Abstract
We have demonstrated that curcumin is an amyloid-specific dye in E. coli. Curcumin binds to curliated whole cells and to isolated curli amyloid fibers. Similar to Congo red, curcumin exhibits a red-shift in absorbance and a significant increase in fluorescence upon binding to isolated curli.
Since the late 1800s, staining of cells and cell structures by small molecule dyes has proven to be an indispensible tool in microbiology1. Today, dyes are crucial tools in the study of bacterial communities called biofilms. Biofilms consist of bacterial cells plus their secreted extracellular matrix comprising a complex mixture of proteins, polysaccharides, and sometimes DNA2. Due to their chemical and structural complexity and insolubility, biofilms are difficult to analyze. However, indicator dyes such as crystal violet, Calcofluor White, and Congo red enable rapid screening of biofilm production and qualitative identification of biofilm components3–5.
The anionic diazo dye, Congo red (CR), is well known for its amyloid binding properties6. Indeed, the definitive diagnosis of Alzheimer’s disease is made upon post-mortum CR staining of brain tissue7. The use of CR as an indicator dye also has a rich history of applications in microbiology and has been used extensively to detect the production of various extracellular polysaccharides and proteins in bacteria and in yeast8–13. In the case of E. coli and Salmonella, these organisms produce extracellular fibers termed curli14, 15, involved in adhesion and biofilm formation, that were discovered to be amyloid in 200216. Since the discovery of curli as amyloid, an increasing number of microbial functional amyloids have been identified and are being investigated to understand their function in microbial ecology and to dissect amyloid-assembly processes17.
CR has been routinely employed to score the production of curli among E. coli and Salmonella species18, 19. CR-binding phenotypes have been valuable in studies of curli biogenesis in E. coli strain MC4100, a strain frequently used to manipulate curli assembly16. When grown on CR-containing agar medium, curliated whole cells bind CR and deplete the dye from the underlying agar. However, because CR can bind to other cellular features in some strains, the curli-dependence of dye binding must first be established.
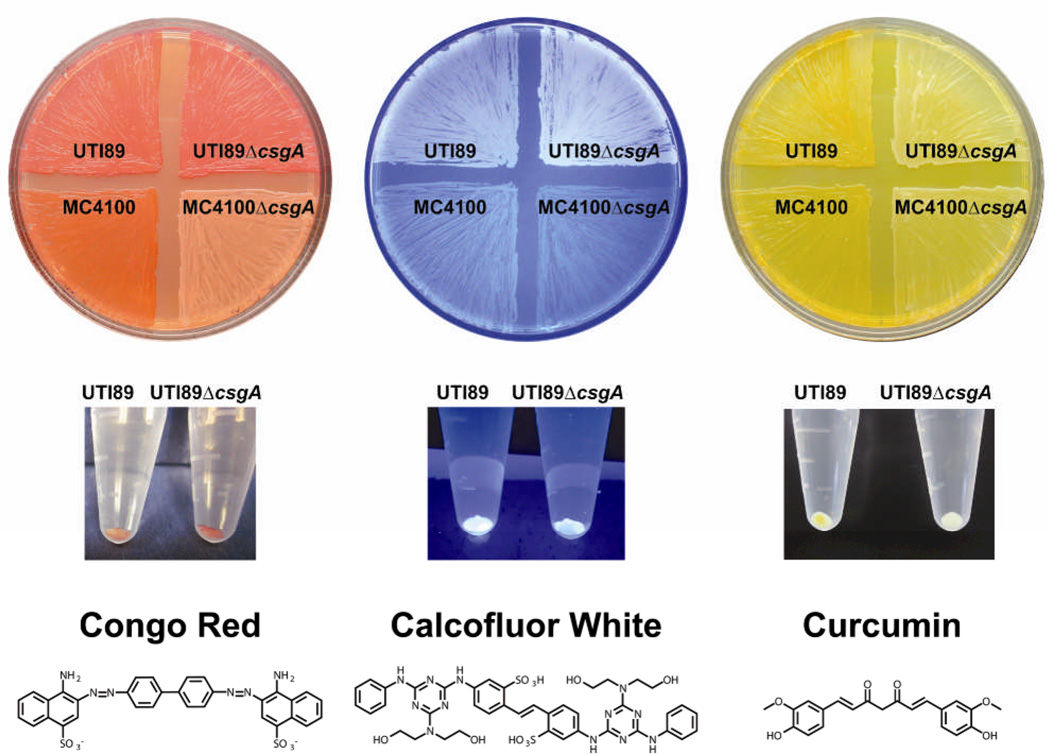
Here, we demonstrate that CR binding does not exhibit curli specificity in the biofilm-forming uropathogenic E. coli strain UTI89, a well-studied strain in models of urinary tract bacterial pathogenesis. We also report our discovery that the natural product curcumin binds E. coli cells in a curli-dependent manner. We grew two strains, MC4100 and UTI89 along with their respective curli knock-out strains, MC4100ΔcsgA and UTI89ΔcsgA, on YESCA agar supplemented with 25 µg/mL CR for 60 hours at 26 °C, typical growth conditions that promote curli expression13. MC4100 does not form biofilms as MC4100 can produce curli but does not produce cellulose, a determinant of biofilm formation in E. coli. UTI89 produces both curli and cellulose which contribute to its ability to form biofilms on agar, on plastic, and at the air-liquid interface20. As expected, CR binds to the curli-producing strains MC4100 and UTI89 and does not bind to MC4100ΔcsgA, a curli mutant lacking the major fiber subunit protein (Figure 1). However, the UTI89 curli mutant, UTI89ΔcsgA, binds CR, although it does not produce curli (Figure 1). Each strain was also grown in the absence of CR on standard YESCA agar and then harvested, resuspended in PBS, and normalized to yield an OD600 1.0 suspension in 1 mL PBS.
Figure 1.
Dye binding to MC4100 and UTI89 wild-type and curli-mutant strains during growth on YESCA agar plates supplemented with the indicated molecules (top row) and after growth on YESCA agar without compound supplementation via a solution-based pull-down assay (middle row). CR binding is not curli specific in UTI89. Calcofluor-based fluorescence indicates the production of cellulose by UTI89 and UTI89ΔcsgA. Curcumin exhibits curli-specific binding in UTI89 even in the presence of cellulose.
CR from a concentrated stock solution was added to each sample to yield a final concentration of 10 µg/mL CR. Cells were incubated for ten minutes in a microcentrifuge tube, centrifuged at 9600 g, and photographed. As illustrated in Figure 1, CR is also able to bind to both UTI89 and UTI89ΔcsgA whole cells in this assay. Both assays illustrate the lack of specificity and problems associated with using CR staining as an amyloid dye for many microorganisms. Indeed, CR is known to bind to cellulose21, 22, a common biofilm component. UTI89 produces both curli and cellulose in the extracellular matrix, whereas MC4100 produces only curli. Thus, CR binding is curli-specific in MC4100 because it does not also produce cellulose. The presence of cellulose is often identified qualitatively through Calcofluor binding and fluorescence. As shown in Figure 1, both UTI89 and UTI89ΔcsgA exhibit Calcofluor-based fluorescence when grown on a YESCA agar plate containing 20 µg/mL Calcofluor. MC4100 and MC4100ΔcsgA, which do not produce cellulose, do not exhibit significant Calcofluor-based fluorescence. Interestingly, for the UTI89 pair, the fluorescence of Calcofluor is reproducibly somewhat greater in the absence of curli. It is possible that Calcofluor binding to cellulose is inhibited by curli-cellulose interactions or that the environment in the presence of curli leads to reduced fluorescence.
Naturally, more specific amyloid dyes as alternatives to CR would be valuable for use in microbiological assays. We discovered that the natural product curcumin, also studied for its ability to interact with amyloid β 23, was amyloid-specific and served as a reliable indicator of curli production in UTI89 (Figure 1). Curucmin uptake is obvious among MC4100 and UTI89 grown on YESCA agar medium containing 18 µg/mL curcumin, whereas MC4100ΔcsgA and UTI89ΔcsgA do not bind curcumin. It is conceivable that the lack of curcumin binding on agar could be attributed to possible inhibition of cellulose production during growth in the presence of curcumin. Thus, the solution-based pull-down assay using cells grown in the absence of curcumin is important to confirm curli-binding specificity. The assay was also performed using Calcofluor. The molecule concentrations were 0.1 µg/mL Calcofluor and 1.8 µg/mL curcumin. The results of these pull-down assays paralleled what was observed in the cells grown in compound-supplemented agar. Notably, UTI89 binds curcumin in the pull-down assay and appears yellow, whereas UTI89ΔcsgA does not and appears white. Thus, curcumin binding is curli specific, even in the presence of cellulose and any other extracellular components in UTI89ΔcsgA.
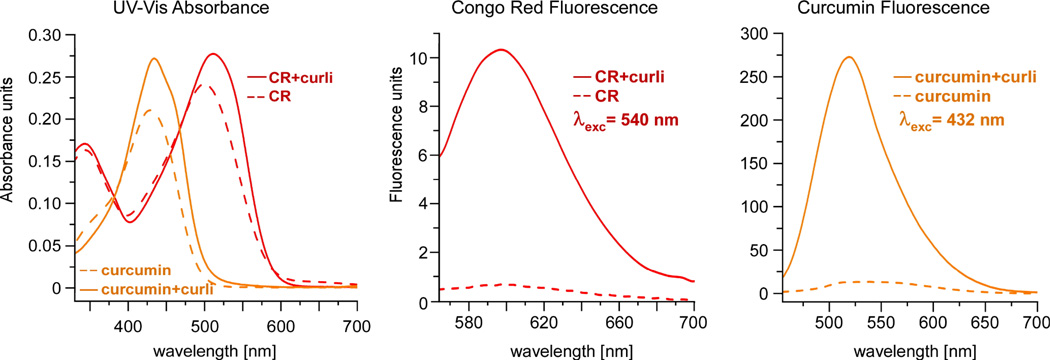
CR has also been examined in many biophysical studies of amyloid fibers in vitro, and is noted for its red-shift in absorbance as well as its characteristic fluorescence when bound to amyloid fibers 24. Thus, we examined the ability of curcumin to interact with curli fibers in vitro and whether curli-bound curcumin would exhibit similar properties. CR or curcumin were individually added from a 0.5 mM stock solution in DMSO to a solution of 1 mg/mL of isolated curli, to yield a final dye concentration of 25 µM. Each sample was examined by UV-Vis spectrophotometry. The fluorescence emission spectra of each dye alone and in the curli-bound samples were also obtained upon excitation at 540 nm for CR and at 432 nm for curcumin. As shown in Figure 2, the UV- Vis spectrum of curli-bound curcumin is red-shifted from a λmax of 423 nm to 432 nm. Furthermore, although curcumin itself exhibits some detectable fluorescence, there is a significant increase in the fluorescence emission, centered at 521 nm, when bound to curli, similar to the behavior of Congo red (Figure 2). In addition, the fluorescence intensity is greater for curcumin than for CR. Thus, curcumin also exhibits the useful spectral properties ascribed to CR when bound to isolated curli16.
Figure 2.
Binding of curcumin to curli results in a red shift in the absorbance spectrum, similar to the spectral shift observed for Congo red and commonly used to characterize amyloids (left). Like Congo red, curcumin exhibits significantly enhanced fluorescence when bound to curli (middle and right).
Since the discovery of curli as amyloid in 2002, there is an increasing number of functional amyloids being identified among microbial species and the identification of amyloid-specific dyes for use in the chemically complex background of microbial biofilm communities will be of value. We have demonstrated that curcumin is an amyloid-specific dye in E. coli and may prove to be valuable in examining amyloid production among other environmental and host-associated microbial species.
Acknowledgments
The authors acknowledge funding from the NIH Director’s New Innovator Award (DP2 OD007488), the Stanford Terman Fellowship, and Stanford University. LC is the recipient of a Burroughs Wellcome Career Award at the Scientific Interface. XZ is the recipient of a Stanford Interdisciplinary Graduate Fellowship.
Footnotes
This article is part of the ‘Emerging Investigators 2013’ themed issue for ChemComm.
References
- 1.Clark G, Kasten FH. History of Staining. Williams and Wilkins; Baltimore: 1983. [Google Scholar]
- 2.Flemming HC, Wingender J. Nature Reviews Microbiology. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 3.Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, Melton DM, Beachey EH. Journal of Clinical Microbiology. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Toole GA, Kolter R. Molecular Microbiology. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 5.Bokranz W, Wang XD, Tschape H, Romling U. Journal of Medical Microbiology. 2005;54:1171–1182. doi: 10.1099/jmm.0.46064-0. [DOI] [PubMed] [Google Scholar]
- 6.Frid P, Anisimov SV, Popovic N. Brain Res Rev. 2007;53:135–160. doi: 10.1016/j.brainresrev.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Picken MM. Arch Pathol Lab Med. 134:545–551. doi: 10.5858/134.4.545. [DOI] [PubMed] [Google Scholar]
- 8.Qadri F, Hossain SA, Ciznar I, Haider K, Ljungh A, Wadstrom T, Sack DA. Journal of Clinical Microbiology. 1988;26:1343–1348. doi: 10.1128/jcm.26.7.1343-1348.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freeman DJ, Falkiner FR, Keane CT. Journal of Clinical Pathology. 1989;42:872–874. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishiguro EE, Ainsworth T, Trust TJ, Kay WW. Journal of Bacteriology. 1985;164:1233–1237. doi: 10.1128/jb.164.3.1233-1237.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stugard CE, Daskaleros PA, Payne SM. Infection and Immunity. 1989;57:3534–3539. doi: 10.1128/iai.57.11.3534-3539.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collinson SK, Emody L, Trust TJ, Kay WW. J Bacteriol. 1992;174:4490–4495. doi: 10.1128/jb.174.13.4490-4495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 14.Olsen A, Jonsson A, Normark S. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 15.Collinson SK, Clouthier SC, Doran JL, Banser PA, Kay WW. J Bacteriol. 1996;178:662–667. doi: 10.1128/jb.178.3.662-667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR. Trends in Microbiology. 2012;20:66–73. doi: 10.1016/j.tim.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collinson SK, Emody L, Trust TJ, Kay WW. Journal of Bacteriology. 1992;174:4490–4495. doi: 10.1128/jb.174.13.4490-4495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Molecular Microbiology. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 20.Cegelski L, Pinkner JS, Hammer ND, Cusumano CK, Hung CS, Chorell E, Aberg V, Walker JN, Seed PC, Almqvist F, Chapman MR, Hultgren SJ. Nat Chem Biol. 2009;5:913–919. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morey DR. Textile Research Journal. 1934;5:105–109. [Google Scholar]
- 22.Puchtler H, Sweat F, Levine M. Journal of Histochemistry & Cytochemistry. 1962;10:355. -&. [Google Scholar]
- 23.Yang F, Lim GP, Begum AN, Ubeda OJ, Simmons MR, Ambegaokar SS, Chen PP, Kayed R, Glabe CG, Frautschy SA, Cole GM. J Biol Chem. 2005;280:5892–5901. doi: 10.1074/jbc.M404751200. [DOI] [PubMed] [Google Scholar]
- 24.Klunk WE, Pettegrew JW, Abraham DJ. J Histochem Cytochem. 1989;37:1273–1281. doi: 10.1177/37.8.2666510. [DOI] [PubMed] [Google Scholar]