study demonstrates that the CR of cTnT modulates Tm activity to tune XB recruitment dynamics. The activity of CR itself is synergistically modulated by the actions of the N-terminal end region of cTnT. The cardiac-specific T2 region of cTnT enables it to tune the Ca2+ regulation of thin filament activation in the cardiac muscle. The combined effects of CR-Tm interactions and the tuning effect of the N-terminal end of cTnT on CR-Tm interactions may manifest as an essential feature that plays a role in tuning the contractile dynamics to heart rates. The newly-acquired novel description of cTnT-Tm interactions is used to refine the model of Tn-mediated regulation of cardiac thin filaments.
The N-terminal extended region of cTnT has a synergistic effect on the ability of the CR to modulate thin filament activation
Previous observations have shown that the N-terminal end region of cTnT is essential for maximal Ca2+ activation 27; 32; 33; 34; 35. Despite such extensive studies, how the N-terminal end region modulates cardiac thin filament activation has remained elusive. What is interesting to note in our study is that RfsT1-RcT2 reconstituted fibers, which contained all of the N-terminal segments of RfsTnT, attenuated both Ca2+-activated maximal tension (~40 %; Fig. 4a) and ATPase activity (~44%; Fig. 4b), leading to the conclusion that the N-terminal residues of cTnT (for example, 1–76) are essential for maximal Ca2+-activation of cardiac thin filaments. Attenuation of thin filament activation leads to the expectation that the number of strongly-bound XBs is lower in RfsT1-RcT2 reconstituted fibers. Consistent with this notion, the infinite frequency stiffness, E∞, was significantly lower in RfsT1-RcT2 reconstituted fibers (Fig. 6b). Because E∞ is proportional to the number of strongly-bound XBs, a significant drop in E∞ is suggestive of a reduction in the number of strongly-bound XBs. An important question is: how does the N-terminal end region of cTnT modulate thin filament activation, such that its absence causes attenuation of activation and a reduction in the number of strongly-bound XBs?
Given the lack of interaction of the N-terminal end region of cTnT with TnI, TnC, Tm, and actin, the only plausible way it can affect thin filament activation is by modulating the activity of its adjoining segment, the CR. Thus, as shown in Fig. 9, we propose that the N-terminal end region of cTnT may interact with the CR to modulate CR-Tm interaction. Consistent with our proposal, affinity chromatography data demonstrated that RfsT1-RcT2 and RfsTnT bound relatively strongly to α-Tm when compared to either RcTnT or RcT1-RfsT2 (Fig. 8a and b, respectively). The removal of the N-terminal 1–76 residues of RcTnT may have caused stronger CR-Tm interactions, leading to a stiffer Tm. Consistent with our observations, Pan et al. 4 and Chandra et al. 27 reported that the deletion of the N-terminal residues of TnT caused a significant increase in the binding of TnT to Tm and subsequent attenuation of myofilament activation. Collectively, these observations provide strong evidence to suggest that the N-terminal end region of cTnT synergistically modulates the effect of CR-Tm interactions on the thin filaments. Such a mechanism for cTnT action has major implications for the cardiac muscle for the following reasons: 1) both the N-terminal end region and CR of cTnT show major differences when compared to those of fsTnT; 2) the T1, which includes both the N-terminal end region and the CR, is known to affect the size of the functional unit (that is, how many actin molecules are turned on per activated Tn) 36; and 3) the T1 is known to modulate off/on states of regulatory units (RU; Tm-Tn) 13; 14.
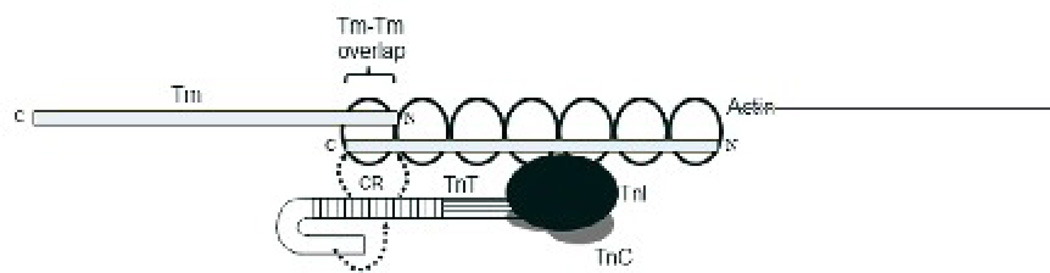
Figure 9. Schematic representation of the refined model of Tn-mediated regulation of cardiac thin filaments.
The horizontal stripes in cTnT represent the T2 domain, which interacts with TnC and TnI. The vertical stripes in cTnT correspond to the CR (residues 77–193). The CR of cTnT is known to participate in strong interactions with Tm near the head-to-tail overlap region of two contiguous Tm molecules 37; 38. The unshaded portion of the cTnT molecule corresponds to the N-terminal end region (residues 1–76 of rat cTnT). Given its lack of interaction with cTnI, cTnC, Tm or actin, the only plausible way the N-terminal end region of cTnT can affect thin filament activation is by modulating the activity the CR. Consistent with this idea, affinity chromatography data demonstrated that RfsT1-RcT2 bound relatively strongly to immobilized α-Tm when compared to either RcTnT or RcT1-RfsT2 (Fig. 8a–d). The N-terminal end region of cTnT (horseshoe-shaped ends of cTnT) may synergistically modulate the interaction of CR with Tm.
Cardiac-specific CR sequence is important for both Ca2+ and cooperative activation of the cardiac thin filaments
RfsT1-RcT2 reconstituted fibers demonstrated a significant decrease in Ca2+ sensitivity (Fig. 5b and Table 1). The Hill coefficient was also significantly lower in RfsT1-RcT2 reconstituted fibers, suggesting that myofilament cooperativity was attenuated by RfsT1-RcT2 (Table 1). It is noteworthy that neither the deletion of N-terminal 1–76 residues nor the substitution of RfsTnT for RcTnT resulted in any change in myofilament Ca2+ sensitivity or cooperativity 8; 27. The effect of RfsT1-RcT2 is consistent with our earlier assertion that sequence variation in the CR affects the Ca2+-mediated activation of thin filaments via its impact on the CR-Tm interaction. One possible mechanism by which myofilament Ca2+ sensitivity is attenuated in RfsT1-RcT2 reconstituted muscle fibers is via an increase in the rate of XB detachment. However, our approximations of both the rate constant of XB distortion dynamics (c) and tension cost showed no significant effect on the rate of XB detachment (Table 2). Thus, the attenuation of Ca2+ sensitivity and cooperativity in RfsT1-RcT2 reconstituted muscle fibers must be due to the altered interaction between CR and Tm, caused by either a lack of cardiac-specific N-terminal extension 33 or due to sequence variation in the CR.
The CR of TnT is known to participate in strong interactions with Tm near the head-to-tail overlap region of two contiguous Tm molecules 37; 38. Previous studies have shown that residues 92–136 of cTnT are important for sufficient helical stability and flexibility of the CR: this is considered to be an essential requirement for the normal interaction of cTnT with Tm 10; 11. Collectively, these observations suggest that sequence variations − caused by the substitution of RcT1 by RfsT1 − may have resulted in a rigid CR-Tm that is ineffective in shifting the azimuthal position of Tm on the actin filament 10. A rigid CR-Tm may thus impact XB-RU and XB-XB interactions such that myofilament cooperativity is attenuated in RfsT1-RcT2 fibers. This attenuation of cooperativity acts to reduce the number of cooperatively recruited XBs in RfsT1-RcT2 fibers at submaximal activations. Thus, our observations demonstrate that altered XB activity results from an effect of altered CR-Tm interaction on allosteric/cooperative mechanisms in the thin filament.
The CR of cTnT is important for tuning XB recruitment dynamics and the frequency of minimum stiffness, fmin, in the heart muscle
Our data shows that while the magnitude of the length-mediated increase in stiffness, E0, decreased significantly by ~33% (Fig. 6d), the rate of length-mediated XB recruitment, b, increased dramatically by ~100% in both RfsT1-RcT2 and RfsTnT reconstituted fibers (Fig. 7d). RfsT1-RcT2 and RfsTnT reconstituted fibers also showed a ~20% increase in the rate of tension redevelopment, ktr (Fig. 7b). Because our recent findings have demonstrated that the N-terminal 1–44, 45–74, and 1–76 residues do not affect the lengthmediated XB recruitment-distortion dynamics and force redevelopment 27; 34, significant changes in ktr and b of RfsT1-RcT2 and RfsTnT fibers must be attributed to sequence variations in the CR of cTnT. Attenuation of myofilament cooperativity is consistent with our observations that both ktr and b increased significantly in RfsT1-RcT2 reconstituted fibers. Campbell’s modified two-state model 39 predicts that decreased cooperativity in RfsT1-RcT2 fibers would reduce the cooperative feedback of strong XBs on the thin filament, thereby reducing the rise of force to a steady-state level that is smaller in magnitude as compared with that of RcTnT fibers. Since catching up with this reduced steady-state force requires shorter time, the net effect is to speed up strong XB turnover rate in a less cooperative myofilament system, as in RfsT1-RcT2 fibers. Consistent with this notion, not only E0 was lower by 33% in RfsT1-RcT2 fibers (Fig. 6d) but both ktr and b were faster because the initial conditions from which XB recruitment occurs was altered by RfsT1-RcT2, more specifically by the RfsT1 domain. This negative impact of fsT1 on E0 is strongly suggestive of the idea that the cardiac-specific CR-Tm interaction is essential for the normal functioning of the allosteric/cooperative processes that modulate the length-dependent activation of the cardiac muscle.
One of the consequences of an increase in the speed of the length-mediated XB recruitment, b, in RfsT1-RcT2 and RfsTnT reconstituted fibers is that the frequency of minimum stiffness, fmin, is shifted to higher frequencies. We have previously demonstrated that the fmin, the delimiter for the dominance of recruitment and distortion phases of the force response, has an important bearing on b 8. Because the fmin was shifted to higher frequencies in both RfsT1-RcT2 and RfsTnT fibers −which lacked the cardiac-specific CR − our data suggests that the effects on b and fmin in these fiber groups are brought about by the sequence heterogeneity in the CR, but not in the T2 region of cTnT. Thus, inferences drawn from these observations suggest that lower values of b and fmin − brought about by the cardiac-specific CR − may be important for matching and tuning the dynamics of myocardial contraction to the heart rate 8. These observations further substantiate the physiological significance of the cardiac-specific CR in the T1 domain.
Cardiac-specific T2 sequence is important for both Ca2+ regulation and the length-mediated activation of thin filaments
RcT1-RfsT2 had no effect on Ca2+-activated maximal tension (Fig. 4a), ATPase activity (Fig. 4b), and E∞ (Fig. 6b). However, RcT1-RfsT2 significantly attenuated both myofilament Ca2+ sensitivity (Fig. 5a and Table 1) and E0 (magnitude of length-mediated increase in the newly-recruited XBs; Fig. 6d). Crystallography studies have shown that T2 directly binds to TnC and that the complex, T2-TnC-TnI, acts as a Ca2+ sensor 40; 41. It has been shown previously that the Ca2+ regulation of actin-myosin ATPase activity is achieved through direct TnT-TnC interactions 3. Previous deletion studies of the T2 region have demonstrated direct effects on both inhibition and Ca2+ sensitive activation of thin filaments 1; 17; 42. In conjunction with these previous findings, our observations suggest that the sequence variations in the T2 region may have reduced Ca2+ sensitive TnT-TnC or TnT-TnI interactions, leading to an attenuation of Ca2+ sensitivity in RcT1-RfsT2 reconstituted fibers. Because the length-mediated activation has an important bearing on the Ca2+ sensitivity 22, a significant decrease in E0 of RcT1-RfsT2 reconstituted fibers (Fig. 6d) also substantiates that TnT-TnC or TnT-TnI interactions are impaired due to the substitution of RcT2 by RfsT2. We believe that the effect of the T2 substitution is localized to its impact on either TnT-TnI or TnT-TnC interactions − but has no effect on the CR − because the affinity of RcT1-RfsT2 to Tm was similar to that of RcTnT (Fig. 8a–b). These observations also demonstrate that the cardiacspecific T2 is essential for strong XB-mediated effect on the troponin complex.
Summary
This is the first explicit study that assigns novel biological roles to the cardiacspecific CR and T2 domain in cTnT. In combination with its synergistic interactions with the N-terminal end region of cTnT, the cardiac CR may exert its role on both inhibition and activation of actin-myosin interactions. The cardiac-specific CR − via its effect on the Tm-Tm overlap region − may “fine tune” myocardial response by slowing XB recruitment dynamics such that the heart beats at a rate commensurate with fmin . The T2 region of cTnT − via its direct effect on either TnC or TnI − may affect allosteric/cooperative processes that modulate Ca2+ , as well as the length-mediated effects on the thin filament. Our findings have significant implications for understanding the effects of cardiomyopathy-related mutations, as well as functional differences associated with tissue-specific expression of different TnT isoforms.
MATERIALS AND METHODS
Animal protocols
All animals used in this study received proper care and treatment and all experiments were carried out in accordance with the guidelines laid down by the Washington State University Institutional Animal Care and Use Committee.
PCR cloning of TnT chimeras
Adult RcTnT (a gift from Dr. J. J. Lin, University of Iowa) and RfsTnT 8 DNA clones were used to create two chimeric DNA clones termed RcT1-RfsT2 and RfsT1-RcT2. The RcT1-RfsT2 DNA clone was generated by splicing the RcT1 region (residues 1–193) with the RfsT2 region (residues 162–259), while the RfsT1-RcT2 DNA clone was generated by splicing the RfsT1 region (residues 1–161) with the RcT2 region (residues 194–289) using a standard high fidelity two-step PCR protocol (Expand High Fidelity system, Roche Applied Science, Indianapolis, IN) as detailed below.
Generation of the RcT1-RfsT2 DNA clone
In step 1, the DNA fragments for the RcT1 and the RfsT2 regions were isolated as follows. The nucleotide sequence coding for RcT1 was amplified from the full-length RcTnT DNA clone using the following oligonucleotides:
Primer 1. 5′ GCAGAATTCAGGCATATGTCTGACGCCGAGGAAGAGGTG 3′
Primer 2. 5′ CTGTTTCTTGCCTCTCTTCTGGTCAGCCTTCTGGATGTACCCTCCAAA 3′
The nucleotide sequence coding for RfsT2 was amplified from the full-length RfsTnT DNA clone using the following oligonucleotides:
Primer 3. 5′ TTTGGAGGGTACATCCAGAAGGCTGACCAGAAGAGAGGCAAGAAACAG 3′
Primer 4. 5′ TGCTGGAATTCAGGATCCTTACTTCCAGCGCCCGCCGACTTT 3′
Generation of the RfsT1-RcT2 DNA clone
In step 1, the DNA fragments for the RfsT1 and the RcT2 regions were isolated as follows. The nucleotide sequence coding for RfsT1 was amplified from the full length RfsTnT DNA clone using the following oligonucleotide primers:
Primer 1. 5′ GCAGAATTCAGGCATATGTCTGACGCCGAGGAAGAGGTG 3′
Primer 2. 5′ CTTCCCACTCTTCCGCTCTGTCTGAGCCTTGGCCAGGTAGCTGCTGTA 3′
The nucleotide sequence coding for RcT2 was amplified by PCR from the full length RcTnT DNA clone using the following oligonucleotides:
Primer 3: 5′ TACAGCAGCTACCTGGCCAAGGCTCAGACAGAGCGGAAGAGTGGGAAG 3′
Primer 4: 5′ TGCTGGAATTCAGGATCCCTATTTCCAACGCCCGGTGAC 3′
In step 2, for both RcT1-RfsT2 and RfsT1-RcT2 clones, the PCR products (RcT1, RfsT2 or RfsT1, RcT2) were gel-extracted, purified (Qiagen, Valencia, CA), and were mixed in equal proportions. The full-length chimeric DNA clones were amplified using the appropriate oligonucleotides.
Expression and purification of recombinant proteins
Rat cTnI (RcTnI) 43, rat cTnC (RcTnC) 44, and rat α-Tm 27 were purified as previously described. RcT1-RfsT2 and RfsT1-RcT2 chimeras were expressed and purified as previously described for RfsTnT and RcTnT 8 with the following modifications: RcT1-RfsT2 was purified from the pellet produced by a 60% ammonium sulfate cut, RfsT1-RcT2 was purified from the pellet produced by a 70% ammonium sulfate cut. All pure protein fractions were extensively dialyzed against deionized water containing 15 mM β–mercaptoethanol, lyophilized, and stored at −80°C.
Affinity chromatography - TnT-Tm binary interaction assay
Recombinant α-Tm was immobilized on to a Sepharose 4B matrix (Sigma-Aldrich, St. Louis, MO) using the standard manufacturer’s protocol and packed into 1×10 cm columns (Econo-columns, Bio-Rad, Hercules, CA) before overnight equilibration with column buffer (20 mM MOPS, 2 mM MgCl2, 150 mM NaCl, 1 mM DTT, and a protease inhibitor/bactericide cocktail). 1.0 mg of the recombinant TnT protein of interest was dissolved in 10 ml of 6 M urea, 50 mM Tris-HCl (pH 8.0), 1 M KCl, 1 mM DTT, and a protease inhibitor/bactericide cocktail and was allowed to equilibrate by gentle rocking for 2 hours at room temperature. Urea and KCl were gradually removed from the dialysate by extensive dialysis at 4°C against three different 50 mM Tris-HCl buffers containing decreasing quantities of urea and KCl (4 M urea, 0.7 M KCl; 2 M urea, 0.5 M KCl; 1 M urea, 300 mM KCl). All these three buffers contained 1 mM DTT and a protease inhibitor/bactericide cocktail. The recombinant TnT solution was then dialyzed against a buffer constituting 20 mM MOPS (pH 7.0), 2 mM MgCl2, 300 mM NaCl, 1 mM DTT, and a protease inhibitor/bactericide cocktail. The dialyzed TnT solution was diluted by adding 5 ml of 20 mM MOPS (pH 7.0), 2 mM MgCl2, 1 mM DTT, and a protease inhibitor/bactericide cocktail. The TnT solution was then applied to the α-Tm column at a flow rate of 0.1 ml·min−1 and eluted with a 150–700 mM NaCl gradient at a flow rate of 0.2 ml·min−1. The eluent was collected in 10 minute fractions and the conductivity of each fraction was recorded. Samples of each fraction were analyzed by standard SDS-PAGE and protein bands were visualized by staining using the Bio-Rad Silver Stain Plus kit (Bio-Rad, Hercules, CA). ImageJ software (from the Word-Wide Web of NIH) was used to determine the optical band density of the gel bands corresponding to TnT protein in the eluted fractions.
Estimation of overall secondary structure in recombinant TnT proteins using far-UV CD spectroscopy
Far-UV CD spectra for RcTnT, RfsTnT, RcT1-RfsT2, and RfsT1-RcT2 proteins were collected in the range of 200–250 nm using an Aviv 202 SF CD spectrometer 45. A urea buffer (6 M urea, 50 mM Tris base, 1 M KCl, and 1 mM DTT (pH 8.0)) to which a fresh cocktail of protease inhibitors (5 mM benzamidine-HCl, 0.4 mM PMSF, 10 µM Leupeptin, 1 µM Pepstatin, 5 µM Bestatin, and 2 µM E-64) added, was used to dissolve the purified proteins. The dissolved protein samples were dialyzed overnight at 4°C using a phosphate buffer (0.2 M sodium phosphate monobasic, 0.2 M sodium phosphate dibasic, 0.5 M KCl, and 0.1 mM DTT (pH 7.0)). The final concentration of each dialyzed protein sample was adjusted to 10 µM by diluting the samples with the phosphate buffer. CD spectral data were collected for each individual mixture containing both the phosphate buffer and the TnT protein sample. The net CD signals for the TnT protein samples were determined by deducting the CD signals of the phosphate buffer from the total CD signals. A K2D algorithm available on DICHROWEB was used to analyze the far-UV CD spectral data and to estimate the secondary-structural content of the recombinant TnT proteins 46; 47; 48.
Reconstitution of TnT chimeras into detergent-skinned rat cardiac muscle fibers
Isolation, dissection, and chemical skinning of left ventricular papillary muscle fibers from rat hearts were as previously described 8. Details of troponin reconstitution into detergent-skinned cardiac myofibers was as described previously 8. Briefly, muscle fibers were treated with an extraction solution containing - RcTnT, RcT1-RfsT2 or RfsT1-RcT2 - and RcTnI for approximately 3–4 hours at room temperature with stirring. The extraction buffer contained 50 mM BES (pH 7.0 at 20°C), 180 mM KCl, 10 mM BDM, 5 mM EGTA, 6.27 mM MgCl2, 1.0 mM DTT, 0.01% NaN3, 5 mM MgATP2−, and a cocktail of protease inhibitors. This was followed by overnight reconstitution with RcTnC (3 mg∙ml−1) at 4°C to complete the reconstitution procedure. In the text to follow, we will refer to detergent-skinned fibers reconstituted with RcT1-RfsT2 + RcTnI + RcTnC as “RcT1-RfsT2 fibers” and those reconstituted with RfsT1-RcT2 + RcTnI + RcTnC as “RfsT1-RcT2 fibers”. Fiber bundles reconstituted with wild-type RcTnT + RcTnI + RcTnC are referred to as “RcTnT fibers” and served as controls in this study. Data for RfsTnT reconstituted fibers from our previous study 8 are used for comparisons in this study, where fibers reconstituted with RfsTnT + RcTnI + RcTnC are termed “RfsTnT fibers.”
SDS-PAGE and Western blot
We ran 10% SDS-PAGE to determine the incorporation of chimeric TnT proteins in the rat cardiac myofibers. For this experiment, reconstituted fibers were first digested in 2% SDS solution, as described previously 27. The total protein concentration of each SDS-digested sample was determined by Nanodrop (ND-1000, Nanodrop Products, Wilmington, DE) and the final concentration of each was standardized to 1 mg/ml by diluting the sample with the gel loading buffer (125 mM Tris-HCl (pH 6.8), 20% glycerol, 2% SDS, 0.01% bromophenol blue, and 50 mM β-mercaptoethanol). Following this, equal quantities of protein samples (10 µg of protein per sample) were loaded and run on 10% SDS-PAGE to separate various sarcomeric proteins according to their molecular weights 27. Proteins were visualized by staining the gel with Coomassie brilliant blue (R-250, Bio-rad Laboratories, Hercules, CA). The level of incorporation of various sarcomeric proteins including TnT was assessed by comparing the stained band profiles of each protein on the SDS-gel between various muscle protein preparations.
For Western blot analysis, proteins from 10% SDS-gel were transferred onto a PVDF membrane and TnT was probed using an anti-TnT primary antibody (Clone JLT-12, Sigma-Aldrich, St. Louis, MO) followed by an anti-mouse secondary anti-body (NIF825, Amersham Pharmacia, Piscataway, NJ). Densitometric scanning of the resulting protein profiles from the Western blot was performed to assess the extent of incorporation of exogenous TnT mutants in the reconstituted myofibers as follows. We first determined the optical band intensities of the native cTnT and recombinant TnT protein profiles from the Western blot of each protein preparation using ImageJ software. The total optical band intensity (i.e., the total amount of TnT expressed) in each preparation was assumed to be the sum of the optical band intensities of native cTnT and recombinant TnT protein profiles. The expression level of recombinant TnT was determined by dividing the optical band intensity of the recombinant TnT protein profile with the total band intensity.
Simultaneous measurement of steady-state isometric force and ATPase activity in reconstituted detergent-skinned rat cardiac muscle fibers
A system described by de Tombe and Stienen 49; 50 was used to simultaneously measure tension and ATPase activity in muscle fibers. After 2 cycles of full activation and relaxation, the resting sarcomere length (SL) was adjusted to 2.2 µm using a He-Ne laser diffraction system 49; 50. The muscle fiber length corresponding to the SL of 2.2 µm was termed initial ML. Tension and ATPase activity (20°C) of muscle fibers were measured in various pCa solutions ranging from pCa 4.3 to 9.0. The compositions of different pCa (-log of free Ca2+ concentration) solutions were calculated using the program developed by Fabiato and Fabiato 51. The pH of each pCa solution was adjusted to 7.0 and the ionic strength to 180 mM. ATPase activity was measured based on an enzyme-coupled assay, as described previously 49; 50. Tension cost was estimated as the slope of the ATPase-tension relationship, as described previously 8; 49; 50.
Mechano-dynamic studies
Dynamic force-length relationships were measured by applying sinusoidal ML changes of constant amplitude (± 0.5% of ML) to maximally-activated muscle fibers in the steady state 8; 25. Two chirps were administered to emphasize low- and high-frequencies: one with frequencies ranging from 0.1 to 4 Hz for a time period of 40 s and the other with frequencies ranging from 1 to 40 Hz for a time period of 5 s. The RD model was fitted to the overall force response (including both low- and high-frequency components) to estimate four important model parameters (E0, b, E∞ , c), as described previously 8; 25. These parameters represent the following: E0 − the magnitude of stiffness increase due to the length-mediated increase in the number of newly recruited strong XBs; b − the rate by which new strong XBs were recruited due to a change in ML; E∞ − the magnitude of the stiffness increase due to strain of the strong XBs, following a sudden change in ML; and c − the rate by which the ML-induced strain of the strong XBs is dissipated. Changes in (b, E0) and (c, E∞ ) were useful in determining the impact of sequence variation in TnT on the recruitment and distortion components of the force responses, respectively 8; 25; 52. For more details on the RD model formulation and the significance of model parameters, please see the Supporting Material.
Rate of tension redevelopment, ktr
We measured ktr in maximally-activated reconstituted muscle fibers (pCa 4.3) using a modification to the original large release protocol, designed by Brenner and Eisenberg 53. Briefly, the maximally-activated muscle fiber was rapidly slackened by 10% of its ML in a step-like fashion and was held at this reduced length for 25 ms. The motor arm was then commanded to stretch the fiber past its preset ML by 10% (for 1 ms) to break any residual number of strongly-bound XBs. The muscle fiber was rapidly brought back to its initial length in a step-like fashion and was allowed to redevelop force. ktr was determined by fitting a mono-exponential function to this rise of force using the following equation:
| (Eq. 1) |
where F(t) is force at time t, Fobs is observed steady-state force, F0 is force from which the fiber starts to redevelop force, and ktr is the rate constant of tension redevelopment.
Data analysis
pCa50 ((-log of [Ca2+ ]free) required for half maximal activation) and the Hill coefficient (nH) were derived by fitting the Hill’s equation to the normalized pCa-tension and pCa-ATPase relationships using a nonlinear least squares regression procedure. pCa50 and nH were individually estimated for each fiber data, and the values from several such experiments were averaged within each group. Our data included four different TnT reconstituted fiber groups: RcTnT (control), RcT1-RfsT2, RfsT1-RcT2, and RfsTnT fibers. Statistical differences in various measurements between the four fiber groups were analyzed by one-way ANOVA, using the Tukey-Kramer multiple comparison post-hoc tests. The criterion for statistical significance was P < 0.05. Data are expressed as mean ± SE.
Supplementary Material
-
▪
Biological significance of sequence heterogeneity in cardiac TnT (cTnT) is unknown
-
▪
Myofilament cooperativity is modulated by the central region (CR) of cTnT
-
▪
CR of cTnT modulates speed of crossbridge (XB) recruitment dynamics
-
▪
N-terminus (NT) of cTnT affects the ability of CR to interact with tropomyosin
-
▪
Unique heart-specific feature emerges from the synergistic effect of NT on CR
ACKNOWLEDGEMENTS
We thank Matthew L. Tschirgi for the technical assistance.
This work was supported by Grant HL-075643 from the National Institute of Health (to Murali Chandra).
Abbreviations used
- Tn
troponin
- TnT
troponin T
- TnC
troponin C
- TnI
troponin I
- Tm
tropomyosin
- fsTnT
fast skeletal TnT
- cTnT
cardiac TnT
- T1
N-terminal of TnT
- T2
C-terminal of TnT
- CR
central region
- RcTnT
rat cTnT
- RfsTnT
rat fsTnT
- XB
crossbridge
- RcT1
T1 of RcTnT
- RcT2
T2 of RcTnT
- RfsT1
T1 of RfsTnT
- RfsT2
T2 of RfsTnT
- UV
ultra violet
- CD
circular dichroism
- ML
muscle length
- RD
recruitment-distortion
- RU
regulatory unit
- RcTnC
rat cTnC
- RfsTnC
rat fsTnC
- PCR
polymerase chain reaction
- MOPS
4-morpholinepropanesulfonic acid
- Tris
Tris(hydroxymethyl) aminomethane
- DTT
dithiothreitol
- BDM
butanedione monoxime
- PMSF
phenylmethylsulfonylfluoride
- EDTA
ethylenediaminetetraacetic acid
- EGTA
ethyleneglycol-bis(β-aminoethyl)-N,N,N′,N′-tetraacetic acid
- BES
N, N-bis[2-hydroxymethyl]-2-amino ethanesulfonic acid
- SL
sarcomere length
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
BIBLIOGRAPHY
- 1.Malnic B, Farah CS, Reinach FC. Regulatory properties of the NH2- and COOH-terminal domains of Troponin T. ATPase activation and binding to troponin I and troponin C. J. Biol. Chem. 1998;273:10594–10601. doi: 10.1074/jbc.273.17.10594. [DOI] [PubMed] [Google Scholar]
- 2.Oliveira DM, Nakaie CR, Sousa AD, Farah CS, Reinach FC. Mapping the domain of troponin T responsible for the activation of actomyosin ATPase activity. Identification of residues involved in binding to actin. J. Biol. Chem. 2000;275:27513–27519. doi: 10.1074/jbc.M002735200. [DOI] [PubMed] [Google Scholar]
- 3.Potter JD, Sheng Z, Pan B-S, Zhao J. A direct regulatory role for troponin T and a dual role for troponin C in the Ca2+ regulation of muscle contraction. J. Biol. Chem. 1995;270:2557–2562. doi: 10.1074/jbc.270.6.2557. [DOI] [PubMed] [Google Scholar]
- 4.Pan BS, Gordon AM, Potter JD. Deletion of the first 45 NH2-terminal residues of rabbit skeletal troponin T strengthens binding of troponin to immobilized tropomyosin. J. Biol. Chem. 1991;266:12432–12438. [PubMed] [Google Scholar]
- 5.Heeley DH, Golosinska K, Smillie LB. The effects of troponin T fragments T1 and T2 on the binding of nonpolymerizable tropomyosin to F-actin in the presence and absence of troponin I and troponin C. J. Biol. Chem. 1987;262:9971–9978. [PubMed] [Google Scholar]
- 6.Pearlstone JR, Smillie LB. The binding site of skeletal alpha-tropomyosin on troponin-T. Can. J. Biochem. 1977;55:1032–1038. doi: 10.1139/o77-154. [DOI] [PubMed] [Google Scholar]
- 7.Manning EP, Guinto PJ, Tardiff JC. Correlation of molecular and functional effects of mutations in cardiac troponin T linked to familial hypertrophic cardiomyopathy: an integrative in silico/in vitro approach. J Biol Chem. 2012;287:14515–14523. doi: 10.1074/jbc.M111.257436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandra M, Tschirgi ML, Rajapakse I, Campbell KB. Troponin T modulates sarcomere length-dependent recruitment of cross-bridges in cardiac muscle. Biophys. J. 2006;90:2867–2876. doi: 10.1529/biophysj.105.076950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehrer SS, Geeves MA. The muscle thin filament as a classical cooperative/allosteric regulatory system. J Mol Biol. 1998;277:1081–1089. doi: 10.1006/jmbi.1998.1654. [DOI] [PubMed] [Google Scholar]
- 10.Hinkle A, Tobacman LS. Folding and function of the troponin tail domain. Effects of cardiomyopathic troponin T mutations. J. Biol. Chem. 2003;278:506–513. doi: 10.1074/jbc.M209194200. [DOI] [PubMed] [Google Scholar]
- 11.Palm T, Graboski S, Hitchcock-DeGregori SE, Greenfield NJ. Disease-causing mutations in cardiac troponin T: Identification of a critical tropomyosin-binding region. Biophys. J. 2001;81:2827–2837. doi: 10.1016/S0006-3495(01)75924-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Regnier M, Rivera AJ, Wang CK, Bates MA, Chase PB, Gordon AM. Thin filament near-neighbour regulatory unit interactions affect rabbit skeletal muscle steady-state force-Ca(2+) relations. J Physiol. 2002;540:485–497. doi: 10.1113/jphysiol.2001.013179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maytum R, Geeves MA, Lehrer SS. A modulatory role for the troponin T tail domain in thin filament regulation. J. Biol. Chem. 2002;277:29774–29780. doi: 10.1074/jbc.M201761200. [DOI] [PubMed] [Google Scholar]
- 14.Tobacman LS, Nihli M, Butters C, Heller M, Hatch V, Craig R, Lehman W, Homsher E. The troponin tail domain promotes a conformational state of the thin filament that suppresses myosin activity. J. Biol. Chem. 2002;277:27636–27642. doi: 10.1074/jbc.M201768200. [DOI] [PubMed] [Google Scholar]
- 15.Pearlstone JR, Smillie LB. Effects of troponin-I plus-C on the binding of troponin-T and its fragments to alpha-tropomyosin. Ca2+ sensitivity and cooperativity. J Biol Chem. 1983;258:2534–2542. [PubMed] [Google Scholar]
- 16.Montgomery DE, Tardiff JC, Chandra M. Cardiac troponin T mutations: correlation between the type of mutation and the nature of myofilament dysfunction in transgenic mice. J Physiol. 2001;536:583–592. doi: 10.1111/j.1469-7793.2001.0583c.xd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stelzer JE, Patel JR, Olsson MC, Fitzsimons DP, Leinwand LA, Moss RL. Expression of cardiac troponin T with COOH-terminal truncation accelerates cross-bridge interaction kinetics in mouse myocardium. Am. J. Physiol. Heart Circul. Physiol. 2004;287:H1756–H1761. doi: 10.1152/ajpheart.00172.2004. [DOI] [PubMed] [Google Scholar]
- 18.Szczesna D, Zhang R, Zhao J, Jones M, Guzman G, Potter JD. Altered regulation of cardiac muscle contraction by troponin T mutations that cause familial hypertrophic cardiomyopathy. J Biol Chem. 2000;275:624–630. doi: 10.1074/jbc.275.1.624. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi-Yanaga F, Ohtsuki I, Morimoto S. Effects of troponin T mutations in familial hypertrophic cardiomyopathy on regulatory functions of other troponin subunits. J Biochem. 2001;130:127–131. doi: 10.1093/oxfordjournals.jbchem.a002950. [DOI] [PubMed] [Google Scholar]
- 20.Gillis TE, Martyn DA, Rivera AJ, Regnier M. Investigation of thin filament near-neighbour regulatory unit interactions during force development in skinned cardiac and skeletal muscle. J Physiol. 2007;580:561–576. doi: 10.1113/jphysiol.2007.128975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol. Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 22.Konhilas JP, Irving TC, de Tombe PP. Length-dependent activation in three striated muscle types of the rat. J Physiol. 2002;544:225–236. doi: 10.1113/jphysiol.2002.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell KB, Razumova MV, Kirkpatrick RD, Slinker BK. Nonlinear myofilament regulatory processes affect frequency-dependent muscle fiber stiffness. Biophys J. 2001;81:2278–2296. doi: 10.1016/S0006-3495(01)75875-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell KB, Razumova MV, Kirkpatrick RD, Slinker BK. Myofilament kinetics in isometric twitch dynamics. Ann Biomed Eng. 2001;29:384–405. doi: 10.1114/1.1366669. [DOI] [PubMed] [Google Scholar]
- 25.Campbell KB, Chandra M, Kirkpatrick RD, Slinker BK, Hunter WC. Interpreting cardiac muscle force-length dynamics using a novel functional model. Am. J. Physiol. Heart Circ. Physiol. 2004;286:H1535–H1545. doi: 10.1152/ajpheart.01029.2003. [DOI] [PubMed] [Google Scholar]
- 26.Huang XQ, Miller W. A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math. 1991;12:337–357. [Google Scholar]
- 27.Chandra M, Montgomery DE, Kim JJ, Solaro RJ. The N-terminal region of troponin T is essential for the maximal activation of rat cardiac myofilaments. J. Mol. Cell. Cardiol. 1999;31:867–880. doi: 10.1006/jmcc.1999.0928. [DOI] [PubMed] [Google Scholar]
- 28.Brenner B. Effect of Ca2+ on cross-bridge turnover kinetics in skinned single rabbit psoas fibers: implications for regulation of muscle contraction. Proc. Natl. Acad. Sci. U. S. A. 1988;85:3265–3269. doi: 10.1073/pnas.85.9.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishii Y, Lehrer S. Two-site attachment of troponin to pyrene-labeled tropomyosin. J. Biol. Chem. 1991;266:6894–6903. [PubMed] [Google Scholar]
- 30.Pearlstone J, Smillie L. Binding of troponin-T fragments to several types of tropomyosin. Sensitivity to Ca2+ in the presence of troponin-C. J. Biol. Chem. 1982;257:10587–10592. [PubMed] [Google Scholar]
- 31.Pearlstone JR, Smillie LB. Identification of a second binding region on rabbit skeletal troponin-T for α-tropomyosin. FEBS Lett. 1981;128:119–122. doi: 10.1016/0014-5793(81)81095-2. [DOI] [PubMed] [Google Scholar]
- 32.Communal C, Sumandea M, de Tombe P, Narula J, Solaro RJ, Hajjar RJ. Functional consequences of caspase activation in cardiac myocytes. Proc. Natl. Sci. Acad. U.S.A. 2002;99:6252–6256. doi: 10.1073/pnas.092022999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gollapudi SK, Mamidi R, Mallampalli SL, Chandra M. The N-Terminal Extension of Cardiac Troponin T Stabilizes the Blocked State of Cardiac Thin Filament. Biophys. J. 2012 doi: 10.1016/j.bpj.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mamidi R, Mallampalli SL, Wieczorek DF, Chandra M. Divergent effects of N-Terminus of cardiac troponin T and isoforms of tropomyosin on myofiber tension and Ca2+ sensitivity. J. Physiol. (Lond) ((Unpublished results)) [Google Scholar]
- 35.Sumandea MP, Vahebi S, Sumandea CA, Garcia-Cazarin ML, Staidle J, Homsher E. Impact of cardiac troponin T N-terminal deletion and phosphorylation on myofilament function. Biochemistry. 2009;48:7722–7731. doi: 10.1021/bi900516n. [DOI] [PubMed] [Google Scholar]
- 36.Schaertl S, Lehrer SS, Geeves MA. Separation and characterization of the two functional regions of troponin involved in muscle thin filament regulation. Biochemistry. 1995;34:15890–15894. doi: 10.1021/bi00049a003. [DOI] [PubMed] [Google Scholar]
- 37.Mak AS, Smillie LB. Structural interpretation of the two-site binding of troponin on the muscle thin filament. J Mol Biol. 1981;149:541–550. doi: 10.1016/0022-2836(81)90486-1. [DOI] [PubMed] [Google Scholar]
- 38.White SP, Cohen C, Phillips GN., Jr Structure of co-crystals of tropomyosin and troponin. Nature. 1987;325:826–828. doi: 10.1038/325826a0. [DOI] [PubMed] [Google Scholar]
- 39.Campbell K. Rate constant of muscle force redevelopment reflects cooperative activation as well as cross-bridge kinetics. Biophys J. 1997;72:254–262. doi: 10.1016/S0006-3495(97)78664-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeda S, Yamashita A, Maeda K, Maeda Y. Structure of the core domain of human cardiac troponin in the Ca2+-saturated form. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 41.Vinogradova MV, Stone DB, Malanina GG, Karatzaferi C, Cooke R, Mendelson RA, Fletterick RJ. Ca2+-regulated structural changes in troponin. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5038–5043. doi: 10.1073/pnas.0408882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montgomery DE, Tardiff JC, Chandra M. Cardiac troponin T mutations: correlation between the type of mutation and the nature of myofilament dysfunction in transgenic mice. J. Physiol. (Lond.) 2001;536:583–592. doi: 10.1111/j.1469-7793.2001.0583c.xd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo X, Wattanapermpool J, Palmiter KA, Murphy AM, Solaro RJ. Mutagenesis of cardiac troponin I. Role of the unique NH2-terminal peptide in myofilament activation. J. Biol. Chem. 1994;269:15210–15216. [PubMed] [Google Scholar]
- 44.Pan BS, Johnson RG., Jr Interaction of cardiotonic thiadiazinone derivatives with cardiac troponin C. J Biol Chem. 1996;271:817–823. [PubMed] [Google Scholar]
- 45.Placek BJ, Gloss LM. The N-terminal tails of the H2A-H2B histones affect dimer structure and stability. Biochemistry. 2002;41:14960–14968. doi: 10.1021/bi026283k. [DOI] [PubMed] [Google Scholar]
- 46.Andrade MA, Chacon P, Merelo JJ, Moran F. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 1993;6:383–390. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- 47.Merelo JJ, Andrade MA, Prieto A, Moran F. Proteinotopic Feature Maps. Neurocomputing. 1994;6:443–454. [Google Scholar]
- 48.Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 49.de Tombe PP, Stienen GJ. Protein kinase A does not alter economy of force maintenance in skinned rat cardiac trabeculae. Circ. Res. 1995;76:734–741. doi: 10.1161/01.res.76.5.734. [DOI] [PubMed] [Google Scholar]
- 50.Stienen GJ, Zaremba R, Elzinga G. ATP utilization for calcium uptake and force production in skinned muscle fibres of Xenopus laevis. J. Physiol. 1995;482(Pt 1):109–122. doi: 10.1113/jphysiol.1995.sp020503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fabiato A, Fabiato F. Calculator programs for computing the composition of the solutions containing multiple metals and ligands used for experiments in skinned muscle cells. J. Physiol. (Paris) 1979;75:463–505. [PubMed] [Google Scholar]
- 52.Chandra M, Tschirgi ML, Ford SJ, Slinker BK, Campbell KB. Interaction between myosin heavy chain and troponin isoforms modulate cardiac myofiber contractile dynamics. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1595–R1607. doi: 10.1152/ajpregu.00157.2007. [DOI] [PubMed] [Google Scholar]
- 53.Brenner B, Eisenberg E. Rate of force generation in muscle: correlation with actomyosin ATPase activity in solution. Proc. Natl. Acad. Sci. U. S. A. 1986;83:3542–3546. doi: 10.1073/pnas.83.10.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chandra M, Tschirgi ML, Tardiff JC. Increase in tension-dependent ATP consumption induced by cardiac troponin T mutation. Am J Physiol Heart Circ Physiol. 2005;289:H2112–H2119. doi: 10.1152/ajpheart.00571.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.