Abstract
Patient-specific induced pluripotent stem cells (iPSCs) represent a potential source for developing novel drugand cell- therapies. Although increasing numbers of disease-specific iPSCs have been generated, there has been limited progress in iPSC-based drug screening/discovery for liver diseases, and the low gene targeting efficiency in human iPSCs warrants further improvement. Using iPSC lines from patients with alpha-1 antitrypsin (AAT) deficiency, for which there is currently no drug- or gene- therapy available, we established a platform to discover new drug candidates and to correct disease-causing mutation with a high efficiency. A high-throughput format screening assay based on our hepatic differentiation protocol was implemented to facilitate automated quantification of cellular AAT accumulation using a 96-well immunofluorescence reader. To expedite the eventual application of lead compounds to patients, we conducted drug screening utilizing our established library of clinical compounds, the Johns Hopkins Drug Library, with extensive safety profiles. Through a blind large-scale drug screening, five clinical drugs were identified to reduce AAT accumulation in diverse patient iPSC-derived hepatocyte-like cells. In addition, using the recently developed transcription activator-like effector nuclease (TALEN) technology, we achieved high gene targeting efficiency in AAT-deficiency patient iPSCs with 25–33% of the clones demonstrating simultaneous targeting at both diseased alleles. The hepatocyte-like cells derived from the gene-corrected iPSCs were functional without the mutant AAT accumulation. This highly efficient and cost-effective targeting technology will broadly benefit both basic and translational applications. Conclusions: Our results demonstrated the feasibility of effective large-scale drug screening using an iPSC-based disease model and highly robust gene targeting in human iPSCs; both of which are critical for translating the iPSC technology into novel therapies for untreatable diseases.
Introduction
Some of the biggest challenges modern medicine faces are the long timeline (>12 years), high failure rate (~95%) and cost (>$1 billion) associated with developing a single new drug (1, 2). The development of novel compounds has been accelerating due to the genome-driven discovery of new drug targets, the expansion of natural and synthetic chemistry compound collections and the development of high-throughput screening (HTS) technologies (3, 4). Despite these advances, frequent attrition of a lead series occurs due to unfavorable drug absorption, distribution, metabolism, excretion and/or toxicity (ADMET) (1, 2, 5), indicating a lack of sufficient predictability of traditional drug screening tools such as cancer cell lines and animal models. To avoid such high failure rate in late-stages of the drug developmental process, more patient-relevant screening platforms need to be developed for early stage drug screens.
The emergence of patient-specific iPSC technology and disease models established from these cells, which may provide renewable sources for a highly patient-relevant and powerful throughput screening platform, has brought high enthusiasm in the field; not only could a patient’s iPSCs be used to generate cells for transplantation to repair damaged tissues, but the differentiated progeny of such cells could also be used to recapitulate disease phenotypes and enable more efficient drug screening to find new treatment of the disease (6–14). To realize such potential of iPSCs, we and others have generated patient-specific iPSCs from various human tissues and differentiated these cells into different somatic cell types including blood and liver cells in the past few years (6–8, 10–13). More recently, we and others have demonstrated that iPSCs derived from patients with multiple metabolic liver diseases including alpha-1 antitrypsin (AAT) deficiency could indeed be utilized for disease modeling after differentiation into hepatocyte-like cells (6, 7, 15, 16). However it remains elusive whether these cellular models of liver diseases can be effective for drug screening and discovery.
AAT-deficiency is one of the common genetic disorders of the liver (17). Importantly AAT-deficiency can progress to severe liver diseases including liver cirrhosis and hepatocellular carcinoma (HCC) (17–19). Currently there is no drug- or gene- therapy available to treat the liver disease or prevent its progression into cirrhosis and HCC. The most common clinical form of AAT-deficiency is associated with the PiZ variant of this protein which is caused by a (G>A) point mutation at codon 342 (Glu342Lys) in exon 5 of the AAT gene (19). The mutation promotes spontaneous polymerization and retention of the polymers in the endoplasmic reticulum (ER) of hepatocytes, resulting in protein overload that in turn causes the liver diseases (18). The deficiency of AAT in plasma predisposes the affected individuals to chronic pulmonary diseases as well. Augmentation therapy has been given for treatment of lung disease, but there is no therapy available other than liver transplantation for individuals with AAT-deficiency related liver disease. Since it is unlikely that current AAT augmentation therapy will alter the course of AAT-liver disease and the organ supply for transplantation is limited, alternative therapeutic strategies are required to prevent/treat both liver and lung failure by tackling the cause rather than the symptoms. The potential of human iPSC-based therapy is therefore very attractive for diseases like AAT-deficiency; it will, therefore, be of value to develop therapeutic strategies: to 1) pharmacologically decrease the mutant AAT accumulation or 2) correct the disease-causing mutation for geneand cell- based therapy. Using our established iPSCs derived from AAT deficiency-patients (6, 7), we explored the feasibility of developing both therapeutic strategies.
To expedite the eventual application of lead compounds to patients, we have employed our established clinical ready drug library, the Johns Hopkins Drug Library (JHDL) (20) for iPSC based drug screening. The JHDL currently consists of 3,131 clinical drugs (including 2800 drugs which either have been approved by US FDA/foreign counterparts or have entered phase II clinical trials) (21). Therefore any hit from this library can bypass early stages of preclinical and clinical studies before clinical testing in parents. Although a few reports including our own have shown the feasibility of testing several candidate drugs with iPSC based models (7, 22, 23), there have been no reports of large-scale drug screening in a blind manner using a patient iPSC-based disease model. To our knowledge this is the first report of a large-scale drug screening using an iPSC based disease model.
To develop potential gene and cell therapy, there have also been efforts to enhance the low efficiency of site-specific gene correction in human iPSCs, including the demonstration of zinc finger nuclease (ZFN) –mediated gene targeting for various genes including the high efficiency correction of AAT gene (24–29). Although the application of ZFNs represents a significant improvement over the traditional targeting technologies, the design of ZFNs has been a formidable engineering challenge, preventing its broad applications in research laboratories. We therefore assessed the efficacy of the recently developed transcription activator-like effector nuclease (TALEN) technology (30–34) for targeted gene correction of liver disease mutation in patient-specific iPSCs. Here we report the application of patient-specific iPSCs in drug screening (and the discovery of new uses of already approved clinical drugs) as well as for highly efficient gene targeting.
Materials and Method
Drug screen using a high throughput immunofluorescence analyzer
All human iPSCs were cultured on matrigel (BD) using mTeSR (Stem Cell Technologies) and differentiated into hepatic cells as we described previously (6, 7, 10) with some modification (see supplemental materials for details). This study was done in accordance with Johns Hopkins ISCRO regulations and following a protocol approved by the Johns Hopkins IRB.
An initial screen of all compounds from the Johns Hopkins Drug Library (JHDL) (20), which includes 3,131 clinical compounds, was conducted using one of our AAT deficiency patient iPSC lines (iAAT2), propagated using the differentiation method described above in 96-well imaging plates. The iPSC derived hepatic cells were treated with JHDL compounds, 1 per well, at a final concentration of 5 µM for 4 days at the end of hepatic maturation. After drug treatment, AAT immunofluorescence (IF) staining was performed and the total AAT fluorescence intensity of each well was measured using a Safire2 microplate reader. The results obtained from this fluorescence based high throughput assay were normalized with signals from DAPI-labeled nuclei.. The percentages of changes of AAT signals were calculated by dividing with that of DMSO-treated samples.
For confirmatory screening, we carefully selected 43 compounds without major side effects from 262 compounds that inhibited AAT accumulation by >50%. We then further tested these drugs using 4 different AAT-deficiency patient-iPSC lines (iAAT2, iAAT3, iAAT45, and iAAT25) and freshly prepared drugs rather than using the stock. The experiments were repeated four times with consistent results. Using the same IF assay, we obtained 5 hits which consistently show the effect in multiple patient iPSCs.
See Supplemental Materials for Hepatic differentiation, TALEN-mediated AAT gene-targeting, PASD, CytochromeP450 Assay, ELISA, Q-PCR and Statistics.
Result
Optimization of the iPSC-based hepatic differentiation protocol for large-scale drug screening
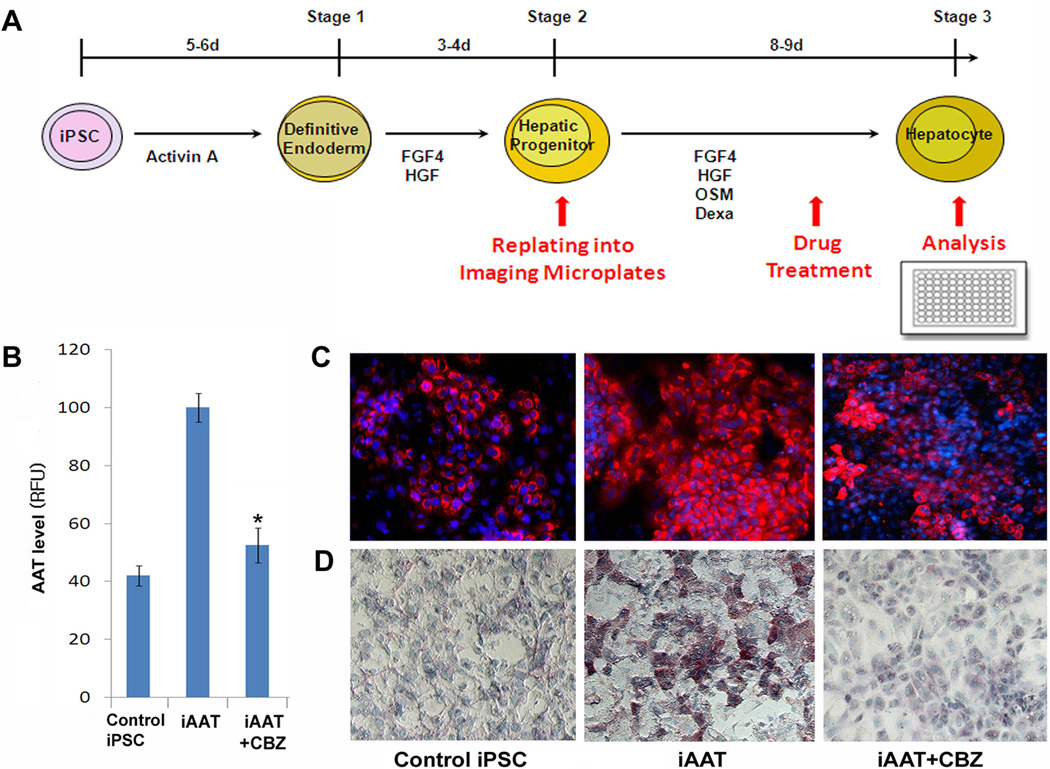
The molecular basis of AAT-deficiency has been shown to be the accumulation of AAT protein as ordered polymers within the ER of hepatocytes (19, 35). We have previously demonstrated the formation of intracellular globules that are formed by the polymers of mutant AAT proteins within AAT-deficiency iPSC-derived mature hepatocyte-like cells by PAS with diastase digestion (PASD) (7). We also showed that carbamazepine (CBZ), which has been shown to decrease the hepatic load of mutant AAT protein and hepatic fibrosis in a mouse model of AAT-deficiency-associated liver disease (35), could reduce the AAT accumulation in AAT-deficiency patient iPSC-derived hepatocyte-like cells using the PASD assay (7). This assay however does not permit automated quantification of readout using a high-throughput format immunofluorescence (IF)/luminescence reader; therefore it is not optimal for a large-scale compound screening. To perform efficient and reliable screening using a high-throughput format IF reader and our iPSC model of AAT-deficiency liver disease (7), we have modified our hepatic differentiation protocol to be compatible with a 96-well format, followed by IF staining with a specific anti-human AAT antibody, permitting visualization and quantitative detection of AAT accumulation within the hepatic cells (Fig 1, Fig S1). To achieve this goal, one replating step was added at the end of the hepatic progenitor (HP) stage to evenly distribute the iPSC-derived hepatic cells into 96-well plates without losing viability or functionality (Fig 1A, Fig S2, S3). Both the definitive endoderm (DE)-cells at an early differentiation stage and the fully mature hepatocyte (MH) like-cells were excluded for this purpose due to their lower viability associated with enzymatic dissociation and the replating step. Of note, no animal feeder cells were used throughout the iPSC expansion and differentiation processes. With future application of the hits in clinics in mind, we purposely selected a relatively low concentration (5µM), from the reported doses of 5–20 µM employed for other studies using the same drug library, for this screening (36–39). Our selected screening dose is within a physiological concentration of a large number of drugs in this library (36–39). Using CBZ, with which we previously demonstrated the reduction of AAT accumulation in patient iPSC-derived hepatocyte-like cells (7), we determined whether the modified differentiation protocol and selected dosage can reproduce similar results when analyzed by a high-throughput format IF reader. Compared to non-patient iPSC normal controls, the patient iPSCs (PiZZ, the most common and severe form of AAT-deficiency) consistently generated a higher AAT signal within mature hepatic cells after differentiation (Fig 1B, C) indicating intracellular AAT accumulation. When treated with CBZ, the hepatocyte-like cells derived from patient iPSCs exhibited significant reduction in the intracellular retention of the AAT proteins, determined by both high-throughput format IF reader and microscopy (Fig 1B, C). This result was indeed consistent with the PASD result of these iPSC-derived hepatocyte-like cells further confirming the validity of the IF based assay (Fig 1D).
Figure 1. Optimization of the iPSC based hepatic differentiation protocol for high-throughput format drug screening.
A A diagram summarizing our modified hepatic differentiation protocol used for the high-throughput format drug screening. Human iPSCs that had been maintained in a feeder free condition were differentiated into hepatic progenitor cells before replating into collagen-1 treated 96-well imaging plates which is specifically designed for a throughput immunofluorescence reader. B Detection of intracellular AAT with a high throughput format immunofluorescence reader. Control iPSC (iH71)- or patient iPSC (iAAT2)- derived mature hepatic cells (105/well) in the 96-well microplates were treated with carbamazepin (CBZ) 5µM for 4 days. AAT staining was performed on the 96-well plates and the AAT fluorescence was obtained using a Safire2 microplate reader. RFU: relative fluorescence unit. C Immunofluorescence images of AAT expression obtained from differentiated hepatocyte-like cells of control iPSCs (iH10, left panel) and an AAT-deficiency patient iPSC line (iAAT3, middle panel). Compared to control iPSCs, increased amount of AAT/AAT globules (red) were observed in iAAT cells after hepatic maturation. The expression level was significantly decreased after CBZ treatment (5µM for 4 days, right panel). (×100). D PASD images of these iPSC derived hepatocyte-like cells. Numerous PASD-positive inclusion bodies (pink) were detected within mature hepatocyte-like cells derived from the AAT patient iPSCs (iAAT3, middle panel) but not in the control iPSC (iLC2)-derived hepatocyte-like cells (left panel). The level of PASD+ inclusion body/globules was decreased with CBZ treatment (5µM for 4 days, iAAT3, right panel). (×100).
Drug screening using patient-specific iPSCs
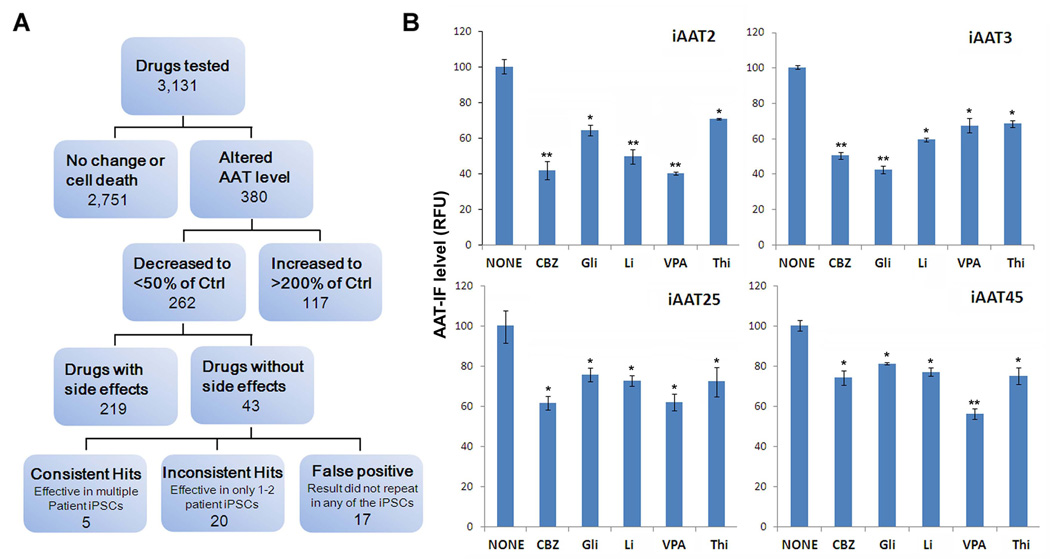
Blind screening of the clinical-ready drug library (JHDL) was initiated with our AAT-deficiency patient iPSC-derived hepatocyte-like cells using the optimized screening assay (Fig 2).
Figure 2. Patient-specific iPSC based drug screening process and results.
A Flowchart depicting the drug screening process and result summary. After the first screening of all the JHDL drugs (3,131 clinical compounds) with the immunofluorescence based screening assay (Fig 1), two hundred sixty-two compounds which significantly decreased accumulation of AAT to less than 50% of the non-treated controls, within the hepatocyte-like cells derived from one AAT-deficient patient iPSC (iAAT2), were selected for further testing. Of the 262 compounds we carefully selected 43 drugs which are relatively safer, without significant unwanted effects to any of the major organs. For the confirmatory screening for the 43 drugs in multiple iPSC lines including iAAT2, iAAT3, iAAT45, and iAAT25, we used independently prepared drugs rather than using the JHDL stock. Of the 43 compounds we discovered 5 final drug hits which consistently showed the AAT reducing effect in the multiple patient derived hepatocyte-like cells. The rest of the drugs did not repeat the results with either one or multiple iPSC lines. B Representative results obtained from multiple independent iPSC lines are shown to demonstrate the AAT reduction effects of the final 5 drug hits in the confirmatory screening (5 µM each for 4 days). *P<0.05, **P<0.01. RFU: relative fluorescence unit. Thi; Thiamine, CBZ; Carbamazepine, Gli; Glipizide, Li; Lithium, VPA; Valproic acid
The initial screen of the entire drug library at a final concentration of 5 µM of each drug yielded 263 hits (Table S1). The drugs that decreased the average total fluorescence intensity within the AAT-deficiency patient iPSC-derived hepatocyte-like cells by more than 50% were considered as hits (Fig 2). A majority of these were antidepressant, anticonvulsant and antibiotics (Table S1). We then performed an extensive and systematic literature search on the hits (such as mechanisms of action, target proteins, pharmacokinetics, etc), and prioritized the hits based on approved status, main effects and side effects. Among these 262 compounds, 43 drugs, which are FDA approved or have a history of clinical application internationally and without significant unwanted/side effects to any of the major organs, were chosen for further study (Fig 2, Table S2). Drugs that have known cytotoxicity (such as anti-cancer drug) or predicted side effects related to their intended use (such as anti-hypertension drugs) on patients were excluded from the further screening.
To confirm the effects of the selected hits and to further identify a more universal candidate drug(s) that is effective in diverse patients, we used separate preparations of the drugs than the original stock of the JHDL and tested the selected 43 hits on hepatocyte-like cells derived from four different patient-iPSC lines (iAAT2, iAAT3, iAAT45, and iAAT25; Fig 2, Fig S2). After confirmatory screening steps with these multiple iPSC lines, we were able to discover the final 5 hits which consistently showed the similar effects in multiple patient derived hepatocyte-like cells (Fig 2 and Table 1). Similar effects were observed in the range of 2.5 to 10µM (Fig S4). The rest of the drugs failed to show the effects in one or more iPSC lines (Fig 2).
Interestingly, one of the hits, after the blind screening, turned out to be CBZ, which strongly validated functionality of our drug screening assay. Importantly, three of the 5 hits were well known clinical mood-stabilizing drugs sharing a similar mechanism of action (Table 1). Inositol depletion seems to be a common mechanism for these 3 drugs; lithium (Li), carbamazepine (CBZ) and valproic acid (VPA) (35, 40, 41). Consistent with a role for inositol depletion in autophagy regulation, these three drugs have been known to enhance the clearance of aggregate-prone proteins in several non-human conditions associated with either liver or neurodegenerative diseases (19, 40–42).
Table 1.
Characterization of the final hits.
| Drug Name | Indication | Approval Status | Reported mechanism of action |
|---|---|---|---|
| Carbamazepine* | Anticonvulsant | FDA | Reduce inositol and IP3 levels; autophagy-enhancer; reduce ATZ accumulation in mice. (47), (41) |
| Glipizide | Antidiabetic | FDA | ATP-dependent K+ channel blocker increase insulin release from pancreatic beta cells |
| Lithium* | Antipsychotic | FDA | Inhibit IMPase and reduce inositol and IP3 levels; (41) Induces autophagy by inhibiting inositol monophosphatase (42), (40) Antimanic and hematopoietic activities. Interferes with transmembrane sodium exchange in nerve cells by affecting Na+, K+-ATPase; affects cAMP concentrations |
| Valproic acid* | Anticonvulsant | FDA | Anticonvulsant that also has efficacy as a mood stabilizer in bipolar disorder (44). Reduce inositol and IP3 levels (41) |
| Thiamine | Vitamin | FDA | Vitamin B1, antioxidant, erythropoietic, mood modulating, and glucose-regulating activities. Reacts with ATP to form an active coenzyme, thiamine pyrophosphate. Thiamine pyrophosphate is necessary for the actions of pyruvate dehydrogenase and alpha-ketoglutarate in carbohydrate metabolism and for the actions of transketolase, an enzyme that plays an important role in the pentose phosphate pathway. |
These drugs share a common mechanism.
To figure out the mechanisms underlying the observed effects of these drugs, we measured AAT levels in cells and media using ELISA (Fig S5), and analyzed the protein expression of an autophagy marker (LC3) and AAT gene expression (Fig S6). We found that intracellular-AAT protein levels were indeed decreased after drug treatments while the protein level in the media and the gene expression did not change after the treatments (Fig S5, Fig S6). Interestingly, LC3 was increased in the cells treated with the 5 drugs (Fig S6) including two drugs (Gli and Thi) which have not previously reported as autophagy enhancers (Table 1). These data collectively suggest that the mechanism of the final drug candidates is likely due to the autophagy-mediated degradation of folded-AAT proteins rather than decreasing synthesis of AAT.
TALEN-mediated efficient gene correction of AAT z mutation in patient-specific iPSCs
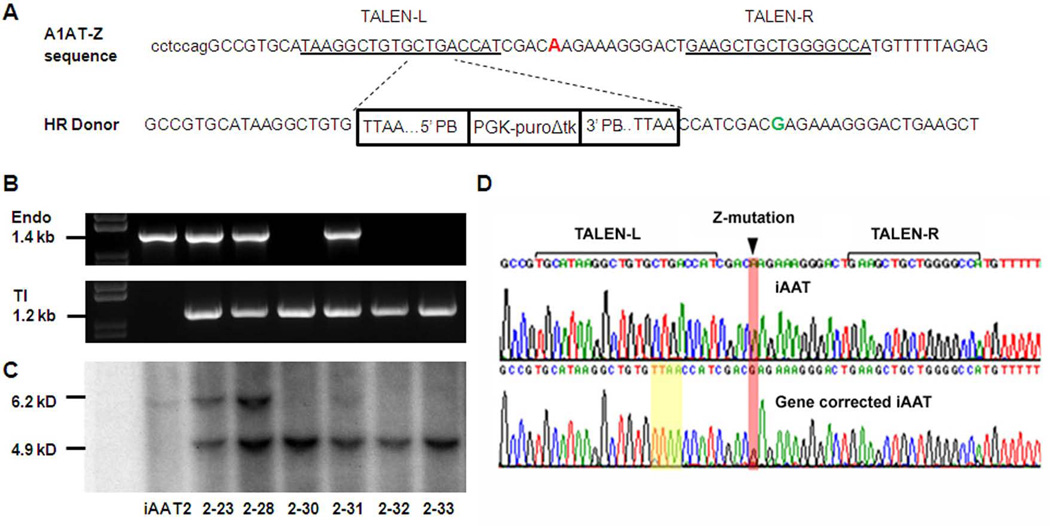
Future autologous cell replacement therapy for AAT-deficiency will require the gene correction of the Z mutation in patient cells. To assess the efficacy of the TALEN technology in gene targeting of liver disease mutations, we designed and constructed a pair of TALEN expression vectors that specifically recognize the flanking sequences of the Z-mutation of AAT gene (Fig 3). In order to compare this approach to the reported ZFN technology, we utilized a previously reported donor construct (24), which also allows the clean excision of integrated DNA from the genomes, in combination with the TALEN vectors (Fig 3A).
Figure 3. TALEN-mediated gene correction of AAT mutation in patient-specific iPSCs.
A The strategy for precise genome modification using TALEN. Genomic DNA sequence around the Z-mutation (“A” in red) in human AAT gene is shown. A pair of TALEN were designed to specifically recognize the sequences flanking the point mutation with a 16-bp spacer. The targeting vector [homologous recombination (HR) donor] (24) contains the wild-type sequence (“G” in Green) in the homology arm. Selection marker expression cassette PGK-puroΔtk is flanked by piggyBac inverted terminal repeat sequences which allow precise excision upon transient expression of piggyBac transposase. Note that, in order to facilitate this transpose-mediated seamless DNA excision, the endogenous sequence CTGA was changed to TTAA without alteration of the amino acid sequence. Confirmation of gene targeting events by PCR B and Southern blot C. B Genomic DNAs isolated from the picked puromycin-resistant clones after transfection of TALENs and targeting vector were screened by PCR. A 1.2 kb PCR product indicates the targeted events. Using a separate pair of primers, the endogenous allele without targeting event would yield a 1.4 kb band. Lack of such endogenous band would suggest that both alleles were successfully targeted. C Selected clones based on PCR results were confirmed by Southern blot. Parental iPSC (iAAT2) was used as a control. After BamHI digestion of genomic DNA, the endogenous allele without targeting produces a 6.2 kD band. A 4.9-kD band indicates the targeting by the HR donor. D Sequence analysis showing correction of the Z mutation in AAT patient-specific iPSCs after homologous recombination and piggyBac transposase-mediated seamless excision of integrated donor vector. Z mutation and TALEN recognition sequences were shown. While the iPSCs derived from AAT patient showed the G>A mutation (iAAT3), Sanger sequencing of the clone (iAAT3-2) after HR-based gene correction showed wild-type sequence.
Puromycin-resistant human iPSC colonies obtained after co-nucleofection of TALEN expression vectors and the targeting vector were screened for targeted events by polymerase chain reaction (PCR) analyses (Fig 3B) and verified by Southern blot (Fig 3C). AAT-iPSC lines derived from two different patients were used in this study and both experiments demonstrated high efficiencies of gene targeting (Table 2). Among 66 puromycin-resistant iPSC clones that had been expanded and analyzed, strikingly, all of them showed the targeted integration of the donor vector based on PCR results (Fig 3B, Table 2). In addition, 25–33% of these clones showed the lack of endogenous allele, suggesting the result of simultaneous targeting of both alleles (Table 2). Six out of six candidate clones were confirmed for bi-allelic gene targeting by Southern blot analysis (Fig 3C, Table 2).
Table 2.
A summary of the TALEN-mediated homologous recombination in two independent AAT deficiency patient-specific iPSC lines (iAAT2 and iAAT3).
| Patient iPSC lines with Z mutation |
Puro-resistant clones expanded |
Positive for Targeted integration (#positive/#analyzed) |
Simultaneously Targeted in Both Alleles (#positive/#analyzed) |
||
|---|---|---|---|---|---|
| PCR | Southern Blot | PCR* | Southern Blot | ||
| iAAT2 | 30 | 30/30 | 6/6 | 11/30 | 3/3 |
| iAAT3 | 36 | 36/36 | 6/6 | 9/36 | 3/3 |
the numbers are the clones that are positive for targeted integration and negative for endogenous allele.
To achieve a clean gene correction at the AAT locus, we removed the piggyBac-flanked drug-selection cassette from two of the homozygously targeted iPSC clones (iAAT3-2 and iAAT2-33) by transient transfection of a piggyBac transposase-expressing vector (24) followed by drug (FIAU) selection. The genotype of the resulting colonies was analyzed by PCR (not shown) and DNA sequencing (Fig 3D). Sequence analyses of selected clones demonstrated that the Z mutation was corrected on both alleles (Fig 3D).
Functional characterization of gene-corrected iPSCs
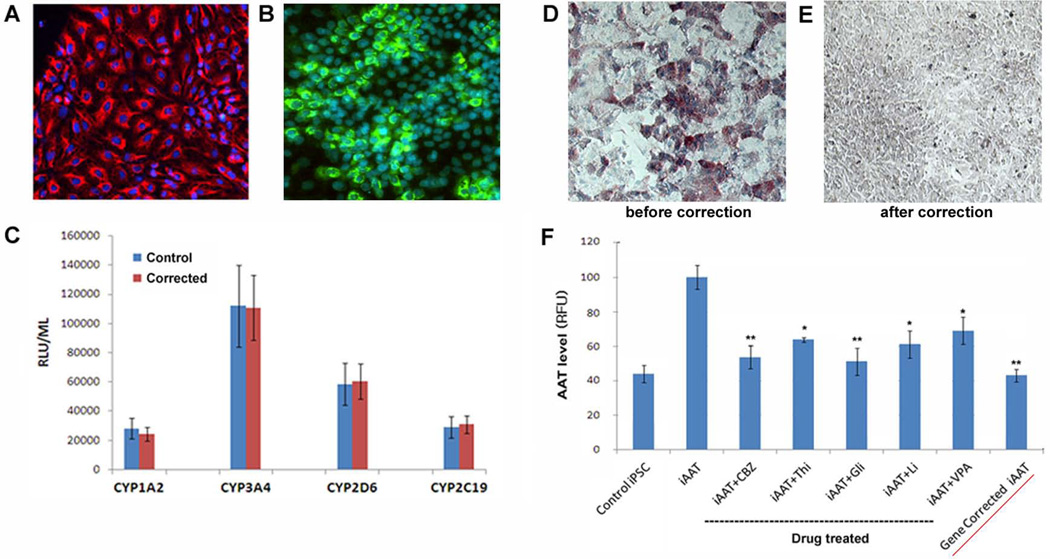
To confirm that the genetic correction of AAT-iPSCs resulted in phenotypic correction, these iPSC clones were differentiated into multistage hepatic cells. The corrected iPSCs could efficiently differentiate to mature hepatocyte-like cells (Fig 4A-C) and there were no significant changes in the growth pattern or differentiation kinetics after the gene modification process. Gene corrected iPSC clones were able to differentiate into late stage hepatic cells expressing mature hepatocyte markers such as CK18 and ALB (Fig 4A, B). These mature stage hepatocyte-like cells derived from the gene corrected iPSCs also exhibited metabolic capabilities as measured by the activities of 4 major cytochrome P450 enzymes (CYP3A4, CYP1A2, CYP2C19, and CYP2D6, Fig 4C), indicating the in vitro functionality of these cells.
Figure 4. Functional restoration of gene-corrected iPSCs.
A, B Hepatic differentiation of gene-corrected iPSCs. Gene-corrected iPSC clones were able to differentiate into mature hepatic cells expressing cytokeratin 18 (CK18, A) and albumin (ALB, B). The representative images of a gene-corrected clone (iAAT3-6) derived mature hepatic cells are shown. (×100) C These mature stage hepatic cells derived from the gene corrected iPSCs also exhibit comparable in vitro functional capabilities to healthy control iPSCs as measured by the activities of 4 major cytochrome P450 enzymes (CYP1A2, CYP3A4, CYP2D6 and CYP2C19). D, E PASD staining of mature hepatocyte-like cells derived from a patient iPSC line (iAAT3, D) and the gene-corrected iPSC clone (iAAT3-2, E). (×100) F The levels of AAT detected within the hepatocyte-like cells derived from control-iPSC (iH71), patient-iPSC (iAAT3), and gene-corrected iPSCs (iAAT3-2, red-underlined). The dotted-underlined are the effects of the final drug hits in the screening condition (i.e., 5µM for 4 days) on hepatocyte-like cells derived from the same patient iPSCs.
Importantly, as predicted, the mutant AAT accumulation was no longer detectable in the mature hepatocyte-like cells derived from the gene-corrected iPSCs (Fig 4D, E). The numerous PASD-positive inclusion bodies/globules were observed within hepatocyte-like cells derived from the AAT patient iPSCs whereas these were not detected within the hepatocyte-like cells derived from the gene-corrected iPSCs (Fig 4D, E), indicating the restored cellular function after gene correction.
In addition, we measured the intracellular AAT levels in the mature hepatocyte-like cells derived from the gene-corrected iPSCs (Fig 4E) using the same IF based AAT assay used for the drug screening process. The AAT level detected within the hepatocyte-like cells derived from the gene-corrected iPSCs was as low as that of control (healthy donor derived) iPSCs and also comparable to some of the drug- treated (without gene correction) cells, further confirming functional correction of the gene-corrected iPSCs (Fig 4F).
Therefore both approaches employed in this study (i.e. gene correction and pharmacological treatment) demonstrated significant reduction of the AAT accumulation within the AAT-deficiency patient iPSC-derived hepatocyte-like cells.
Discussion
Using our patient-specific iPSCs, we screened the clinical ready drug library (the JHDL) and identified multiple validated hits for novel treatment of AAT-deficiency (Table 1), demonstrating the feasibility of iPSC-based drug screening (Fig 1, 2). These findings have great implications for developing similar drug screening platforms for other diseases. Specifically, this proof of principle study with AAT-deficiency iPSCs will provide a foundation for iPSC-based preclinical drug discovery and development of novel therapeutics not only for its associated diseases such as liver cirrhosis and cancer, but also for other complex diseases such as neurodegenerative disorders caused by pathologic accumulation of misfolded proteins (43).
Interestingly, three drugs (Li, CBZ, and VPA) among the final 5 hits were previously implicated as enhancers of autophagy—a physiological process involved in the clearance of aggregate-prone cytosolic proteins (40, 44). In the case of lithium, we found out after the blind screening that there were multiple different forms of lithium (i.e. Li-Br, Li-OH, and Li-Cl) within the drug library and all were detected as hits. In addition, one of the compounds, which significantly increased the AAT level (Fig 2) (i.e. bovinocidin or 3-nitropropionic acid), has been previously shown to cause brain lesions similar to those of Huntington′s disease which is also caused by protein misfolding (45). Together three results support that our disease-specific iPSC based assay is suitable for drug screening as well as further pathogenesis research, and that our findings are not due to bias or chance. Our screening results, based upon AAT-deficiency patient iPSCs, also indirectly suggest the mammalian target of rapamycin (mTOR)- independent autophagy as a potential action mechanism of these drugs, because rapamycin, which inhibits mTOR and a negative regulator of autophagy, did not alter the AAT levels in our screening assay system. In addition, we did not observe significant effects of GSK3β inhibitors such as CHIR99021 and HDAC inhibitors such as sodium phenylbutyrate and sodium butyrate (not shown). Therefore the effects of Li (a known GSK3β inhibitor) and VPA (a known HDAC inhibitor) can not be accounted for by GSK3β or HDAC inhibition either. It has been recently reported that another HDAC inhibitor Suberoylanilide Hydroxamic Acid (SAHA) mediated correction of AAT-deficiency in part through HDAC7 silencing (46); therefore the contribution of HDAC inhibition in VPA’s effects may require further investigation. Together our data (Fig 2, S5, S6) suggest that the mechanisms by which the final 5 drugs influence AAT accumulation within hepatocytes are likely to be associated with mTOR-independent autophagy-mediated degradation of folded-AAT proteins rather than decreasing synthesis of the AAT. However we cannot rule out the possibility of other mechanisms including unconventional autophagy/proteasomal degradation. Thus, the iPSC-based drug screening and discovery strategy outlined in this report may be applicable to various protein misfolding disorders including Alzheimer's disease, Parkinson's disease, Huntington's disease, and amyotrophic lateral sclerosis in addition to AAT-deficiency (43).
Importantly, it has been recently shown that the autophagy-enhancing drug CBZ decreased the hepatic load of mutant AAT accumulation and hepatic fibrosis in a mouse model of AAT-deficiency-associated liver disease (47). This in vivo finding is consistent with our drug screening result based upon the human iPSC model of the disease. Together, these results provide a strong basis for testing autophagy enhancers for the therapeutic use. Given that most of our drug candidates are already FDA approved and have extensive clinical safety profiles (we also confirmed that these drugs do not influence functionality or viability of hepatocyte-like cells derived from patient-and control-iPSCs; Fig S7–S9), there will be no need for further safety tests, which are a main impediment in moving a “hit” to a clinical drug. However, considering their existing use for these drugs and non-toxic therapeutic ranges, it may be necessary to re-adjust the therapeutic range of certain drugs including Li, before their new applications (i.e. treating or preventing AAT-deficiency-associated liver disease). The new applications should also avoid unwanted drug interactions including mutual antagonism between these drugs (Fig S10).
Efficient gene targeting is essential for future iPSC-based gene and cell therapy. Towards this goal, technologies such as ZFN-mediated enhancement of homologous recombination rates in iPSCs have been developed, including gene correction at the AAT locus (24, 25, 27–29, 48). Although it can be highly efficient, the broad application of ZFNs has been limited due to the highly specialized knowledge and tools that are required for designing functional ZFNs in addition to the high cost. In comparison, the TALEN design has been much more flexible and less costly. Although it is still in the early developmental stage, this technology has shown great potential for many applications including gene targeting in human stem cells (30–34, 49, 50). Our study with multiple patient-specific iPSC lines demonstrate that TALEN-mediated targeting of disease-causing mutations can be a broadly applicable approach to generate isogenic and disease-free sources for cell replacement therapy. Our results also demonstrated that the TALEN we used in this study can achieve comparable or higher gene targeting efficiencies (100% efficiency with 25–33% biallelic targeting) than that observed with ZFNs (54% efficiency with 4% of biallelic targeting) using the exact same targeting vector (24). Because of the different spacer requirements for ZFN and TALEN binding sequences, it is not feasible to design the TALEN pairs that recognize the exact same sequences as ZFNs, therefore a precise comparison of the targeting efficiency may not be practical. Nevertheless, our TALENs have significant overlap with the reported ZFNs in their recognition sequences and the targeting results using the same donor vector suggest that TALEN can achieve a similar targeting efficiency as ZFNs. The results from a reporter assay (Fig S11) further support TALEN as an efficient, robust and economic alternative to ZFN technology. Importantly, our data demonstrate a high efficiency of biallelic gene correction using TALEN, which is fast and cost effective. Therefore this approach may be highly compatible with large-scale production of corrected patient-specific iPSCs for many other monogenic disorders. In addition to the application for gene therapy, it will be widely useful for basic gene targeting applications such as creating ideal (i.e. isogenic) controls for iPSC based disease modeling.
In summary, with emerging new tools and technologies including patient-specific iPSCs, a clinical ready drug library, and TALEN, we demonstrated proof of principles for the feasibility of iPSC-based large-scale drug screening and highly efficient gene correction. Integration of patient-specific iPSC based screening in early stages of drug development will help to more accurately predict drug effects in humans thereby significantly shortening the timeline and reducing the costs associated with clinical trials and high failure rates. While many basic or preclinical applications can immediately benefit from our gene targeting study, gene therapy still warrants further extensive safety studies before translation into clinic. Nonetheless, our findings have great implications for developing iPSC based novel therapeutics for the treatment or prevention of currently incurable diseases including AAT-deficiency associated liver diseases and other complex disorders which would benefit from drug screening or gene targeting.
Supplementary Material
Acknowledgements
This work was supported by NIH grant AA020020 and by MSCRF grants 2010-MSCRFII-0101-00, 2011-MSCRFE-0087 and 2012-MSCRFF-0152-00. This work is also supported by NIH Center for Regenerative Medicine. Dr. Leach is supported by NIH grant DK61215 and the Paul K. Neumann Professorship at Johns Hopkins University. The maintenance of JHDL was supported in part by NCI (R01CA122814), FAMRI and by Grant Number UL1 RR 025005 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research (to Johns Hopkins School of Medicine).
References
- 1.Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat Rev Drug Discov. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 2.Munos B. Lessons from 60 years of pharmaceutical innovation. Nat Rev Drug Discov. 2009;8:959–968. doi: 10.1038/nrd2961. [DOI] [PubMed] [Google Scholar]
- 3.Frearson JA, Collie IT. HTS and hit finding in academia--from chemical genomics to drug discovery. Drug Discov Today. 2009;14:1150–1158. doi: 10.1016/j.drudis.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayr LM, Fuerst P. The future of high-throughput screening. J Biomol Screen. 2008;13:443–448. doi: 10.1177/1087057108319644. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, Skolnik S. Recent advances in physicochemical and ADMET profiling in drug discovery. Chem Biodivers. 2009;6:1887–1899. doi: 10.1002/cbdv.200900117. [DOI] [PubMed] [Google Scholar]
- 6.Choi SM, Kim Y, Liu H, Chaudhari P, Ye Z, Jang YY. Liver engraftment potential of hepatic cells derived from patient-specific induced pluripotent stem cells. Cell Cycle. 2011;10:2423–2427. doi: 10.4161/cc.10.15.16869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi SM, Liu H, Chaudhari P, Kim Y, Cheng L, Feng J, Sharkis S, et al. Reprogramming of EBV-immortalized B-lymphocyte cell lines into induced pluripotent stem cells. Blood. 2011;118:1801–1805. doi: 10.1182/blood-2011-03-340620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun YS, Chaudhari P, Jang YY. Applications of patient-specific induced pluripotent stem cells; focused on disease modeling, drug screening and therapeutic potentials for liver disease. Int J Biol Sci. 2010;6:796–805. doi: 10.7150/ijbs.6.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jang YY, Ye Z, Cheng L. Molecular Imaging and Stem Cell Research. Molecular Imaging. 2011 in press. [PubMed] [Google Scholar]
- 10.Liu H, Kim Y, Sharkis S, Marchionni L, Jang YY. In vivo liver regeneration potential of human induced pluripotent stem cells from diverse origins. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002376. 82ra39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye Z, Liu CF, Jang YY. Hematopoietic cells as sources for patient-specific iPSCs and disease modeling. Cell Cycle. 2011;10:2840–2844. doi: 10.4161/cc.10.17.17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Ye Z, Kim Y, Sharkis S, Jang YY. Generation of endoderm-derived human induced pluripotent stem cells from primary hepatocytes. Hepatology. 2010;51:1810–1819. doi: 10.1002/hep.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye Z, Zhan H, Mali P, Dowey S, Williams DM, Jang YY, Dang CV, et al. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood. 2009;114:5473–5480. doi: 10.1182/blood-2009-04-217406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharkis SJ, Jones RJ, Civin C, Jang YY. Pluripotent stem cell-based cancer therapy: promise and challenges. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3003920. 127ps129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cayo MA, Cai J, Delaforest A, Noto FK, Nagaoka M, Clark BS, Collery RF, et al. JD induced pluripotent stem cell-derived hepatocytes faithfully recapitulate the pathophysiology of familial hypercholesterolemia. Hepatology. 2012 doi: 10.1002/hep.25871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rashid S, Corbineau S, Hannan N, Marciniak S, Miranda E, Alexander G, Huang-Doran I, et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perlmutter DH. Liver injury in alpha 1-antitrypsin deficiency. Clin Liver Dis. 2000;4:387–408. doi: 10.1016/s1089-3261(05)70115-x. vi. [DOI] [PubMed] [Google Scholar]
- 18.Eriksson S, Hägerstrand I. Cirrhosis and malignant hepatoma in alpha 1-antitrypsin deficiency. Acta Med Scand. 1974;195:451–458. doi: 10.1111/j.0954-6820.1974.tb08170.x. [DOI] [PubMed] [Google Scholar]
- 19.Perlmutter DH. Autophagic disposal of the aggregation-prone protein that causes liver inflammation and carcinogenesis in alpha-1-antitrypsin deficiency. Cell Death Differ. 2009;16:39–45. doi: 10.1038/cdd.2008.103. [DOI] [PubMed] [Google Scholar]
- 20.Chong CR, Chen X, Shi L, Liu JO, Sullivan DJ. A clinical drug library screen identifies astemizole as an antimalarial agent. Nat Chem Biol. 2006;2:415–416. doi: 10.1038/nchembio806. [DOI] [PubMed] [Google Scholar]
- 21.Kim J, Tang JY, Gong R, Lee JJ, Clemons KV, Chong CR, Chang KS, et al. Itraconazole, a commonly used antifungal that inhibits Hedgehog pathway activity and cancer growth. Cancer Cell. 2010;17:388–399. doi: 10.1016/j.ccr.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egawa N, Kitaoka S, Tsukita K, Naitoh M, Takahashi K, Yamamoto T, Adachi F, et al. Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3004052. 145ra104. [DOI] [PubMed] [Google Scholar]
- 23.Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA, Ganat YM, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yusa K, Rashid ST, Strick-Marchand H, Varela I, Liu PQ, Paschon DE, Miranda E, et al. Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou J, Maeder M, Mali P, Pruett-Miller S, Thibodeau-Beganny S, Chou B, Chen G, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou C, Chou BK, Dowey SN, Tsang K, Huang X, Liu CF, Smith C, et al. Efficient Derivation and Genetic Modifications of Human Pluripotent Stem Cells on Engineered Human Feeder Cell Lines. Stem Cells Dev. 2012 doi: 10.1089/scd.2011.0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver R, Katibah G, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sebastiano V, Maeder ML, Angstman JF, Haddad B, Khayter C, Yeo DT, Goodwin MJ, et al. In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells. 2011;29:1717–1726. doi: 10.1002/stem.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou J, Mali P, Huang X, Dowey SN, Cheng L. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood. 2011;118:4599–4608. doi: 10.1182/blood-2011-02-335554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 33.Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 36.Shim JS, Matsui Y, Bhat S, Nacev BA, Xu J, Bhang HE, Dhara S, et al. Effect of nitroxoline on angiogenesis and growth of human bladder cancer. J Natl Cancer Inst. 2010;102:1855–1873. doi: 10.1093/jnci/djq457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Platz EA, Yegnasubramanian S, Liu JO, Chong CR, Shim JS, Kenfield SA, Stampfer MJ, et al. A novel two-stage, transdisciplinary study identifies digoxin as a possible drug for prostate cancer treatment. Cancer Discov. 2011;1:68–77. doi: 10.1158/2159-8274.CD-10-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rovira M, Huang W, Yusuff S, Shim JS, Ferrante AA, Liu JO, Parsons MJ. Chemical screen identifies FDA-approved drugs and target pathways that induce precocious pancreatic endocrine differentiation. Proc Natl Acad Sci U S A. 2011;108:19264–19269. doi: 10.1073/pnas.1113081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, Cook LJ, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renna M, Jimenez-Sanchez M, Sarkar S, Rubinsztein DC. Chemical inducers of autophagy that enhance the clearance of mutant proteins in neurodegenerative diseases. J Biol Chem. 2010;285:11061–11067. doi: 10.1074/jbc.R109.072181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, Zhu H, et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci U S A. 2007;104:19023–19028. doi: 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gosai SJ, Kwak JH, Luke CJ, Long OS, King DE, Kovatch KJ, Johnston PA, et al. Automated high-content live animal drug screening using C. elegans expressing the aggregation prone serpin α1-antitrypsin Z. PLoS One. 2010;5:e15460. doi: 10.1371/journal.pone.0015460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams RS, Cheng L, Mudge AW, Harwood AJ. A common mechanism of action for three mood-stabilizing drugs. Nature. 2002;417:292–295. doi: 10.1038/417292a. [DOI] [PubMed] [Google Scholar]
- 45.Leventhal L, Sortwell CE, Hanbury R, Collier TJ, Kordower JH, Palfi S. Cyclosporin A protects striatal neurons in vitro and in vivo from 3-nitropropionic acid toxicity. J Comp Neurol. 2000;425:471–478. doi: 10.1002/1096-9861(20001002)425:4<471::aid-cne1>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 46.Bouchecareilh M, Hutt DM, Szajner P, Flotte TR, Balch WE. Histone Deacetylase inhibitor (HDACi) Suberoylanilide Hydroxamic Acid (SAHA) Mediated Correction of Alpha-1 Antitrypsin Deficiency. J Biol Chem. 2012 doi: 10.1074/jbc.M112.404707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 48.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 49.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug Ii RG, Tan W, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012 doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, Joung JK, et al. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012;40:8001–8010. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.