Abstract
Upon illumination the visual receptor rhodopsin (Rho) transitions to the activated form Rho*, which binds the heterotrimeric G protein, transducin (Gt) causing GDP to GTP exchange and Gt dissociation. Using succinylated concanavalin A (sConA) as a probe, we visualized native Rho dimers solubilized in 1 mM n-dodecyl-β-D-maltoside (DDM) and Rho monomers 5 mM in DDM. By nucleotide depletion and affinity chromatography together with crosslinking and size exclusion chromatography, we trapped and purified nucleotide-free Rho*•Gt and sConA-Rho*•Gt complexes kept in solution by either DDM or lauryl-maltose-neopentyl-glycol (LMNG). The 3-D envelope calculated from projections of negatively stained Rho*•Gt-LMNG complexes accommodated two Rho molecules, one Gt heterotrimer and a detergent belt. Visualization of triple sConA-Rho*•Gt complexes unequivocally demonstrated a pentameric assembly of the Rho*•Gt complex in which the photoactivated Rho* dimer serves as a platform for binding the Gt heterotrimer. Importantly, individual monomers of the Rho* dimer in the heteropentameric complex exhibited different capabilities to be regenerated with either 11-cis or 9-cis-retinal.
Keywords: G protein-coupled receptor, heterotrimeric G protein, photoactivated rhodopsin dimer, transmission electron microscopy, transducin
INTRODUCTION
G protein-coupled receptors (GPCRs) transduce signals across cellular membranes, and are involved in most physiological processes. Rhodopsin (Rho), the primary molecule in the visual signaling cascade of rod photoreceptor cells, is the most widely studied GPCR and a paradigm for understanding signal transduction. Upon absorption of a single photon Rho transitions to the activated state (Rho*), which catalyzes nucleotide exchange on multiple transducin (Gt) heterotrimers, providing an initial signal amplification (Hofmann et al., 2009; Palczewski, 2006; Sakmar et al., 2002). Down-stream events amplify the photon-induced signal to ultimately produce a detectable electric pulse, the visual signal processed by vertebrate brains. The first crystal structure determined for a GPCR was that of Rho (Palczewski et al., 2000), and Rho crystals have yielded the highest resolution native structure of any GPCR determined to date (Okada et al., 2004). The structure of Rho* has been solved (Salom et al., 2006), the interaction site between Rho and the Gt C-terminus structurally defined (Choe et al., 2011; Scheerer et al., 2008; Standfuss et al., 2011), and Gtα activation by Rho* characterized by site-directed spin labeling and spectroscopy (Van Eps et al., 2011) as well as by hydroxyl radical labeling and deuterium uptake in combination with mass spectrometry (Orban et al., 2012). But an unresolved question concerns the molecular function(s) of Rho dimers and higher oligomers observed in native disc membranes (Fotiadis et al., 2003).
Binding of a single heterotrimeric G protein to GPCR dimers and formation of pentameric complexes has been observed in reconstitution experiments involving the leukotriene B4 receptor BLT1 (Baneres et al., 2003), dopamine D2 receptor (Han et al., 2009), serotonin 5-HT4 receptor (Pellissier et al., 2011), photoactivated Rho (Jastrzebska et al., 2011b), and the metabotropic glutamate receptor (El Moustaine et al., 2012), each with its respective G protein. Extensive evidence indicates the existence of Rho dimers in native disc membranes (Fotiadis et al., 2003; Jastrzebska et al., 2006; Jastrzebska et al., 2004; Suda et al., 2004), and of other GPCRs dimers in living cells (serotonin receptor 5-HT2C (Herrick-Davis et al., 2006; Herrick-Davis et al., 2012), β1-adrenergic receptor (Kobayashi et al., 2009), β2-adrenergic receptor (b2AR) (Angers et al., 2001; Angers et al., 2000)). In contrast, the most detailed structural insight into the receptor-G protein interaction provided by the recently solved crystal structure of T4 lysozyme (T4L)-b2AR•Gs•nanobody complex indicates a binding stoichiometry of one receptor and one G protein (Rasmussen et al., 2011). This is supported by a two-dimensional projection analysis of negatively stained T4L-b2AR•Gs•nanobody complexes purified in lauryl-maltose-neopentyl-glycol (LMNG). The significant size-mismatch between the projection map and the receptor monomer was explained by an unusually large size of the LMNG micelles (Westfield et al., 2011).
Single receptor activation triggering a physiological response in vitro has been observed for some GPCRs (Whorton et al., 2007; Whorton et al., 2008). However, the molecular envelope of negatively stained native Rho*•Gt complexes purified in n-dodecyl-β-D-maltoside (DDM) accommodated two Rho molecules and one Gt, in agreement with biochemical analyses and the mass of the 220±12 kDa complex determined by electron scattering (Jastrzebska et al., 2011b). To resolve uncertainties about the composition of this GPCR-G protein complex, we prepared Rho*•Gt in LMNG and DDM, and chose sConA as a labeling probe for the Rho molecules. Visualization of the negatively stained sConA-Rho complexes solubilized in either 1 or 5 mM DDM, as well as the triple sConA-Rho*•Gt complex unequivocally revealed that photoactivation promotes assembly of a pentameric complex in which a Rho* dimer provides a binding platform for the Gt heterotrimer. Furthermore we show that the two Rho monomers constituting the Rho* dimer in the activated complex have distinct functional properties, as demonstrated by their different capabilities to be regenerated with either 11-cis or 9-cis-retinal.
RESULTS
Formation and visualization of sConA-Rho complexes
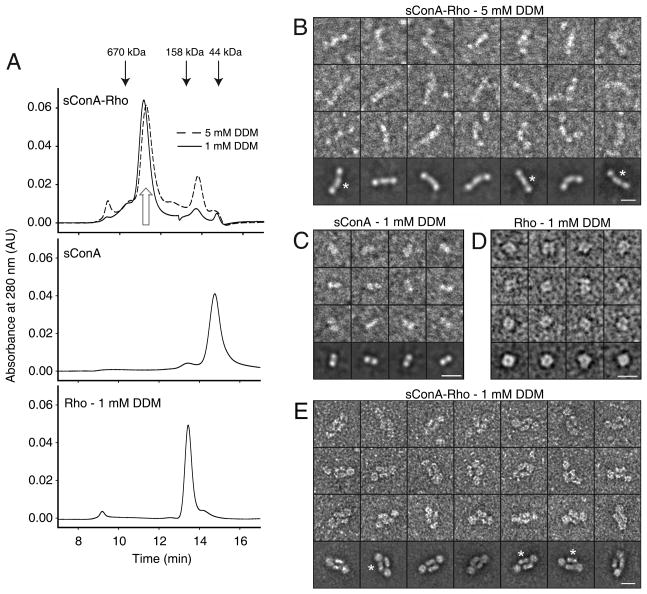
Purified bovine Rho was incubated with sConA to visualize whether the native Rho dimer (Fotiadis et al., 2003) exists in solution. The sConA-Rho complexes formed were submitted to size exclusion chromatography in buffer containing either 1 mM DDM (Fig. 1A, solid line, top panel) or 5 mM DDM (Fig. 1A, broken line, top panel). Gel filtration elution profiles were similar under both conditions with the elution peaks of the sConA-Rho complexes significantly shifted towards larger structures relative to the elution peak of either sConA (Fig. 1A, middle panel) or Rho in 1 mM DDM (Fig. 1A, bottom panel). However, the profile obtained in 1 mM DDM (Fig. 1A, solid line, top panel) revealed a distinctly larger shift than that in 5 mM DDM (Fig. 1A, broken line, top panel).
Figure 1. Purification and visualization of sConA-Rho complexes.
A) Gel filtration elution profile of the sConA-Rho complex in 1 mM DDM (solid line, top panel) and 5 mM DDM (broken line, top panel) compared with sConA (middle panel) and Rho alone (bottom panel). Open arrow indicates the peak fraction used for further characterization. Black arrows on top of the chromatogram indicate the elution profiles of molecular standards: thyroglobulin (670 kDa), IgG (158 kDa) and ovalbumin (44 kDa). Negatively stained and manually selected particles (3 top rows) and class averages (bottom row) obtained from selected particles of sConA-Rho complex purified in 5 mM DDM (B), sConA alone (C), Rho dimer purified in 1 mM DDM (D), and sConA-Rho complex purified in 1 mM DDM (E). sConA exhibits a bi-modal density (C) that is seen in class averages of both types of sConA-Rho complexes (*). Scale bars 100 Å.
TEM analysis of sConA-Rho complexes kept in 5 mM DDM revealed heterogeneous particle populations. Most of the tri-lobed complexes featured a central elongated density with similar spherical domains attached to both ends (Fig. 1B), whereas a minority of particles had a central domain consisting of two elongated particles (not shown). Class averages showed some central domains with a bi-modal appearance (Fig. 1B, asterisks). Size and shape of negatively stained sConA dimers documented that the central domain of the sConA-Rho complexes represents the sConA moiety (Fig. 1C). Negatively stained Rho in 1 mM DDM revealed two elongated bimodal domains separated by a stained groove (Fig. 1D). These dimers exhibited a length of 65±4 Å (n=12) and a width of 56±4 Å (n=12). Apparently, they have the capacity to bind two sConA dimers, as revealed by structures composed of two parallel sConA dimers with domains accounting for Rho on each side (Fig. 1E). Rho domains exhibited a width of 63 ± 4 Å in 1 mM DDM, and 40 ± 2 Å in 5 mM DDM, compatible with the dimensions of Rho dimers shown in Fig. 1D and Rho monomers observed previously (Jastrzebska et al., 2006).
Formation and visualization of native Rho*•Gt complexes
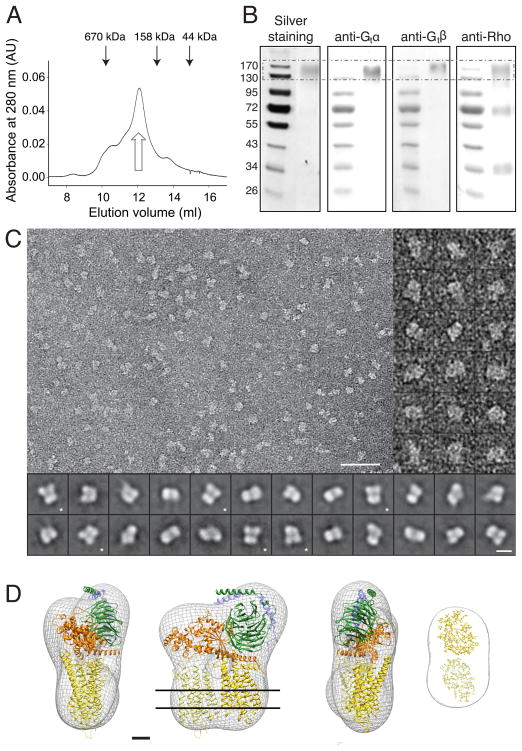
The Rho*•Gt complex was formed and purified by using sConA affinity chromatography as described previously (Jastrzebska et al., 2011b). Solubilized in DDM and bound to the resin, Rho was activated by light and then saturated with Gt. The resulting Rho*•Gt-DDM complex was eluted from the column with α-methyl-D-mannopyranoside and immediately crosslinked with DSG, a short (spacer arm 7.7 Å) Lys residue -Lys residue homobifunctional crosslinker, to prevent complex dissociation. The crosslinked Rho*•Gt complex was separated from free Rho and Gt by size exclusion chromatography and fractions containing the Rho*•Gt complex were gel-filtered again to assure protein homogeneity and/or to exchange DDM for LMNG (Fig. 2A). The Rho*•Gt-LMNG complex migrated faster than the Rho*•Gt-DDM complex (supplementary Fig. S1A), and its calculated molecular mass was ~280 kDa, 60 kDa larger than the mass of the complex in DDM (Jastrzebska et al., 2011b), indicating that LMNG forms larger micelles than DDM. SDS-PAGE analysis of these complexes revealed a protein band that migrated at a molecular mass of ~160 kDa, supporting a 2:1 Rho:Gt stoichiometry (2 Rho - 78 kDa + Gt - 86 kDa = 164 kDa) (Fig. 2B, left panel (LMNG) and supplementary Fig. S1B, left panel (DDM)). Immunoblotting with antibodies against Gtα, Gtβ and Rho confirmed this composition of the Rho*•Gt complexes (Fig. 2B, right panels (LMNG) and supplementary Fig. S1B, right panel (DDM)).
Figure 2. Visualization and 3D reconstruction of the Rho*•Gt-LMNG complex.
A) Gel filtration elution profiles of purified DSG-crosslinked Rho*•Gt complexes in LMNG. Open arrow indicates protein elution fraction used for further analyses. Black arrows on top of the chromatogram indicate the elution profiles of molecular standards: thyroglobulin (670 kDa), IgG (158 kDa) and ovalbumin (44 kDa). B) Silver-stained SDS-PAGE gel and immunoblot analyses of the peak fraction in A incubated with antibodies against Gtα, Gtβ and Rho. Identification of DSG-crosslinked Rho*•Gt complexes is indicated by the broken line frame. C) TEM image of negatively stained Rho*•Gt-LMNG complexes (upper left panel). Scale bar 500 Å. Negatively stained and manually selected particles of Rho*•Gt-LMNG (upper right panel). 2D class averages obtained from particles selected from images taken at 0° tilt (bottom panel). Scale bar 100 Å. Asterisks indicate projections of class averages clearly displaying a tri-lobed morphology of the complex. D) Semi-empirical model of the Rho*•Gt complex fitted into 3D maps calculated from projections of DSG-crosslinked Rho*•Gt complex purified in LMNG. Based on the Fourier shell correlation function, the resolution of the map obtained for the Rho*•Gt-LMNG was determined to 29 Å. The 20 Å thick slice on the right shows the volume occupied by the detergent. Scale bar 20 Å. In the Rho*•Gt complex, the activated Rho monomer in the dimer is depicted in dark yellow and the second Rho is shown in light yellow. Gtα, Gtβ, Gtγ are colored orange, green and pale blue, respectively.
Negatively stained Rho*•Gt-LMNG complexes revealed ~130 Å long particles with a variable width (Fig. 2C, left and right panels). Similar particles were found when Rho*•Gt was purified in DDM (supplementary Fig. S2), confirming our previous results (Jastrzebska et al., 2011b). Image pairs from the same area of untilted and tilted (45°) grids were used to select particles based on their dimensions in the untilted sample, yielding class averages that showed a tri-lobed complex (Fig. 2C, bottom panel). The 3D map calculated by refining the initial model against 12950 projections of Rho*•Gt-LMNG complexes accommodated the model constructed previously based on the 3D envelope of Rho*•Gt-DDM, taking into account all structural and chemical information available ((Jastrzebska et al., 2011b); Fig. 2D).
Visualization of purified sConA-Rho*•Gt complexes
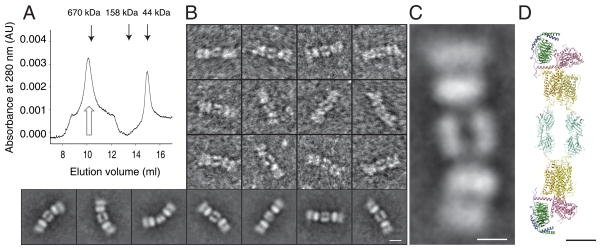
Binding of DSG-crosslinked Rho*•Gt-LMNG complexes to sConA yielded triple sConA-Rho*•Gt complexes, which were isolated by size exclusion chromatography (Fig. 3A). The triple complex elution peak was dramatically shifted compared to the elution peak of Rho*•Gt complexes (Fig. 2A) indicating the formation of larger structures. TEM analysis of negatively stained sConA-Rho*•Gt complexes (Fig. 3B, upper panel) and calculation of 2D class averages from manually selected particles revealed elongated structures with lengths of 325±12 Å (Fig. 3B, bottom panel). These complexes were composed of two elongated densities lying parallel to each other in the middle of the complex and facing two bi-modal densities on each side (Fig. 3C). The bi-modal densities correspond to the Rho*•Gt-LMNG complex displayed in Fig. 2D, in which Gt (wider domain) binds Rho (narrower domain) on the C-terminal side. The N-terminal side of Rho, however, binds to sConA to form the elongated triple complex. The width of the Rho densities was smaller in triple complexes than in some class averages of Rho*•Gt-LMNG complexes (Fig. 2C, bottom panel; Table 1), indicating a flexible arrangement of the Rho*•Gt pentamer with respect to sConA consistent with the flexibility of the mannose chain. A semi empirical model of the sConA-Rho*•Gt complex built based on views obtained from 2D projections and using the crystal structure of a ConA dimer (pdb: 1DQ4) and the model of the Rho*•Gt complex (Jastrzebska et al., 2011b) supports our model of the triple complex composition (Fig. 3D).
Figure 3. Purification and visualization of the triple sConA-Rho*•Gt complex.
A) Gel filtration elution profile of sConA-Rho*•Gt-LMNG complex. Open arrow indicates peak fraction used for further characterization. B) Negatively stained and manually selected particles (3 top rows) and class averages (bottom row) obtained from selected particles of sConA-Rho*•Gt complex. C) Magnified 2D projection of the sConA-Rho*•Gt complex. D) Semi-empirical model of the sConA-Rho*•Gt complex composed of two Rho*•Gt complexes and two sConA dimers. This model was built based on views obtained from 2D projections and using a known crystal structure of a sConA dimer (pdb: 1DQ4) and a model of the Rho*•Gt complex (Jastrzebska et al., 2011b). Scale bars 50 Å. In the Rho*•Gt complex, activated Rho in the dimer is depicted in dark yellow and the second Rho is shown in light yellow. Gtα, Gtβ, Gtγ are colored dark salmon, light green and pale blue, respectively. sConA dimers are shown in cyan.
TABLE 1.
Statistical analyses of particle sizes.
| Preparation | Length of the complex (Å) | n of particles | Width of Rho(Å) | Detergent belt thickness (Å)e |
|---|---|---|---|---|
| sConA-Rho_5 mM DDM a, c | 191 ± 13 | 14 | 40 ± 2 | - |
| sConA-Rho_1 mM DDM b, c | 195 ± 10 | 12 | 63 ± 4 | - |
| sConA a, c | 84 ± 3 | 14 | - | - |
| Rho*•Gt_LMNG a, d | 129 ± 7 | 15 | 107 ± 6 | 14 (15±5) |
| Rho*•Gt_DDM a, d | 130 ± 7 | 15 | 91 ± 4 | 6 (11±4) |
| sConA-Rho*•Gt_LMNG a, c | 325 ± 12 | 14 | 93 ± 5 | - |
Images were collected with a aTf20 and a bCM10 transmission microscope. cParticles or dparticle pairs from images collected at 0° and 45° tilt were selected manually by using the boxer or the pair-picking features of EMAN2. eThe detergent belt thickness, Tbelt, was calculated from the width of Rho, WRho, measured on a micrograph as Tbelt=(WRho-80)/2, where the width of the protein is 80 Å. Tbelt also was estimated from the difference of the iso-contoured 3D map and the fitted model measured on 3 sections (in brackets). The discrepancy between the width of Rho dimers in sConA-Rho-DDM and Rho*•Gt-DDM is due to the preferential orientation of the latter after adsorption to a carbon film.
Asymmetry of Rho monomers in the Rho*•Gt complex
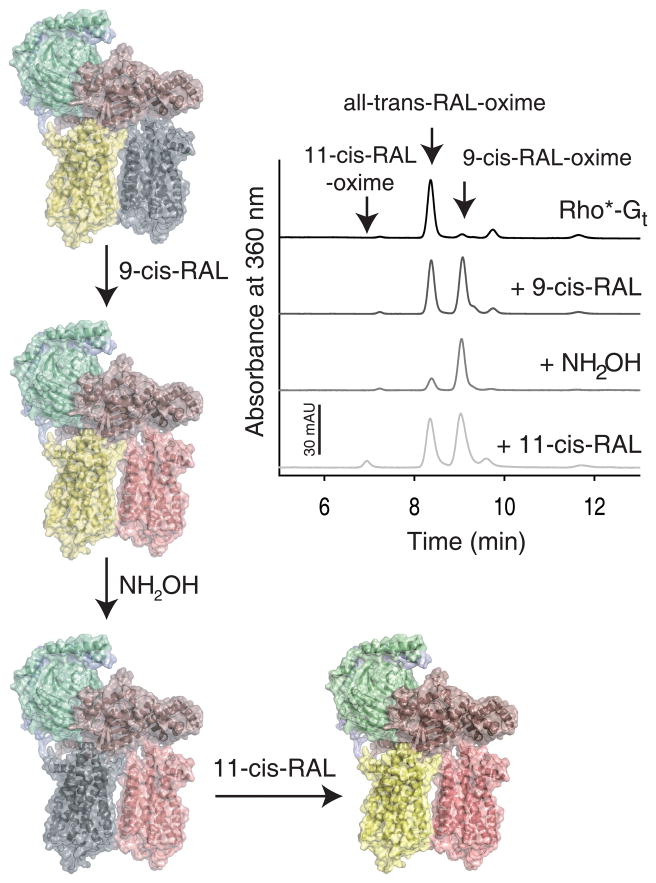
Our TEM analysis revealed that photoactivation promotes binding of heterotrimeric Gt to a Rho dimer. As a result of light activation, Rho’s 11-cis-retinylidene chromophore isomerizes to all-trans-retinylidene followed by Schiff base linkage hydrolysis and release of all-trans-retinal (Jastrzebska et al., 2011a) unless a Rho*•Gt complex is formed. As shown previously (Jastrzebska et al., 2011b), this complex exhibits a UV-visible maximum absorption peak at 380 nm and contains only all-trans-retinal bound, indicating that Gt stabilizes the activated conformation of Rho and inhibits all-trans-retinal release from the retinal-binding pocket. Taking into account an asymmetric nature of the Rho*•Gt complex (the C-terminus of Gtα binds only one Rho) it is unlikely that both Rho monomers in this complex are structurally and functionally equal. To validate this hypothesis, we quantified the retinoid content in the Rho*•Gt complex and in the complex incubated with 9-cis-retinal. Because chromophore regeneration was performed while the Rho*•Gt was still bound to the sConA resin, excess retinoid was washed out and did not affect this analysis. Before 9-cis-retinal treatment the Rho*•Gt complex contained only all-trans-retinal (Fig. 4, top black chromatogram and model of yellow-grey Rho dimer with bound Gt) but the complex regenerated with 9-cis-retinal contained both all-trans-retinal and 9-cis-retinal in ~ 1:1 stoichiometry (Fig. 4, upper middle dark grey chromatogram and model of yellow-pink Rho dimer with bound Gt). To achieve chromophore release from the Rho molecule stabilized by Gt, a wash with the strong nucleophile hydroxylamine (NH2OH) was required to promote Schiff base hydrolysis. The resulting complex was depleted of all-trans-retinal but still contained 9-cis-retinal, because ground-state Rho and 9-cis-retinal regenerated opsin called isoRho are not affected by this NH2OH exposure (Fig. 4, lower middle grey chromatogram and model of grey-pink Rho dimer with bound Gt). Surprisingly, incubation of such complexes with 11-cis-retinal produced complexes containing both all-trans-retinal and 9-cis-retinal in a 1:1 molar ratio, but virtually lacking 11-cis-retinal (Fig. 4, bottom light grey chromatogram and model of yellow-pink Rho dimer with bound Gt).
Figure 4. Asymmetry of Rho monomers in Rho*•Gt complex.
The Rho*•Gt complex prepared on a sConA affinity resin (shown as a yellow-grey dimer bound to Gt) was first regenerated with 9-cis-retinal (shown as a yellow-pink dimer bound to Gt) followed by treatment with NH2OH (modeled as a grey-pink dimer bound to Gt) and then regenerated with 11-cis-retinal (shown as a yellow-pink dimer bound to Gt). Isocratic analyses of retinoid oximes extracted from Rho*•Gt (black line), either regenerated with 9-cis-retinal (dark grey line), treated with NH2OH (grey line) or regenerated with 11-cis-retinal (light grey line) are shown. Though only all-trans-retinal was detected in the Rho*•Gt complex, a mixture of all-trans-retinal and 9-cis-retinal with about a 1:1 stoichiometry was found in the Rho*•Gt complex incubated with 9-cis-retinal. Treatment of this regenerated complex with NH2OH caused a significant reduction in all-trans-retinal. However, incubation of this complex with 11-cis-retinal resulted in formation a complex carrying a mixture of all-trans-retinal and 9-cis-retinal in about a 1:1 molar ratio.
DISCUSSION
Recent work attests the existence of pentameric complexes comprised of a GPCR dimer and a heterotrimeric G protein (Baneres et al., 2003; El Moustaine et al., 2012; Han et al., 2009; Jastrzebska et al., 2011b; Pellissier et al., 2011). GPCR dimers such as the serotonin receptor 5-HT2C (Herrick-Davis et al., 2006; Herrick-Davis et al., 2012), the β1-adrenergic receptor (Kobayashi et al., 2009), and the b2AR (Angers et al., 2001; Angers et al., 2000) have also been identified in living cells. Atomic force microscopy revealed Rho to be packed in rows of dimers in the native murine disc membrane (Fotiadis et al., 2003; Fotiadis et al., 2004; Liang et al., 2003) and biochemical analyses confirmed the existence of native Rho dimers (Jastrzebska et al., 2006; Jastrzebska et al., 2004; Suda et al., 2004). Finally, in vivo studies of the luteinizing hormone receptor provided compelling functional evidence of intermolecular communication between ligand-binding deficient and G protein activation deficient mutants (Rivero-Muller et al., 2010). In contrast, some GPCR monomers were found to induce a physiological response in vitro (Whorton et al., 2007; Whorton et al., 2008), and the crystal structure of the T4 lysozyme (T4L)-b2AR•Gs•nanobody complex indicates a binding stoichiometry of one receptor and one G protein (Rasmussen et al., 2011).
For understanding the molecular signal transduction mechanisms of GPCRs knowledge of their minimal functional unit is indispensable. Here we present results from experiments designed to validate that Rho exists as a dimer in native disc membranes and to demonstrate that the native Rho*•Gt complex is a heteropentamer. In addition, we show that each Rho molecule of the heteropentamer has different structural and functional properties (Jastrzebska et al., 2013). We used the carbohydrate-binding protein Concanavalin A to assemble the Rho*•Gt complex ‘on column’, and as tag to decorate the heteropentamer.
Rho contains two sugar groups that modify the opsin structure through an asparagine linkage: mannose (Man)3 attached to Asp15 and N-acetyl glucosamine (GlcNAc)3 attached to Asp2 (Fukuda et al., 1982; Salom et al., 2012). Though ConA itself forms tetramers, succinylated ConA exists in dimeric form and provides two sugar binding sites (Gunther et al., 1973). Moreover, sConA only binds mannose and not N-acetyl-glucosamine (Lis and Sharon, 1973). Therefore only one Rho can bind to one sugar-binding site in sConA, and a single sConA dimer will bind two Rho monomers in 5 mM DDM, as illustrated in Fig. 1B. The elongated central domain of the sConA-Rho complex has the size and shape of negatively stained sConA (Fig. 1C), whereas domains at its ends have a size (40 ± 2 Å) that corresponds to a Rho monomer (Jastrzebska et al., 2006).
In 1 mM DDM, however, Rho exists as dimer as previously documented by blue native gel experiments, and images of negatively stained samples (Suda et al., 2004). Fig. 1D reveals particles exhibiting a length of 65±4 Å, a width of 56±4 Å, and a central stained cleft, features compatible with the structure of two Rho molecules in a side-by-side arrangement (Palczewski et al., 2000). Rho dimers have the capability not only of binding two sConA dimers in solution but also of keeping them in a side-by-side configuration by binding at both ends (Fig. 1E). This configuration is only possible if the Rho monomers are parallel, and directly proves that native Rho exists in a dimeric form which can be purified at low DDM concentrations.
DDM solubilized Rho dimers bound to a sConA column and activated by light present a platform to bind Gt. As previously demonstrated, this complex retains full biological activity, can be eluted from the column, and exhibits dimensions, shape and mass of a Rho*•Gt heteropentamer (Jastrzebska et al., 2011b). Here we have exchanged DDM with LMNG and determined the 3D map of Rho*•Gt-LMNG to understand the difference of the heteropentameric Rho*•Gt-LMNG and the (T4L)-b2AR•Gs•nanobody-LMNG described in (Westfield et al., 2011). As illustrated in Fig. 2 negatively stained particles, their class averages, and the new 3D map are similar to those of Rho*•Gt-DDM published previously (Jastrzebska et al., 2011b). The major difference is the stain exclusion volume resulting from the detergent (Fig. 2D, right): LMNG forms a larger belt than DDM (Table 1). This is compatible with a larger mass of the Rho*•Gt-LMNG than the Rho*•Gt-DDM complex (Jastrzebska et al., 2011b) estimated from gel filtration (Fig. 2A), and in agreement with the belts different detergents form to keep a plant aquaporin tetramer in solution (Vahedi-Faridi et al., 2012). However, the size of the LMNG micelle we observed is much smaller than what has been reported in (Westfield et al., 2011), a discrepancy that we cannot resolve.
To complete our structural analysis, a triple sConA-Rho*•Gt complex was formed by incubating Rho*•Gt complexes in 1 mM DDM with sConA and isolating the resulting triple complex by gel filtration (Fig. 3A). Images of negatively stained complexes confirmed the finding that Rho dimers arrange sConA as side-by-side dimers, and further demonstrate the native arrangement of the Rho dimer (Figs. 3B and C). A rough model was assembled of the triple sConA-Rho*•Gt complex to illustrate our interpretation of these findings (Fig. 3D).
Our protocol to isolate the native functional Rho*•Gt complex lends itself to probe the capacity of Rho to bind different retinoid chromophores. Excess or weakly bound chromophore can be washed away before elution of the complex and chromophore extraction for quantitative HPLC analysis. While a full account of these experiments is presented elsewhere (Jastrzebska et al., 2013), the binding of 9-cis- and 11-cis-retinal to the Rho*•Gt complex is described here. As Fig. 4 demonstrates, all-trans-retinal is not released from the binding pocket of Rho*, probably as result of the bound Gtα C-terminus, whereas the other Rho releases all-trans-retinal. Regeneration with 9-cis-retinal consequently leads to a complex that houses equal amounts of all-trans-retinal and 9-cis-retinal, the latter bound to the Rho monomer without bound Gtα. Because subsequent treatment with NH2OH removes all-trans-retinal but not 9-cis-retinal, regeneration with 11-cis-retinal affects only the Rho* monomer with the bound Gtα. Unexpectedly, the regenerated heteropentamer again houses equal amounts of all-trans-retinal and 9-cis-retinal bound, suggesting that Rho* is stabilized by Gt in its active conformation even after chromophore depletion. The rigid constraints of this protein promote isomerization of 11-cis-retinal to its all-trans-retinal conformation because only the latter can be accommodated in the chromophore-binding pocket of Meta II-like Rho.
In conclusion, visualization of a sConA-Rho complex and a triple sConA-Rho*•Gt complex presents direct evidence that the native complex formed between activated Rho and Gt, i.e., the minimal functional unit, is a Rho dimer bound to a single Gt heterotrimer. Moreover, once coupled to Gt each Rho molecule of the dimer features different structural and functional properties (Fig. 4; (Jastrzebska, 2012)). Although the physiological significance of this arrangement is still elusive, GPCR dimerization appears to be the rule rather than the exception (Gonzalez-Maeso, 2011; Khelashvili et al., 2010; Nemoto et al., 2011; Pandit et al., 2010). Compelling evidence for the existence of b2AR dimers in living cells (Angers et al., 2001; Angers et al., 2000) suggests that the solved T4L-b2AR•Gs•nanobody complex, which consists of one receptor and one G protein (Rasmussen et al., 2011), could be promoted by specific crystallization conditions and bound nanobody.
MATERIALS AND METHODS
Chemicals
N-dodecyl-β-D-maltoside and lauryl-maltose-neopentyl-glycol were purchased from Affymetrix (Santa Clara, CA). Guanosine 5′-3-O-(thio)triphosphate (GTPγS) and 9-cis-retinal was from Sigma (St. Louis, MO). 11-cis-retinal was a generous gift from Dr. R. Crouch (Medical University of South Carolina, Charleston, USA).
Purification of Rho
Bovine ROS membranes were prepared from fresh retinas under dim red light as described in (Papermaster, 1982). ROS were solubilized in DDM and Rho was purified by the ZnCl2-opsin precipitation method (Okada et al., 2004). ZnCl2 was removed by dialysis against 10 mM BTP, pH 7.5, 100 mM NaCl and 0.2 mM DDM. Rho concentrations were determined with a Cary 50 UV-visible spectrophotometer (Varian, Palo Alto, CA) by absorption at 500 nm using the absorption coefficient ε = 40,600 M−1cm−1 (Wald, 1957). The dialyzed preparation with ~ 10 mg/ml Rho also contained ~ 16 mM DDM as determined by measuring the contact angle of a sessile drop (Kaufmann et al., 2006). Therefore the ratio of DDM/Rho (w/w) was kept in a range 0.8–1.2 to preserve the oligomeric organization of Rho (Suda et al., 2004).
Purification of Gt
Bovine ROS membranes were prepared from fresh retinas and Gt was purified by extraction with hypotonic buffer from 200 dark-adapted bovine ROS membranes as described in (Goc et al., 2008). Briefly, ROS membranes were re-suspended in 30 ml of isotonic buffer (20 mM HEPES, pH 7.5, 5 mM MgCl2, 1 mM DTT, and 100 mM NaCl) and soluble proteins were removed by gentle homogenization followed by centrifugation at 25,000g at 4°C for 15 min. Gt then was extracted from the pellet in 30 ml of hypotonic buffer (5 mM HEPES, pH 7.5, 0.1 mM EDTA, and 1 mM DTT). Membranes were pelleted by centrifugation at 25,000g at 4°C for 30 min and the supernatant was saved for further purification. This extraction procedure was repeated twice. Combined supernatants were centrifuged at 25,000g for 60 min to remove ROS membrane contaminants, and then HEPES, pH 7.5, was added to a final concentration of 10 mM and MgCl2 to a final concentration of 2 mM, each from 1 M stock solutions. The resulting sample was applied at a flow rate of 15 ml/h to a 10 × 100 mm column containing 5 ml of pentyl-agarose resin pre-equilibrated with buffer containing 10 mM HEPES, pH 7.5, 2 mM MgCl2 and 1 mM DTT and the column was washed with 10 column volumes of the equilibrating buffer. Bound proteins were subsequently eluted with a 50 ml linear gradient of 0 to 0.5 M NaCl in the equilibrating buffer at a flow rate of 15 ml/h, and 1 ml fractions were collected. Fractions containing Gt were pooled and concentrated with a 30,000 NMWL Centricon device (Millipore, Billerica, MA). Gt then was purified to homogeneity on a tandem Superdex 200 gel filtration column equilibrated with buffer containing 10 mM HEPES, pH 7.5, 100 mM NaCl, 2 mM MgCl2, and 1 mM DTT at 4°C (flow rate 0.4 ml/min with collection of 0.5 ml fractions). Fractions containing Gt were combined and concentrated with a 30,000 NMWL Centricon device (Millipore, Bilerica, MA) to ~ 10 mg protein/ml as determined by the Bradford assay (Bradford, 1976).
Purification of the Rho*•Gt complex
The Rho*•Gt complex was purified by sConA affinity chromatography followed by gel filtration as described in (Jastrzebska et al., 2011b). Briefly, the sConA affinity resin was prepared by coupling sConA (Vector Laboratories, CA) to CNBr-activated agarose (Santa Cruz Biotechnology Inc., Santa Cruz, CA) at a density of 8 mg sConA/ml of resin. Purified Rho was diluted to ~ 0.2 mg/ml in equilibration buffer and loaded onto the sConA column pre-equilibrated with equilibrating buffer (20 mM BTP, pH 6.9, containing 120 mM NaCl, 1 mM MnCl2, 1 mM CaCl2, 1 mM MgCl2, 1 mM DTT and 0.5 mM DDM) at 0.5 ml/min and room temperature. The resin was washed with 5 column volumes of the same ice cold buffer and then illuminated for 10 min with a 150 Watt fiber light (Dolan Jenner Industries Inc., Boxborough, MA) delivered through a 480–520 nm band pass filter. Purified native Gt, diluted to ~ 0.2 mg/ml with equilibrating buffer, was applied to the column immediately after light exposure at 0.5 ml/min and washed with 10 column volumes of the above buffer. The complex then was eluted with the same buffer containing 200 mM α-methyl-D-mannoside at 0.2 ml/min. One mM DSG (disuccinimidyl glutarate) crosslinker (Thermo Scientific, Rockford, IL) was added immediately to the eluted fractions (0.5 ml) containing Rho*•Gt and the crosslinking reaction was allowed to proceed for 2 h on ice. The reaction was terminated by 1 M Tris, pH 8.0, added to a final concentration of 50 mM and incubated for 15 min. Fractions then were pooled and concentrated to 200 μl with a 30,000 NMWL Centricon device and used for size exclusion chromatography. Protein was loaded onto a gel filtation column (SRT™ SEC-300, SEPAX-Technologies, Inc., Newark, DE) equilibrated with 20 mM BTP, pH 6.9, containing 120 mM NaCl, 1 mM MgCl2, 1 mM DTT and 0.5 mM DDM. Fractions containing crosslinked Rho*•Gt complex separate from free Rho and Gt were pooled, concentrated and processed on the same gel filtration column to increase sample homogeneity. The peak fraction was used for analysis by transmission electron microscopy (TEM). To prepare the Rho*•Gt-LMNG complex, fractions containing the Rho*•Gt complex separated by gel filtration were pooled, concentrated to 100 μl and incubated with 100 μl of 10 mM LMNG solubilized in 20 mM BTP, pH 6.9, containing 120 mM NaCl, 1 mM MgCl2, and 1 mM DTT. After 1 h of incubation on ice, protein was processed on the same gel filtration column equilibrated with 20 mM BTP, pH 6.9, containing 120 mM NaCl, 1 mM MgCl2, 1 mM DTT and 0.05 mM LMNG. Peak fractions containing Rho*•Gt-LMNG were then used for TEM analyses.
Purification of sConA-Rho complexes
Purified Rho (200 μg) diluted to 1 mg/ml with 20 mM BTP, pH 6.9, containing 120 mM NaCl, 1 mM MnCl2, 1 mM CaCl2, 1 mM MgCl2 and either 1 mM or 5 mM DDM was mixed with an equimolar concentration of sConA and incubated at 4 °C for 1 h following gel filtration through a SRT™ SEC-300 column (SEPAX-Technologies, Inc., Newark, DE) equilibrated with one of the two above DDM-containing buffers. Peak fractions containing sConA-Rho complexes were analyzed by TEM.
Purification of the sConA-Rho*•Gt complex
The Rho*•Gt complex purified in LMNG was incubated at 4 °C overnight with excess sConA in buffer composed of 20 mM BTP, pH 6.9, containing 120 mM NaCl, 1mM MnCl2, 1 mM MgCl2, 1 mM DTT and 0.05 mM LMNG. The triple sConA-Rho*•Gt complex was separated by size exclusion chromatography on a SRT™ SEC-300 gel filtration column equilibrated with the same buffer as above and peak fractions were analyzed by TEM.
TEM of negatively stained sConA-Rho*•Gt, sConA-Rho, and Rho*•Gt, complexes and sConA
All purified complexes were diluted to ~20 μg/ml. sCon A was resuspended in 20 mM BTP, pH 6.9, containing 120 mM NaCl, 1 mM MnCl2, 1 mM CaCl2, 1 mM MgCl2 and a specified detergent to reach a 5–20 μg/ml concentration. Protein complexes were adsorbed for 1 min to glow-discharged, 400 mesh, carbon-coated grids (Quantifoil Micro Tools GmbH, Germany). Grids were washed with 4 drops of distilled H2O and negatively stained with 2% (w/v) uranyl acetate. Both single film and double film sandwich negative staining methods were used (Cheng et al., 2006). Electron micrographs of Rho dimers were recorded with a Hitachi H-7000 electron microscope operated at 100 kV. sConA-Rho sample images were taken with either a Philips CM10 microscope (Philips, Eindhoven, Netherlands) operated at 80 kV or a FEI TF20 microscope operated at 200 kV (FEI, Eindhoven, Netherlands). Rho*•Gt, sConA-Rho*•Gt and sConA samples were visualized only with a TF20 microscope operated at 200 kV. Electron micrographs of Rho*•Gt complexes were recorded at 0° and 45° tilts on a TF20 Gatan Ultrascan 4k CCD (Gatan Inc, Pleasanton, CA 94588, USA). Magnification was determined by measuring the meridional reflection of co-prepared tobacco mosaic virus (TMV; kindly supplied by Dr. Ruben Diaz-Avalos (Diaz-Avalos and Caspar, 1998)).
Single particle analyses and image processing of sConA-Rho, Rho*•Gt, sConA-Rho*•Gt, and sConA
Image processing was performed with the EMAN2 software package (Tang et al., 2007). Particles of sConA-Rho, sConA-Rho*•Gt, and sConA were selected manually by using the BOXER feature of EMAN2. Class averages were calculated for sConA-Rho in 1 mM DDM from 240 projections, sConA-Rho in 5 mM DDM from 836 projections, sConA from 885 projections and sConA-Rho*•Gt in LMNG from 667 projections. Particles of Rho*•Gt complexes purified in DDM or LMNG were selected manually by using the tilt pair boxing feature of EMAN2. For CTF correction, a Wiener filter approach was implemented in the SEMPER package (Saxton, 1996), and applied to all images before particle selection. For images of tilted samples, the defocus was determined for 64 5122 pixel subregions to define the tilt angle and axis. Strips with a defocus range Δf < 50 nm were selected, CTF-corrected and merged in the CTF-corrected image. This procedure facilitated particle selection (Vahedi-Faridi et al., 2012) and was used throughout the project, although the resolution of the final 3D map (29 Å) indicated that CTF correction could have been omitted. Ninety six class averages were calculated from the Rho*•Gt complexes recorded with TF20 at a tilt of 0° and 45° (12950 projections for Rho*•Gt in LMNG and 8934 projections for Rho*•Gt in DDM). Initial models were calculated from the two class average sets by using the RCT method as well as the statistical method implemented in EMAN2 (e2initialmodel generator), and initial models were refined against the respective data sets.
Modeling procedure
A model of Rho*•Gt previously constructed (Jastrzebska et al., 2011b) was found to fit well to the 3D map of the DSG crosslinked Rho*•Gt -LMNG. As detailed in the supplementary information (Jastrzebska et al., 2011b), this model was built based on the intact Gt heterotrimer from the 1GOT structure, a Rho dimer featuring a two-fold dimer axis in between Helix IV and Helix V of adjacent rhodopsin protamers, and the 3DQB structure of opsin, which contains the C terminus of alpha from Gt bound, allowing Gt to be positioned. The triple sConA-Rho*•Gt complex was built from this model and a crystal structure of a ConA dimer (PDB 1DQ4). Two ConA dimers were arranged to mimic the features of the two elongated domains in the center of the triple complex. Rho*•Gt complexes were then placed at both ends of the sConA dimers in an arrangement dictated by the sugar moieties and respective binding sites.
Retinoid analyses
Rho*•Gt or chromophore-regenerated complexes (~100 μg protein) were denatured for 30 min at room temperature with 50% CH3OH in the presence of 40 mM NH2OH. The resulting retinal oximes were extracted with 300 μl of hexane and their isomeric content was determined after normal phase HPLC with an Ultrasphere-Si, 5 μm; 4.5 × 250 mm column (Beckman, San Ramon, CA). Retinoids, eluted isocratically with 10% ethyl acetate in hexane at a flow rate of 1.4 ml/min, were detected by their absorption at 360 nm or 325 nm (Garwin and Saari, 2000; Van Hooser et al., 2000) and quantified based on areas under corresponding chromatography peaks. Retinal oxime concentrations were calculated based on standard curves representing correlations between areas and peaks of synthetic standards for each retinal oxime isomer.
Supplementary Material
Acknowledgments
We thank Drs. Leslie T. Webster Jr., Phoebe L. Stewart, and members of the Palczewski laboratory for helpful comments on the manuscript, Dr. David Lodowski for his advice about building the model of the triple complex. This work was supported by National Institutes of Health grants EY008061 and EY019478, and the Swiss National Foundation grant 3100A0-108299 to AE who belongs to the Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS), which is a center for membrane protein structure determination funded by the NIH Protein Structure Initiative under grant U54GM094598. KP is John Hord Professor of Pharmacology.
Abbreviations used
- b2AR
β2-adrenergic receptor
- Bis-Tris propane
1,3-bis[tris(hydroxymethyl)-methylamino]propane
- CTF
contrast transfer function
- DDM
n-dodecyl-β-D-maltoside
- DTT
dithiothreitol
- GDP
guanosine diphosphate
- GTP
guanosine 5′-triphosphate
- GTPγS
guanosine 5′-3-O-(thio)triphosphate
- Gt
rod photoreceptor G protein (transducin)
- GPCR
G protein-coupled receptor
- HEPES
N-[2-hydroxyethyl]piperazine-N′-[2-ethanesulfonic acid]
- HPLC
high performance liquid chromatography
- LMNG
lauryl-maltose-neopentyl-glycol
- Rho
rhodopsin
- Rho*
photoactivated rhodopsin or Meta II
- ROS
rod outer segment(s)
- sConA
succinylated concanavalin A
- T4L
T4 lysozyme
- TMV
tobacco mosaic virus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angers S, Salahpour A, Bouvier M. Biochemical and biophysical demonstration of GPCR oligomerization in mammalian cells. Life sciences. 2001;68:2243–2250. doi: 10.1016/s0024-3205(01)01012-8. [DOI] [PubMed] [Google Scholar]
- Angers S, Salahpour A, Joly E, Hilairet S, Chelsky D, Dennis M, Bouvier M. Detection of beta 2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET) Proc Natl Acad Sci U S A. 2000;97:3684–3689. doi: 10.1073/pnas.060590697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneres JL, Martin A, Hullot P, Girard JP, Rossi JC, Parello J. Structure-based analysis of GPCR function: conformational adaptation of both agonist and receptor upon leukotriene B4 binding to recombinant BLT1. J Mol Biol. 2003;329:801–814. doi: 10.1016/s0022-2836(03)00438-8. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Wolf E, Larvie M, Zak O, Aisen P, Grigorieff N, Harrison SC, Walz T. Single particle reconstructions of the transferrin-transferrin receptor complex obtained with different specimen preparation techniques. J Mol Biol. 2006;355:1048–1065. doi: 10.1016/j.jmb.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- Diaz-Avalos R, Caspar DL. Structure of the stacked disk aggregate of tobacco mosaic virus protein. Biophys J. 1998;74:595–603. doi: 10.1016/S0006-3495(98)77818-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Moustaine D, Granier S, Doumazane E, Scholler P, Rahmeh R, Bron P, Mouillac B, Baneres JL, Rondard P, Pin JP. Distinct roles of metabotropic glutamate receptor dimerization in agonist activation and G-protein coupling. Proc Natl Acad Sci U S A. 2012;109:16342–16347. doi: 10.1073/pnas.1205838109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-force microscopy: Rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. The G protein-coupled receptor rhodopsin in the native membrane. FEBS Lett. 2004;564:281–288. doi: 10.1016/S0014-5793(04)00194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda MN, Papermaster DS, Hargrave PA. Structural analysis of carbohydrate moiety of bovine rhodopsin. Methods Enzymol. 1982;81:214–223. doi: 10.1016/s0076-6879(82)81034-3. [DOI] [PubMed] [Google Scholar]
- Garwin GG, Saari JC. High-performance liquid chromatography analysis of visual cycle retinoids. Methods Enzymol. 2000;316:313–324. doi: 10.1016/s0076-6879(00)16731-x. [DOI] [PubMed] [Google Scholar]
- Goc A, Angel TE, Jastrzebska B, Wang B, Wintrode PL, Palczewski K. Different properties of the native and reconstituted heterotrimeric G protein transducin. Biochemistry. 2008;47:12409–12419. doi: 10.1021/bi8015444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Maeso J. GPCR oligomers in pharmacology and signaling. Molecular brain. 2011;4:20. doi: 10.1186/1756-6606-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther GR, Wang JL, Yahara I, Cunningham BA, Edelman GM. Concanavalin A derivatives with altered biological activities. Proc Natl Acad Sci U S A. 1973;70:1012–1016. doi: 10.1073/pnas.70.4.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Moreira IS, Urizar E, Weinstein H, Javitch JA. Allosteric communication between protomers of dopamine class A GPCR dimers modulates activation. Nat Chem Biol. 2009;5:688–695. doi: 10.1038/nchembio.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick-Davis K, Weaver BA, Grinde E, Mazurkiewicz JE. Serotonin 5-HT2C receptor homodimer biogenesis in the endoplasmic reticulum: real-time visualization with confocal fluorescence resonance energy transfer. J Biol Chem. 2006;281:27109–27116. doi: 10.1074/jbc.M604390200. [DOI] [PubMed] [Google Scholar]
- Herrick-Davis K, Grinde E, Lindsley T, Cowan A, Mazurkiewicz JE. Oligomer size of the serotonin 5-hydroxytryptamine 2C (5-HT2C) receptor revealed by fluorescence correlation spectroscopy with photon counting histogram analysis: evidence for homodimers without monomers or tetramers. J Biol Chem. 2012;287:23604–23614. doi: 10.1074/jbc.M112.350249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann KP, Scheerer P, Hildebrand PW, Choe HW, Park JH, Heck M, Ernst OP. A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem Sci. 2009;34:540–552. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Jastrzebska B, Palczewski K, Golczak M. Role of bulk water in hydrolysis of the rhodopsin chromophore. J Biol Chem. 2011a;286:18930–18937. doi: 10.1074/jbc.M111.234583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Orban T, Golczak M, Engel A, Palczewski K. Asymmetry of the rhodopsin dimer in complex with transducin. FASEB J. 2013 doi: 10.1096/fj.12-225383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Fotiadis D, Jang GF, Stenkamp RE, Engel A, Palczewski K. Functional and structural characterization of rhodopsin oligomers. J Biol Chem. 2006;281:11917–11922. doi: 10.1074/jbc.M600422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Maeda T, Zhu L, Fotiadis D, Filipek S, Engel A, Stenkamp RE, Palczewski K. Functional characterization of rhodopsin monomers and dimers in detergents. J Biol Chem. 2004;279:54663–54675. doi: 10.1074/jbc.M408691200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Ringler P, Lodowski DT, Moiseenkova-Bell V, Golczak M, Muller SA, Palczewski K, Engel A. Rhodopsin-transducin heteropentamer: three-dimensional structure and biochemical characterization. J Struct Biol. 2011b;176:387–394. doi: 10.1016/j.jsb.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Orban T, Golczak M, Engel A, Palczewski K. Asymmetry of the rhodopsin dimer in complex with transducin. FASEB J. 2012 doi: 10.1096/fj.12-225383. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann TC, Engel A, Remigy HW. A novel method for detergent concentration determination. Biophys J. 2006;90:310–317. doi: 10.1529/biophysj.105.070193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelashvili G, Dorff K, Shan J, Camacho-Artacho M, Skrabanek L, Vroling B, Bouvier M, Devi LA, George SR, Javitch JA, Lohse MJ, Milligan G, Neubig RR, Palczewski K, Parmentier M, Pin JP, Vriend G, Campagne F, Filizola M. GPCR-OKB: the G Protein Coupled Receptor Oligomer Knowledge Base. Bioinformatics. 2010;26:1804–1805. doi: 10.1093/bioinformatics/btq264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Ogawa K, Yao R, Lichtarge O, Bouvier M. Functional rescue of beta-adrenoceptor dimerization and trafficking by pharmacological chaperones. Traffic. 2009;10:1019–1033. doi: 10.1111/j.1600-0854.2009.00932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Fotiadis D, Filipek S, Saperstein DA, Palczewski K, Engel A. Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. Journal Of Biological Chemistry. 2003;278:21655–21662. doi: 10.1074/jbc.M302536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis H, Sharon N. The biochemistry of plant lectins (phytohemagglutinins) Annu Rev Biochem. 1973;42:541–574. doi: 10.1146/annurev.bi.42.070173.002545. [DOI] [PubMed] [Google Scholar]
- Nemoto W, Fukui K, Toh H. GRIPDB - G protein coupled Receptor Interaction Partners DataBase. Journal of receptor and signal transduction research. 2011;31:199–205. doi: 10.3109/10799893.2011.563312. [DOI] [PubMed] [Google Scholar]
- Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- Orban T, Jastrzebska B, Gupta S, Wang B, Miyagi M, Chance MR, Palczewski K. Conformational dynamics of activation for the pentameric complex of dimeric G protein-coupled receptor and heterotrimeric G protein. Structure. 2012;20:826–840. doi: 10.1016/j.str.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K. G protein-coupled receptor rhodopsin. Annu Rev Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Pandit SB, Brylinski M, Zhou H, Gao M, Arakaki AK, Skolnick J. PSiFR: an integrated resource for prediction of protein structure and function. Bioinformatics. 2010;26:687–688. doi: 10.1093/bioinformatics/btq006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papermaster DS. Preparation of retinal rod outer segments. Methods Enzymol. 1982;81:48–52. doi: 10.1016/s0076-6879(82)81010-0. [DOI] [PubMed] [Google Scholar]
- Pellissier LP, Barthet G, Gaven F, Cassier E, Trinquet E, Pin JP, Marin P, Dumuis A, Bockaert J, Baneres JL, Claeysen S. G protein activation by serotonin type 4 receptor dimers: evidence that turning on two protomers is more efficient. J Biol Chem. 2011;286:9985–9997. doi: 10.1074/jbc.M110.201939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero-Muller A, Chou YY, Ji I, Lajic S, Hanyaloglu AC, Jonas K, Rahman N, Ji TH, Huhtaniemi I. Rescue of defective G protein-coupled receptor function in vivo by intermolecular cooperation. Proc Natl Acad Sci U S A. 2010;107:2319–2324. doi: 10.1073/pnas.0906695106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakmar TP, Menon ST, Marin EP, Awad ES. Rhodopsin: insights from recent structural studies. Annu Rev Biophys Biomol Struct. 2002;31:443–484. doi: 10.1146/annurev.biophys.31.082901.134348. [DOI] [PubMed] [Google Scholar]
- Salom D, Wang B, Dong Z, Sun W, Padayatti P, Jordan S, Salon JA, Palczewski K. Post-translational modifications of the serotonin type 4 receptor heterologously expressed in mouse rod cells. Biochemistry. 2012;51:214–224. doi: 10.1021/bi201707v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salom D, Lodowski DT, Stenkamp RE, Le Trong I, Golczak M, Jastrzebska B, Harris T, Ballesteros JA, Palczewski K. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci U S A. 2006;103:16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WO. Semper: Distortion Compensation, Selective Averaging, 3-D Reconstruction, and Transfer Function Correction in a Highly Programmable System. J Struct Biol. 1996;116:230–236. doi: 10.1006/jsbi.1996.0035. [DOI] [PubMed] [Google Scholar]
- Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- Standfuss J, Edwards PC, D’Antona A, Fransen M, Xie G, Oprian DD, Schertler GF. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda K, Filipek S, Palczewski K, Engel A, Fotiadis D. The supramolecular structure of the GPCR rhodopsin in solution and native disc membranes. Mol Membr Biol. 2004;21:435–446. doi: 10.1080/09687860400020291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Vahedi-Faridi A, Jastrzebska B, Palczewski K, Engel A. 3D imaging and quantitative analysis of small solubilized membrane proteins and their complexes by transmission electron microscopy. Journal of electron microscopy. 2012 doi: 10.1093/jmicro/dfs091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eps N, Preininger AM, Alexander N, Kaya AI, Meier S, Meiler J, Hamm HE, Hubbell WL. Interaction of a G protein with an activated receptor opens the interdomain interface in the alpha subunit. Proc Natl Acad Sci U S A. 2011;108:9420–9424. doi: 10.1073/pnas.1105810108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser JP, Garwin GG, Saari JC. Analysis of visual cycle in normal and transgenic mice. Methods Enzymol. 2000;316:565–575. doi: 10.1016/s0076-6879(00)16750-3. [DOI] [PubMed] [Google Scholar]
- Wald G. The chemistry of visual excitation. Bibliotheca ophthalmologica : supplementa ad ophthalmologica. 1957;12:173–188. [PubMed] [Google Scholar]
- Westfield GH, Rasmussen SG, Su M, Dutta S, DeVree BT, Chung KY, Calinski D, Velez-Ruiz G, Oleskie AN, Pardon E, Chae PS, Liu T, Li S, Woods VL, Jr, Steyaert J, Kobilka BK, Sunahara RK, Skiniotis G. Structural flexibility of the G alpha s alpha-helical domain in the beta2-adrenoceptor Gs complex. Proc Natl Acad Sci U S A. 2011;108:16086–16091. doi: 10.1073/pnas.1113645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton MR, Bokoch MP, Rasmussen SG, Huang B, Zare RN, Kobilka B, Sunahara RK. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci U S A. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton MR, Jastrzebska B, Park PS, Fotiadis D, Engel A, Palczewski K, Sunahara RK. Efficient coupling of transducin to monomeric rhodopsin in a –phospholipid bilayer. J Biol Chem. 2008;283:4387–4394. doi: 10.1074/jbc.M703346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.