Abstract
Chinese rhesus macaques are of particular interest in SIV/HIV research as these animals have prolonged kinetics of disease progression to AIDS, compared to their Indian counterparts, suggesting that they may be a better model for HIV. Nevertheless, the specific mechanism(s) accounting for these kinetics remains unclear. The study of Major Histocompatibility Complex (MHC) molecules, including their MHC:peptide binding motifs, provides valuable information for measuring cellular immune responses and deciphering outcomes of infection and vaccine efficacy. In this study, we have provided detailed characterization of six prevalent Chinese rhesus macaque MHC class I alleles, yielding a combined phenotypic frequency of 29%. The peptide binding specificity of two of these alleles, Mamu-A2*01:02 and -B*010:01, as well as the previously characterized allele Mamu-B*003:01 (and Indian rhesus Mamu-B*003:01), was found to be analogous to that of alleles in the HLA-B27 supertype family. Specific alleles in the HLA-B27 supertype family, including HLA-B*27:05, have been associated with long-term non-progression to AIDS in humans. All six alleles characterized in the present study were found to have specificities analogous to HLA-supertype alleles. These data contribute to the concept that Chinese rhesus macaque MHC immunogenetics is more similar to HLA than their Indian rhesus macaque counterparts, and thereby warrant further studies to decipher the role of these alleles in the context of SIV infection.
Keywords: MHC, non-human primate, Chinese rhesus macaques, MHC:peptide binding motif
Introduction
Indian-origin rhesus macaques and Chinese-origin rhesus macaques have both been utilized extensively in SIV/HIV research, but also as models for other pathogens (Campillo-Gimenez et al. 2010; Chen et al.; Gardner and Luciw 2008; Giraldo-Vela et al. 2008; Hu et al. 2005; Joag et al. 1994; Kanthaswamy et al. 2009a; Kanthaswamy et al. 2009b; Ling et al. 2002; Miller et al. 1989; Wambua et al. 2011; Xia et al. 2010). While these two sets of animals appear to be indistinguishable physiologically, Chinese rhesus macaques have a much more prolonged progression to AIDS as compared to Indian rhesus macaques (Campillo-Gimenez et al. 2010; Hu et al. 2005; Ling et al. 2002; Xia et al. 2010). The mechanism for these differential outcomes has not been deciphered, but may be related to differences in the cellular immune response and the host genetics that influence their development. Recent studies have shown that the MHC gene regions of Indian and Chinese macaques differ appreciably in terms of the degree of polymorphism, the specific allelic variants, and their functional characteristics (Li et al. 2012; Maness et al. 2011; Ouyang et al. 2008; Ouyang et al. 2009; Solomon et al. 2010; Southwood et al. 2011; Wambua et al. 2011; Wiseman et al. 2009). This is particularly relevant in terms of MHC class I molecules, which determine the repertoire of CD8+ T-cell responses that an individual can develop against SIV and/or any other foreign pathogen (Parham 2005).
A number of studies have detailed the specificity of MHC class I molecules expressed in Indian macaques (Allen et al. 2001; Chen et al. 1992; Dzuris et al. 2000; Giraldo-Vela et al. 2008; Kuroda et al. 1998; Letvin et al. 1993; Loffredo et al. 2008; Loffredo et al. 2007c; Loffredo et al. 2005; Loffredo et al. 2004; Miller et al. 1991; Mothe et al. 2002a; Mothe et al. 2002b; Pal et al. 2002; Reed et al. 2011; Sette et al. 2005; Sidney et al. 2000). It is of relevance to note that not all disease protection or susceptibility studies have been linked to MHC genes, but in specific instances, MHC class I alleles have been shown to be associated with disease progression, or to non-progression, to AIDS in HIV/SIV infections. In particular, recent studies have shown that Mamu-B*003:01, a relatively infrequent allele in Indian rhesus macaques, but expressed in Chinese rhesus macaques at a frequency of 5.8% (Kaizu et al. 2007; Solomon et al. 2010), has a peptide binding repertoire analogous to that of the human MHC class I molecule HLA-B*27:05 (Loffredo et al. 2009). Strikingly, the Mamu-B*003:01 allele has a protective effect in the context of SIV infection of Indian rhesus macaques that parallels the protective effect of its human counterpart in HIV infection (Loffredo et al. 2008; Loffredo et al. 2007b; Loffredo et al. 2007c; Loffredo et al. 2009). Mamu-B*008:01, present in Indian rhesus macaques, also has a binding repertoire similar to HLA-B*27:05 (Loffredo et al. 2009). Several studies have shown a protective effect in Mamu-B*008:01 expressing animals in the context of SIV infection in Indian rhesus macaques in terms of a delayed progression to AIDS and superior viral control (Loffredo et al. 2008; Loffredo et al. 2007a; Loffredo et al. 2007b). Most recently, it has been shown that CD8+ T-cell responses specifically can be attributed to protection in the context of SIV-infection in Mamu-B*008:01-positive animals (Mudd et al. 2012). Other MHC class I alleles, such as HLA-B*57 (Chen et al. 2012), have also been implicated in delayed progression to AIDS and therefore should not be ignored.
In this context, it would be insightful to understand the role of other MHC class I alleles expressed in non-human primate animal models that have alleles that are HLA-B27-like, as well as animals possessing other HLA-like alleles. For this to be possible, further investigation and characterization of the MHC class I alleles most commonly expressed in other models, such as Chinese macaques, is necessary. These types of studies will allow better realization of the potential of Chinese-origin macaques to facilitate infectious disease research, and perhaps yield a better understanding of the differences between Indian and Chinese macaques in terms of outcomes of SIV infection. Since no specific MHC class I allele is present in the Chinese population at a frequency greater than 9% (Solomon et al. 2010), the characterization of several MHC class I molecules is needed to understand the complexity of immune responses in these animals.
We have recently elucidated peptide-binding motifs for four common Chinese macaque MHC class I molecules: Mamu-B*039:01 (Sette et al. 2012), Mamu-A1*026:01, Mamu-B*083:01 (Southwood et al. 2011) and Mamu-A1*022:01 (Solomon et al. 2010). Interestingly, the motifs for three of these alleles were found to overlap with those associated with HLA class I supertypes, which describe sets of HLA class I alleles grouped together on the basis of shared main peptide-binding specificity (i.e., the supermotif) (see, e.g., (Lund et al. 2004; Sette and Sidney 1999; Sidney et al. 2008; Sidney et al. 1995; Sidney et al. 1996).). Specifically, the Mamu-A1*026:01 motif overlaps with that of the HLA-A2 supertype, Mamu-B*083:01 with HLA-A3 supertype (Southwood et al. 2011) and Mamu-A1*022:01 with the HLA-B7 supertype (Solomon et al. 2010). The HLA-B7, -A3, and -A2 supertypes are the three most abundant supertypes in the human population, each present with a phenotypic frequency of greater than 40%, averaged across various ethnic groups (Lund et al. 2004; Sette and Sidney 1999; Sidney et al. 2008; Sidney et al. 1995).
In this study, we have characterized a set of six additional alleles that were the next most commonly expressed MHC class I alleles in rhesus macaques of Chinese origin. OStrikingly, we found that the motifs associated with all of the new alleles are also analogous to those of various common HLA alleles. In particular, we found that two of these alleles are associated with HLA B27-like peptide binding motifs. We were interested in developing these motifs to provide additional resources to facilitate future research towards an understanding of the role of MHC class I alleles, and specific responses, in the context of infection.
Materials and Methods
Creation of stable Mamu transfectant lines
Stable MHC class I transfectants were produced in the MHC class I deficient EBV-transformed B-lymphoblastoid cell line 721.221. An expression construct was created for each Chinese rhesus macaque MHC class I allele by sub-cloning a full-length allele transcript into separate pcDNA 3.1 vectors (Invitrogen). These constructs were then used to transfect MHC class I-null 721.221 cells using an Amaxa Nucleofector II transfection instrument (Lonza AG, Walkersville, MD, USA). Each construct was transfected using the specific G-016 program on the Nucleofector II instrument. For each transfection, 1×107 721.221 cells were mixed with 15 micrograms of DNA using the Amaxa Nucleofactor Solution C (Lonza AG, Walkersville, MD, USA).
Elution of naturally bound ligands from common Chinese MHC class I molecules
721.221 cell lines transfected with a single macaque MHC class I allele were expanded and harvested to generate a pellet equivalent to approximately 5 × 109 cells. Cell pellets were lysed for 30 min in a buffer containing 150mM NaCl, 20mM Tris HCl, pH 8.0, 1% CHAPS (Sigma), and a cocktail of protease inhibitors that included 5 μg/ml Aprotinin (Sigma), 10 μg/ml Leupeptin (Sigma), 10 μg/ml Pepstatin A (EMD Millipore), 1 mM PMSF (Fisher), Phosphatase inhibitor cocktail I (Fisher), Phosphatase inhibitor cocktail II (Sigma), and 10 nM Calyculin A (Sigma). The lysate was centrifuged for 1h at 100,000 × g (SW28 rotor at 27,500 rpm). The supernatant containing soluble proteins was decanted and filtered through an Acrodisc PF 0.2 μM syringe filter with 0.8 μM pre-filter (Fisher), retaining the filtrate.
The filtrate was passed over, in series, a Sepharose CL-4B pre-column, an irrelevant MHC class I antibody affinity column to assess non-specific binding (anti-mouse MHC class I MKD6, in this case), and then a W6/32 antibody (anti-primate MHC class I) affinity column to bind MHC class I molecules, as previously described (Sidney et al. 2001b). Affinity columns were then separated and washed with 2 column volumes (cv) of lysis buffer, 20 cv of 20 mM Tris-HCl, pH 8.0, with 150 mM NaCl, 20 cv of 20 mM Tris-HCl, pH 8.0, with 1.0 M NaCl, and 20 cv of 20 mM Tris-HCl, pH 8.0.
Separately, each affinity column was eluted with 4 cv of 0.2 N acetic acid into a 50mL conical vial, at which point glacial acetic acid was added to bring the final concentration to 10% acetic acid. The acid eluate was transferred to the top reservoirs of pre-conditioned centrifugal filtration units with a molecular weight cutoff of 3000 Dalton (Millipore). This threshold excludes β2-microglobulin from passing into the sample of endogenous ligands. The units were then centrifuged at 3500 × g for approximately 3 hours, until the majority of the eluate had been filtered to the collection reservoir, with minimal retentate (~200 μl) remaining.
The filtrate containing the soluble endogenous peptides was then aliquoted into Eppendorf Protein LoBind microcentrifuge tubes (Fisher) and concentrated using vacuum centrifugation. The sample aliquots were pooled and washed with 10% acetic acid as volume decreased, ceasing the process when a final volume of ~250 μl was achieved. The samples were stored at −80°C until analysis by tandem mass spectrometry was performed.
Peptide Sequence Analysis by Tandem Mass Spectrometry
Peptides were analyzed by nanoflow-HPLC/microeletrospray ionization, coupled directly to a Thermo Fisher Scientific Orbitrap or FT-ICR mass spectrometer, equipped with either a home-built, front-end ETD (FETD) source (Earley et al. 2010) or an Orbitrap Velos mass spectrometer equipped with a commercial ETD source.
Data were acquired as previously described (Udeshi et al. 2008). In brief, a pre-column, loaded with 5 × 107 to 1 × 108 cell equivalents of MHC eluted peptides, was connected with polytetrafluoroethylene tubing (0.06-inch o.d. and 0.012-inch i.d.; Zeus Industrial Products) to the end of an analytical HPLC column (360 μm o.d. × 50 μm i.d.) containing 6 – 7 cm of C18 reverse-phase packing material (5-μm particles; YMC). Peptides were eluted through a laser-pulled electrospray tip directly into the mass spectrometer with an Agilent 1100 series binary LC pump at a flow rate of ~60 nl/min. Elution gradients utilized were as follows: solvent A was 0.1 M acetic acid in H2O and solvent B was 70% acetonitrile. For the analyses, CAD and ETD fragmentation were performed sequentially on the same parent ions. The FETD reagent was azulene and the ion-ion reaction time was set to 30 ms, while fluoranthene was used with a 50 ms reaction time on the commercial source. The instrument was operated in a data-dependent mode where a full-scan mass spectrum was acquired with the high-resolution mass analyzer and this was then followed by sequential acquisition of CAD and ETD MS/MS spectra in the linear trap on the top five, most abundant, non-excluded ions observed in the full-scan spectrum.
Data from MS/MS experiments were searched against the SwissProt (Bairoch and Apweiler 2000) human database using the Open Mass Spectrometry Search Algorithm (OMSSA) software (Geer et al. 2004) to generate a list of candidate peptide sequences. Instrument parameters included a precursor mass tolerance of ± 0.005 Da and a monoisotopic fragment ion mass tolerance of ± 0.6 Da. Database search parameters allowed variable modifications for phosphorylation on serine, threonine and tyrosine residues and oxidation of methionine. Database assignments were confirmed by manual interpretation of the corresponding MS/MS spectra and by showing that spectra recorded on synthetic peptides were identical to those obtained from the biological sample.
Positional scanning combinatorial library and peptide synthesis
Positional scanning combinatorial libraries (PSCL) were synthesized as previously described (Pinilla et al. 1999). In the PCSL, each pool in the library contains randomized 9-mer peptides with one fixed residue at a single position. With each of the 20 naturally occurring residues represented at each position along a 9-mer backbone, the entire library consisted of 180 peptide mixtures.
Peptides utilized in screening studies were purchased as crude or purified material from Mimotopes (Minneapolis, MN, USA/Clayton, Victoria, Australia), Pepscan Systems B. V. (Lelystad, Netherlands), A&A Labs (San Diego, CA, USA), Genescript Corporation (Piscataway, NJ, USA), or the Biotechnology Center at the University of Wisconsin-Madison (Madison, WI, USA). Peptides synthesized for use as radiolabeled ligands were synthesized by A&A Labs and purified to >95% homogeneity by reverse-phase HPLC. Peptide purity was determined with analytical reverse-phase HPLC and amino acid analysis, sequencing, and/or mass spectrometry. Peptides were radiolabeled utilizing the chloramine T method (Sidney et al. 2001a). Lyophilized peptides were re-suspended at 20 mg/ml in 100% DMSO, then diluted to required concentrations in PBS+0.05% (v/v) nonidet P40 (Fluka Biochemika, Buchs, Switzerland).
SIV peptide sequences were derived from the SIVmac239 sequence, Genbank accession M33262 (Kestler et al. 1990).
MHC purification and peptide-binding assays
MHC class I purification was performed by affinity chromatography using the W6/32 class I antibody, as previously described (Allen et al. 2001; Loffredo et al. 2009; Sidney et al. 2005). Protein purity, concentration, and depletion efficiency steps were monitored by SDS-PAGE.
Quantitative assays for peptide binding to detergent-solubilized MHC class I molecules were based on the inhibition of binding of a high affinity radiolabeled standard probe peptide and performed as detailed in prior studies (Loffredo et al. 2009; Schneidewind et al. 2008; Sette et al. 2001; Sidney et al. 2005). Peptides were tested at six different concentrations covering a 100,000-fold dose range in three or more independent assays. For each peptide, the concentration of peptide yielding 50% inhibition of the binding of the radiolabeled probe peptide (IC50) was calculated. Under the conditions used, where [radiolabeled probe] < [MHC] and IC50 ≥ [MHC], the measured IC50 values are reasonable approximations of the true Kd values (Gulukota et al. 1997; Sette et al. 1994b). Control wells to measure non-specific (background) binding were also included. In each experiment, a titration of the unlabeled version of the radiolabeled probe was also tested as a positive control for inhibition.
The radiolabeled peptides utilized for developing the peptide-binding assays that represent sequences identified by Edman degradation and mass spectrometry analysis (described above), are presented in Supplemental Table 2 (bolded). For one allele, Mamu-A7*01:03, an analog of a natural ligand was synthesized (RAEDNADYL), in which a threonine substitution was made at position 8 (T->Y) to allow for Iodine125 radiolabeling. For the Mamu-B*090:01 assay, a ligand (sequence MSAPPAEYK) previously identified by direct binding assay, was utilized as the radiolabeled probe. The binding assay for Mamu-B*090:01 was therefore developed empirically prior to the use, in our laboratory, of natural ligand elution methodologies as a means to identifying a ligand of interest.
Bioinformatic analysis
Analysis of the PSCL data was performed as described previously (Sidney et al. 2008). Briefly, IC50 nM values for each mixture were standardized as a ratio to the geometric mean IC50 nM value of the entire set of 180 mixtures and then normalized at each position so that the value associated with optimal binding at each position corresponds to 1. For each position, an average (geometric) relative binding affinity (ARB) was calculated, and then the ratio of the ARB for the entire library to the ARB for each position was derived. We have denominated this ratio, which describes the factor by which the normalized geometric average binding affinity associated with all 20 residues at a specified position differs from that of the average affinity of the entire library, as the specificity factor (SF). As calculated, positions with the highest specificity will have the highest SF value. Primary anchor positions were then defined as those with an SF > 2.4. This criterion identifies positions where the majority of residues are associated with significant decreases in binding capacity.
Secondary anchor designations were based on the standard deviation (SD) of residue specific values at each position. Dominant secondary anchor positions were defined as those where the SD was > 3 and the SF < 2.4, as well as positions associated with an SD > 2 if the SF is between 1.5 and 2.4. Weak secondary anchors have been defined as positions associated with a SD in the 2.5–3 range with an SF < 1.5, or an SF in the 1.5–2.4 range with an SD < 2.
To identify predicted binders, all possible 9-mer peptides in SIVmac239 sequences were scored using the matrix values derived from the PSCL analyses of the six Mamu alleles under investigation in this study (Sidney et al. 2008). The final score for each peptide represents the product of the corresponding matrix values for each peptide residue-position pair. Peptides scoring amongst the top 3.0% (n = 100) were selected for binding analysis.
Results
Selection of a panel of MHC class I alleles commonly expressed in Chinese rhesus macaques
To identify the MHC class I alleles most frequently expressed in Chinese macaques, we previously sequenced alleles from a cohort of 50 Chinese rhesus macaques representing 3 colonies currently utilized in biomedical research: the Scripps Research Institute, the Tulane National Primate Research Center and the Washington National Primate Research Center (Solomon et al. 2010). Using data from that particular study, as well MHC class I allele frequency information from other publications, a panel of the 12 most frequent alleles was generated (Solomon et al. 2010). We have already characterized six of these most common molecules (Dzuris et al. 2000; Loffredo et al. 2005; Sette et al. 2012; Solomon et al. 2010; Southwood et al. 2011), to include Mamu-A1*022:01, -A1*026:01, -B*083:01, and -B*039:01, and Mamu-B*003:01 and -B*001:01:02, which are present in both Indian and Chinese rhesus macaque populations.
In the present study we have undertaken efforts to characterize the six next most common Chinese rhesus alleles (Mamu-A7*01:03, -B*066:01, -B*010:01, -B*087:01, -A2*01:02 and -B*090:01), thereby doubling the number of alleles characterized in this model (Table 1). These six alleles range in phenotypic frequency from 6.1% (Mamu-A7*01:03) to 2.2% (Mamu-B*090:01). The combined phenotypic frequency of these six new alleles is 29%, which, together with the six previously characterized alleles, brings the combined frequency of Chinese rhesus macaque MHC class I alleles studied to date to almost 60% (Table 1).
Table 1.
MHC class I alleles comprising approximately 59% of Chinese macaques studied to date
| Mamu allele | Phenotypic frequency (%) a | HLA homology | Reference |
|---|---|---|---|
| A1*022:01 | 8.9 | B07-like | Solomon et al. 2010 |
| A1*026:01 | 6.7 | A02-like | Southwood et al. 2011 |
| A7*01:03 | 6.1 | A01-like | This study |
| B*083:01 | 5.8 | A03-like | Southwood et al. 2011 |
| B*066:01 | 5.8 | A03-like | This study |
| B*039:01 | 5.8 | -- b | Sette et al. 2012 |
| B*010:01 | 5.8 | B27-like | This study |
| B*087:01 | 5.8 | B44-like | This study |
| B*003:01 | 5.8 | B27-like | Dzuris et al. 2000 |
| B*001:01:02 | 5.8 | B44-like | Loffredo et al. 2005 |
| A2*01:02 | 5.6 | B27-like | This study |
| B*090:01 | 2.2 | A03-like | This study |
Frequency values determined in Solomon et al 2010
Dashes (--) indicate no HLA homology observed
Establishment of a peptide binding assay for Mamu-B*090:01 based on predicted overlap with HLA
Mamu-B*090:01 MHC class I molecules expressed in single allele transfectants of the 721.221 cell line were purified by affinity chromatography as described in the Materials and Methods. As mentioned above, previous studies demonstrated that HLA supertype ligands also bind to MHC molecules expressed in other species, such as chimpanzees (Pan troglodytes), mice (Mus musculus) (Bertoni et al. 1998; McKinney et al. 2000; Sette et al. 2005; Sidney et al. 2006), and Chinese rhesus macaques (Solomon et al. 2010).
Hierarchical clustering analysis (data not shown) predicted that Mamu-B*090:01 might be associated with an HLA-A3 supertypic peptide binding specificity. Indeed, significant binding was detected using the HLA-A3 supertype ligand MSAPPAEYK with as little as 2.0 nM of purified MHC (Supplemental Fig. 1). This binding was specific in that it could be inhibited by unlabeled MSAPPAEYK with an IC50 of approximately 1.1 nM (Fig. 1 and Supplemental Table 1). The specificity of binding was further demonstrated by the fact that the same MHC preparation did not bind the VVMAYVGIK radiolabeled peptide, which is also a HLA-A3 supertype ligand.
Fig. 1. Specificity of competitive MHC:peptide binding assays for six common Chinese rhesus MHC class I molecules.
Endogenous or empirically identified peptide ligands for each MHC class I molecule were radiolabeled and used as probes in direct binding assays. Those strongest signals were obtained with SHSHVGYTL for Mamu-A2*01:02, RAEDNADYL for Mamu-A7*01:03, YFAIAENESK for Mamu-B*066:01, MSAPPAEYK for Mamu-B*090:01, SDIDGDRYV for Mamu-B*087:01, and SHIDRVYTL for Mamu-B*010:01. The affinity of each peptide for their respective class I molecule was determined in inhibition assays, as described in the Materials and Methods. As shown, each peptide was tested at multiple doses over a 5 log range. The binding in each case was specific, as binding by the radiolabeled ligand could be inhibited by the unlabeled version of the respective probe peptide, but not by unrelated control ligands. IC50 values for the reference peptides ranged from 500 pM to 2.0 nM, depending on the exact allele/ligand combination. Representative inhibition curves for each MHC class I molecule are shown.
Determination of natural ligands from common Chinese rhesus macaque MHC class I molecules
Previous studies have demonstrated that the elution and characterization of naturally bound ligands is an effective method for determining the peptide binding specificity of MHC class I molecules (Hickman-Miller et al. 2005; Kubo et al. 1994). Accordingly, naturally bound peptides were identified as described in the Materials and Methods for the remaining five alleles. A total of 18 peptide ligands were sequenced from Mamu-A2*01:02, and for Mamu-A7*01:03, Mamu-B*010:01, Mamu-B*066:01, and Mamu-B*087:01, 5, 30, 14, and 27 ligands, were identified (Supplemental Table 2), respectively.
The majority of ligands for both Mamu-A2*01:02 and -A7*01:03 were 9 residues in length (Supplemental Table 2). Specifically, 14 of the 18 (78%) ligands eluted from Mamu-A2*01:02, and 5 of the 6 (83%) eluted from Mamu-A7*01:03 were 9-mers. In addition, one Mamu-A2*01:02 ligand was an 8-mer, two where 10-mers, and another was 11 residues in length. For Mamu-A7*01:03 one additional identified ligand was 10 residues.
The lengths of endogenous peptides identified for the three Mamu-B alleles were more variable. For Mamu-B*010:01 peptides ranged in size from 8- to 12 residues, with just over half (17 of 30; 57%) being nonamers. The next most frequent size was 11-mers, with 6 peptides identified (20%). For Mamu-B*066:01 the most common peptide size was also 9 residues, with 6 of the 14 ligands being this length. Furthermore, 5 of 14 ligands were 10-mers, suggesting that peptides of both 9 and 10 peptide lengths are commonly bound. Mamu-B*087:01 also displayed ligands of variable sizes. In this case 10-mers comprised the majority of the ligands (41%; 11 of 27), although 9-mers, 12-mers, and 13-mers were all represented, with 6, 4, and 3 ligands each, respectively. Mamu-B*087:01 also bound a peptide 14 residues in length, but it did not bind 8-mer peptides (Supplemental Table 2).
Establishment of peptide binding assays based on naturally processed ligands
We next sought to establish binding assays for the five Chinese MHC class I molecules for which naturally processed ligands were defined. MHC class I molecules were purified by affinity chromatography, and peptides corresponding to naturally bound ligands were synthesized. Peptides containing Y were radiolabeled and their capacity to bind their associated purified MHC was investigated. For natural ligands in which a tyrosine was not present, Y substitutions were introduced by substitution at various traditionally non-main anchor positions.
For each allele, the peptide yielding the strongest signal (cpms) in direct binding assays was selected for further characterization. Those ligands were SHSHVGYTL for Mamu-A2*01:02, RAEDNADYL (T8->Y analog) for -A7*01:03, SHIDRVYTL for -B*010:01, YFAIAENESK for -B*066:01, and SDIDGDYRV for -B*087:01. In all cases, the binding was selective, as other radiolabeled peptides assayed displayed little to no binding (Supplemental Fig. 1). Furthermore, the binding of each was specific at the level of inhibition by unlabeled ligands, with measured IC50 values ranging from 200 pM to 2.0 nM (Fig. 1 and Supplemental Table 1).
The identification of MHC:peptide binding motifs for six common Chinese rhesus macaque class I molecules
To define the peptide binding motifs of the six alleles, we tested the capacity of 9-mer positional scanning combinatorial libraries (PSCL) to bind purified MHC (Supplemental Table 3). The data was analyzed, and anchor positions determined, following the criteria outlined in the Materials and Methods.
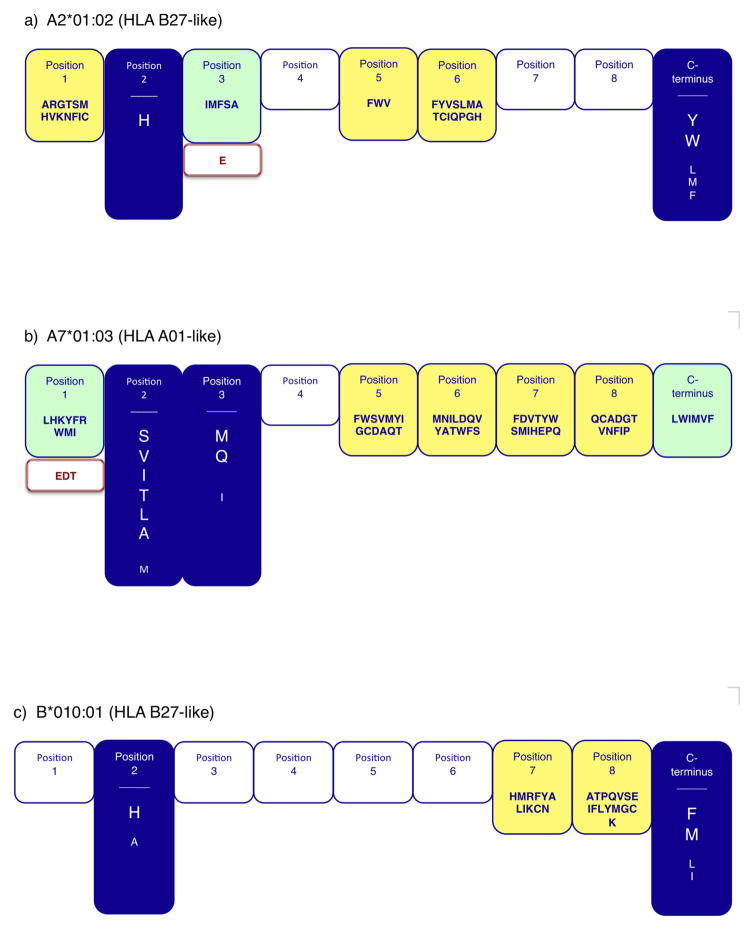
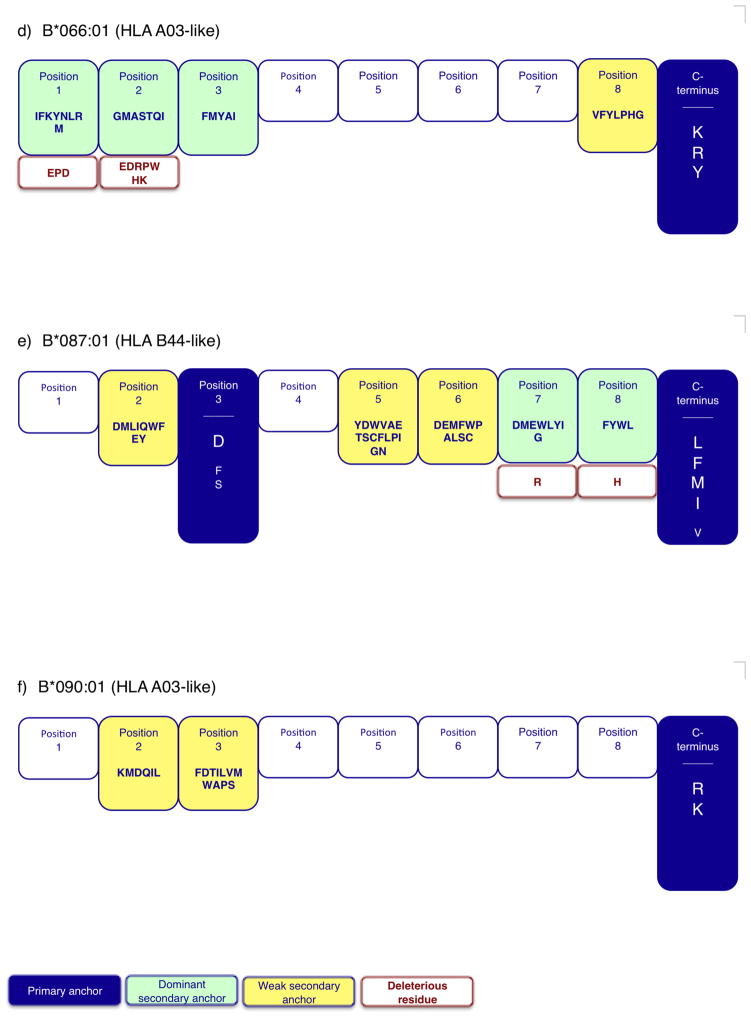
Mamu-B*087:01 was determined to have primary anchors at position 3 and the C-terminus (Fig. 2). Aspartic acid (D) is dominant at position 3, with F and S being tolerated. Interestingly, D is a preferred residue at a number of positions, including positions 2, 3, 5, 6, and 7. At the C-terminal primary anchor a preference for the hydrophobic residues L, F, M and I was found (and V was tolerated). Mamu-B*087:01 also has dominant secondary anchors at positions 7 and 8. Based on the described profile, the Mamu-B*087:01 motif (Fig. 2) closely resembles the motif associated with the HLA-B44 supertype (Sidney et al. 2003), although the HLA molecules have a primary anchor at position 2 as opposed to position 3.
Fig. 2. Summary motifs for six Chinese MHC class I molecules.
Pictorial representation of the respective motifs indicate the residues and positions contributing positive and negative binding potential for a) Mamu-A2*01:02, b) Mamu-A7*01:03, c) Mamu-B*010:01, d) Mamu-B*066:01, e) Mamu-B*087:01, and f) Mamu-B*090:01. Primary anchor positions are indicated by blue shading, with preferred (larger font) and tolerated residues displayed. Dominant (green) and weak (yellow) secondary anchor positions are also highlighted. Residues that are associated with deleterious effects on binding capacity (>100-fold decrease), when present at a given position, are indicated by red font.
For Mamu-A7*01:03, the analysis revealed a motif which parallels the one observed for HLA-A*01:01 (Kondo et al. 1997). Positions 2 and 3 were identified as primary anchor positions. At position 2, residues S, V, I, T, A and L were preferred. At position 3, M and Q were preferred, while I was tolerated. The C-terminus was classified as a dominant secondary anchor, with L, W, I, M, V and F being tolerated. Position 1 was also designated as a dominant secondary anchor (Fig. 2).
The current analysis also revealed that two of the MHC class I molecules were associated with an HLA-A3-like motif (Sidney et al. 1996). In the case of Mamu-B*066:01, the C-terminus primary anchor position was associated with a preference for K, R, and Y. Positions 1, 2, and 3 were identified as dominant secondary anchors positions. For Mamu-B*090:01 the preferred positions and residues were similar to those observed in the case of Mamu-B*066:01. Specifically, the C-terminus was the primary anchor position, with a preference for the positively charged residues R and K. Positions 2 and 3 were identified as weak secondary anchors (Fig. 2).
Most interestingly, two of the Chinese rhesus MHC class I molecules were associated with an HLA B27-like motif (Sette and Sidney 1999). In the case of Mamu-A2*01:02, position 2 and the C-terminus were identified as the primary anchor positions. Histidine (H) was dominant at position 2, and the aromatic residues Y and W were preferred at the C terminus (with M, L and F being tolerated) (Fig. 2). For Mamu-B*010:01, position 2 and the C-terminus were also identified as primary anchors. Similar to Mamu-A2*01:02, H was the dominant residue at position 2, while A was tolerated. At the C-terminus, the aromatic residue F, and aliphatic residues M, L and I, were preferred. Weak secondary anchors were identified as positions 7 and 8 (Fig. 2).
The peptide-binding motifs of all six newly characterized Chinese rhesus macaque class I molecules overlap with HLA class I supertypic motifs
The addition of these alleles yields a total of 12 MHC class I alleles that have been characterized for Chinese rhesus macaques. Out of these 12 alleles, 11 have peptide binding motifs that overlap those defined for several HLA class I supertype (Table 2). The HLA analogy spans the HLA-A1 (1 Chinese rhesus macaque allele), HLA-A2 (1 allele), -A3 (3 alleles), -B7 (1 allele), -B27 (3 alleles), and -B44 (2 alleles) supertypes. This is in contrast to the case with Indian rhesus macaques, where only 7 of the 13 alleles characterized to date have motifs that could be classified as analogous with HLA supertype alleles. Also, only four HLA supertypes are covered by the Indian rhesus macaque alleles: HLA-A1 (1 allele), -B27 (3 alleles), -B44 (2 alleles), -B58 (1 allele).
Table 2.
HLA homology of characterized rhesus macaque MHC class I molecules
| Chinese origin | Indian origin | ||
|---|---|---|---|
| Allele | HLA homology a | Allele | HLA homology |
| A1*022:01 | B07 | A1*001:01 | B58 |
| A1*026:01 | A02 | A1*002:01 | A01 |
| A2*01:02 | B27 | A1*007:01 | B27 |
| A7*01:03 | A01 | A1*011:01 | B44 |
| B*001:01:02c | B44 | B*001:01:02c | B44 |
| B*003:01c | B27 | B*003:01c | B27 |
| B*010:01 | B27 | B*004:01 | -- |
| B*039:01 | -- b | B*008:01 | B27 |
| B*066:01 | A03 | B*017:01 | -- |
| B*083:01 | A03 | B*022:01 | -- |
| B*087:01 | B44 | B*029:01 | -- |
| B*090:01 | A03 | B*048:01 | -- |
| B*052:01 | -- | ||
Indicated homology refers to supertype family of alleles
Dashes (--) indicate no known homology exists.
Low crossreactivity between peptide binding repertoires of Chinese MHC class I analogs of HLA-B27 and HLA-B27
The HLA-B27 motif is characterized as preferring positively charged residues at position 2 and hydrophobic or aliphatic residues at the C-terminus. Two of the macaque MHC class I alleles described herein, Mamu-A2*01:02 and -B*010:01, possess comparable motifs. To evaluate whether these motif similarities would also correspond to overlapping repertoires, we evaluated the capacity of both molecules to bind a panel of previously characterized HLA-B27 epitopes. It was found that Mamu-B*010:01 bound 9 of HLA-B27 epitopes (33%) with an affinity of 500 nM, or better (Table 3), representing a crossreactivity similar to that seen amongst various HLA supertype (Greenbaum et al. 2011). At the same time, however, Mamu-A2*01:02 was associated with a much lower level of crossreactivity, and bound only 3 of the HLA-B27 peptides (11%) with an affinity of 500 nM, or better. These data highlight that, while the motif associated with these molecules are remarkably similar to HLA-B27, the corresponding peptide binding repertoires may be more divergent.
Table 3.
Rhesus macaque MHC class I binding affinity for HLA-B27-specific epitopes
| IC50 nM to purified MHC | ||||||
|---|---|---|---|---|---|---|
| Sequence | Source | HLA-B*27:05b | Mamu-B*03b | Mamu-B*08b | Mamu-A2*01:02 | Mamu-B*010:01 |
| ARKLLLDNL | Chlamydia peptide 138 | 74 | 7004 | 316 | 222 | -- a |
| DRLALLANL | C. trachomatis recC exodeoxyribonuclease-V g chain | 181 | 871 | 110 | 19,102 | -- |
| EREQTLNQL | C. trachomatis ATP-dependent zinc protein | 2694 | 10,195 | 1747 | 13,162 | -- |
| ERFLAQEQL | Chlamydia peptide 196 | 728 | 5839 | 3760 | -- | -- |
| IRMFKILPL | Chlamydia peptide 80 | 13 | 12 | 7.4 | 227 | 194 |
| IRSSVQNKL | C. trachomatis papQ invasin repeat-family-phosphatase | 37 | 462 | 109 | 1921 | 210 |
| KRLAETLAL | Chlamydia peptide 131 | 165 | 29 | 55 | -- | 11,982 |
| MRDHTITLL | C. trachomatis NADH-ubiquinone-oxidoreductase-a-chain | 52 | 380 | 91 | -- | 8.0 |
| NRAKQVIKL | C. trachomatis protease ATPase | 324 | 524 | 282 | -- | 106 |
| NRELIQQEL | Chlamydia peptide 195 | 5789 | 738 | 54 | 3784 | 1110 |
| NRFSVAYML | C. trachomatis pmpC putative outer membrane protein | 9.8 | 2865 | 444 | 5472 | 0.15 |
| YRLLLTRVL | C. trachomatis ygeD efflux protein | 5.6 | 14 | 5.9 | -- | 24 |
| ARLFGIRAK | E. coli derived | 133 | 148 | 6.7 | -- | 1128 |
| FRYNGLIHR | E. coli derived | 18 | 3512 | 799 | -- | 259 |
| RRISGVDRY | E. coli derived | 21 | 148 | 403 | 9605 | 15,610 |
| RRSKEITVR | E. coli derived | 5.5 | 2622 | 2698 | 12,517 | -- |
| RRYQKSTEL | E. coli derived | 6.2 | 6.1 | 4.1 | 6646 | 291 |
| RRIYDLIEL | EBNA 258-266 (EBV) | 6.6 | 5.7 | 7.1 | 78 | 742 |
| GRRGWEALKY | HIV-1 gp41 | 129 | 6503 | 1879 | -- | -- |
| IRLRPGGKK | HIV Gag p17 (LTNP/CI) | 45 | 9358 | 2199 | -- | -- |
| KRWIILGLNK | HIV Gag p24 (LTNP/CI) | 5.4 | 14 | 25 | 3697 | 12,750 |
| RRQDILDLWI | HIV-1 Nef | 99 | 53 | 46 | 17,922 | 1740 |
| RRWIQLGLQK | HIV-1 p24 | 0.0027 | 12 | 14 | -- | -- |
| SRYWAIRTR | influenza NP 383-391 | 6.0 | 347 | 82 | 4964 | -- |
| KRVVINKDT | Y. enterocolitica Hsp60 | 406 | 1055 | 12,467 | -- | -- |
| SRFPEALRL | proteasome 26S non-ATPase subunit 2 variant p256 | 12 | 65 | 8.4 | 15,627 | 100 |
| SRHHAFCFR | Aggrecan precursor p667 | 42 | 1345 | 379 | 2058 | -- |
Dashes (--) indicate binding affinity >20,000 nM
Data from Loffredo et al. 2009
Differences in the size of the peptide repertoire associated with different Mamu MHC class I alleles
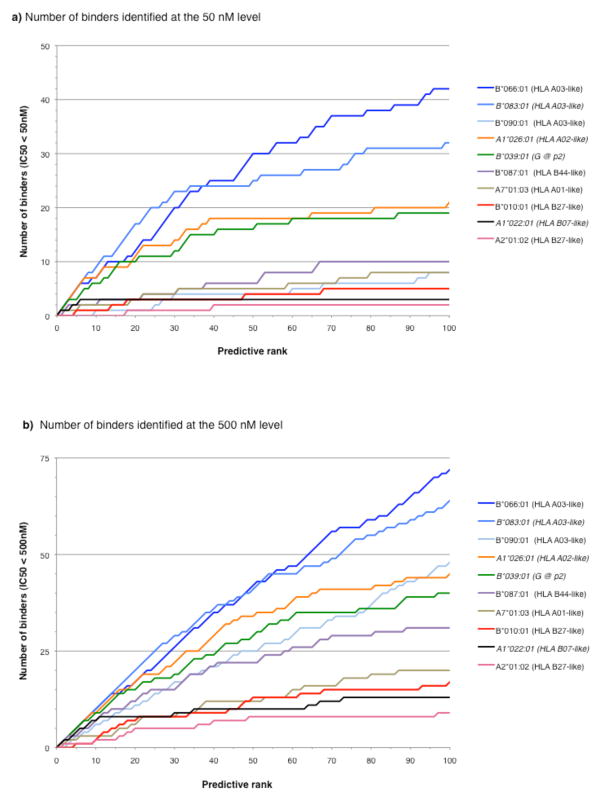
Next, we determined if the PSCL-based motifs could be used to identify MHC class I molecule-specific binding peptides. The PSCL matrices derived for each allele were used to score all 9-mer sequences in the SIVmac239 proteome, as described in the Materials and Methods. Sequences scoring in the upper 3.0% range (n=100) for each individual allele were synthesized and tested for their capacity to bind the corresponding MHC class I molecule. A 500 nM affinity threshold, described previously as being associated with immunogenicity (Allen et al. 2001; Loffredo et al. 2005; Loffredo et al. 2004; Mothe et al. 2002b; Sette et al. 1994a; Sette et al. 2005; Sette et al. 1994c; van der Most et al. 1998; Vitiello et al. 1996) was utilized to define binding. The binding capacity of the predicted peptides is summarized in Table 4. From these data, the average of the predicted peptides that bound with affinities of 50 nM or less was 12.5% (75 peptides bound out of 600), and 33% bound at the 500 nM level (197 peptides out 600). The actual values for the specific alleles varied substantially, however, with as few as 9 of the peptides binding to Mamu-A2*01:02 at the 500 nM level, and as many as 72 of the peptides binding for Mamu-B*066:01 at the 500 nM level. At the 50 nM level, the same 2 alleles represented the extremes of the variability spectrum: 2 for Mamu-A2*01:02 and 42 for Mamu-B*066:01.
Table 4.
Summary of PSCL matrices’ predictive efficiency for frequent Chinese rhesus MHC class I alleles
| Ch-MHC class I allele
|
Mamu binding affinity (IC50 nM) a
|
|||
|---|---|---|---|---|
| Allele | <50 | 50–500 | >500 | |
| Present study | A7*01:03 | 8 | 12 | 80 |
|
| ||||
| B*010:01 | 5 | 12 | 83 | |
|
| ||||
| B*066:01 | 42 | 30 | 28 | |
|
| ||||
| B*087:01 | 10 | 21 | 69 | |
|
| ||||
| A2*01:02 | 2 | 7 | 91 | |
|
| ||||
| B*090:01 | 8 | 40 | 52 | |
|
| ||||
| Peptide totals | 600 total peptides were tested | 75 | 122 | 403 |
|
| ||||
| Previous studies | A1*022:01 | 4 | 9 | 87 |
|
| ||||
| A1*026:01 | 21 | 24 | 55 | |
|
| ||||
| B*039:01 | 19 | 21 | 60 | |
|
| ||||
| B*083:01 | 32 | 32 | 36 | |
All possible 9-mer peptides in the SIVmac239 proteome were scored using the matrix values derived from the allele-appropriate PSCL analysis. The final score for each peptide represents the product of the corresponding matrix values for each peptide residue. Numbers indicate # of peptides, not percentages.
To assess whether the lower number of peptides identified for certain alleles, such as Mamu-A2*01:02 and -A1*022:01, was due to lower predictive capacity of the respective algorithms, or whether they were reflective of the inherent size of the peptide repertoire, the number of binders was plotted as a function of the prediction ranking (Fig. 3a for binders at the 50 nM level and Fig. 3b for binders at the 500 nM level). In both cases, most binders were identified in the highest-ranking predictions. Prediction efficacy leveled off after the top 50 peptides, corresponding to the top 2%. At the 500 nM level for these 2 alleles, 86% of binding peptides (19 of 22) were identified in the highest-ranking peptides (top 2%) (Fig. 3). Similarly, at the 50 nM level for the same 2 alleles, 100% of binding peptides (6 out of 6) were identified within the top 2% of predictions (Fig. 3). These data indicate that the lower number of peptides identified for Mamu-A2*01:02 and -A1*022:01 was not due to a limited predictive capacity of the algorithms, but rather to the smaller size of the available peptide repertoire.
Fig. 3. Different Chinese rhesus macaque MHC class I alleles are associated with different peptide binding repertoire sizes.
For each of the Chinese rhesus MHC class I molecules examined in the present study, as well as several others previously characterized, the cumulative number of binders identified from the predicted top 3% SIV-derived peptides was plotted as a function of the prediction ranking. Shown is the cumulative number of peptides binding with affinities of a) 50 nM or better, or b) 500 nM or better. Alleles characterized in previous studies are indicated by italicized font. The HLA supertype associated with each allele is indicated; G@2 indicates an allele that was associated with a unique motif requiring the presence of glycine at the second position (Sette et al. 2012).
Comparing data generated for the Chinese rhesus alleles characterized herein to previous studies utilizing the same methodology (italicized in the legend of Fig. 3), a similar variation in peptide binding repertoire was noted (Fig. 3). The two of the three alleles associated with lower repertoire size (Mamu-A2*01:02 and -B*010:01) are also the ones associated with HLA B27-like motifs. Conversely, Mamu-B*066:01, -B*083:01, and -B*090:01, which were associated with higher frequencies of binders with affinities <500 nM (Fig. 3b), were all associated with HLA A3-like motifs. Specifically, Mamu-B*090:01 is not one of the best MHC class I molecules for binding predicted peptides as it had only 8 binders <50nM (Table 4). But, as shown in Fig. 3b, we identified an additional 40 binders between 50–500nM, for a total of 48 binders. This total places Mamu-B*090:01 third overall in its ability to bind predicted peptides.
Discussion
In this study, we report the peptide binding motifs associated with six common Chinese-origin rhesus macaque MHC class I molecules. With the characterization of these alleles along with previously characterized Chinese-origin MHC class I alleles, we have determined motifs accounting for approximately 60% of the MHC class I genes expressed in captive-bred Chinese rhesus macaques. Chinese rhesus macaques are unlike their Indian rhesus macaques in that no specific allele is present with greater than 9% phenotypic frequency (Solomon et al. 2010). Therefore, characterization of a substantial number of alleles is required to achieve appreciable coverage.
Through the process of characterizing the most common alleles, we determined that many of the MHC class I alleles in Chinese rhesus macaques are analogous to HLA supertypic alleles. In this study we determined that Mamu-A7*01:03 possesses an HLA A1-like motif, two additional macaque alleles are HLA A3-like (Mamu-B*066:01 and -B*090:01), and Mamu-B*087:01 is HLA B44-like. Additionally, both Mamu-A2*01:02 and -B*010:01 are HLA B27-like. This data should be interpreted in the context of previous studies that had characterized Mamu-A1*022:01 (HLA B7-like), -A1*026:01 (HLA A2-like), and -B*083:01 (HLA A3-like) (Solomon et al. 2010; Southwood et al. 2011).
The identification of Mamu-A2*01:02 and -B*010:01 as having HLA B27-like motifs is striking, given the previously described association, in Indian macaques, of the HLA B27-like alleles Mamu-B*003:01 and -B*008:01 (Loffredo et al. 2009) with elite controller status. Even in HIV infection, it is true that some of the most protective alleles have HLA-B27 supertype properties (i.e. HLA-B*27:05), it is not true for all of them (example B*15:03) (McMichael and Jones 2010). In the context of the Chinese macaque HLA B27-like alleles, while the motifs of these various alleles are similar, the exact epitopes identified in each context do not appear to crossreact between the human and macaque MHC class I molecules. It remains to be determined whether Chinese macaque HLA B27-like alleles confer protection in the context of SIV infection. It is possible that other alleles have a protective role in the context of Chinese rhesus macaque SIV infection. Indeed, in HIV infection, HLA-B*27:05 is not the only protective allele, as other alleles such as HLA-B*57, have been associated with long-term non-progression of the disease (Chen et al. 2012; Matthews et al. 2012). While the focus of the present study has been on MHC class I alleles, it is also possible that other genes play a significant role in disease progression.
A total of 11 out of 12 of Chinese macaque MHC class I molecules studied to date appear homologous to HLA supertype molecules. By comparison, only 7 out of 13 of Indian macaque MHC class I molecules studied to date appear homologous to HLA supertype molecules. Thus, it is possible to hypothesize that the MHC:peptide binding properties of Chinese rhesus macaque MHC class I alleles are more similar to the features of human MHC class I molecules. If this is indeed the case, this would suggest that they may represent a better model of HIV infection than the more extensively studied Indian macaques. However, before reaching this conclusion, several factors need to be considered.
First, there are several alleles that are present in both Indian and Chinese rhesus macaques that are of interest from an HLA analogy perspective. Specifically, Mamu- B*001:01:02 and -B*003:01 are present in both sets of animals with somewhat different phenotypic frequencies. Mamu-B*001:01:02 is present in 5.8% of Chinese rhesus macaques (Solomon et al. 2010), but 26% of Indian rhesus macaques (Loffredo et al. 2005). This allele is of further interest in that it does not seem to present peptides from SIV in Indian rhesus macaques (Loffredo et al. 2005). Mamu-B*003:01, an HLA B27-like allele, is also present in 5.8% of Chinese rhesus macaques (Solomon et al. 2010), yet is only present in 0.7% of Indian rhesus macaques (Kaizu et al. 2007). These allele frequency differences can partially be explained by the continued importation of Chinese rhesus macaques and the ban on importation of Indian rhesus macaques (Southwick and Siddiqi 1988). A further consideration is that in Indian rhesus macaques there are several alleles which are expressed at higher frequencies, such as Mamu-A1*001:01 [22.1%;(Kaizu et al. 2007)] and Mamu-A1*002:01 [20.2%;(Kaizu et al. 2007)]. These two alleles, combined with Mamu-B*001:01:02, illustrate that in Indian rhesus macaques frequencies for specific alleles are much higher than any allele identified in Chinese rhesus macaques thus far. Nevertheless, allele selection for specific studies needs to surpass solely phenotypic frequencies and extend into functional properties such as epitope presentation and/or role in disease. Hence, more functional studies directly addressing potential crossreactivity are clearly necessary required.
While our results indicate a higher incidence of MHC alleles with HLA supertype characteristics in the Chinese macaque population, compared to the Indian population, this observation should be interpreted with caution. First, based on the limited data available the trend is just approaching statistical significance (exact Fisher test p value of 0.07). Furthermore, most alleles of the Mamu-A2 locus likely share the same characteristics as Mamu-A2*01:02, as this locus is oligomorphic. In fact, Mamu-A2*01:01 from Indian rhesus macaques possess the same amino acid sequence as Mamu-A2*01:02 throughout the alpha 1, 2 and 3 extracellular domains (Robinson et al. 2013). As ~60% of Indian rhesus macaques possess at least one haplotype carrying the Mamu-A2 locus, the HLA-B27 supertype property should be rather frequent among Indian rhesus macaques. Indeed, several HLA supertypes present in the Chinese rhesus macaque population that were not also observed in the Indian population, may have been missed in the latter group solely because of the limited number of alleles characterized to date. However, since the alleles studied were selected on the basis of phenotypic frequency, the results should be representative of the most frequently encountered binding specificities. Future analyses of data relating to additional MHC class I alleles, perhaps using hierarchical clustering analyses (Greenbaum et al. 2011), or alternative prediction strategies probing MHC:peptide specificities (Hoof et al. 2009) will address these issues in more detail.
Our study also provides predictive algorithms to assist in the identification of epitopes restricted by MHC class I molecules expressed in Chinese rhesus macaques. The method utilized, based on PSCL matrices, was previously successfully applied to the definition of quantitative motifs for Mamu-A1*022:01, -A1*026:01, -B*083:01, and -B*039:01 (Sette et al. 2012; Solomon et al. 2010; Southwood et al. 2011). In addition, we have identified SIV-derived binding peptides for a total of 10 different Chinese rhesus MHC macaque class I alleles. We anticipate that these algorithms and binding data will be made available in the IEDB database and analysis resource [http://tools.iedb.org; (Zhang et al. 2008)].
The data presented also highlight a significant variation in the peptide-binding repertoire associated with different allelic molecules. The basis of this phenomenon is presently unclear. However, it seems that the type of motif associated with a given allele might play a role in determining its corresponding repertoire size. For example, Mamu-B*066:01, -B*083:01, and -B*090:01, all three of which are associated with the HLA-A3 supertype, have relatively large peptide binding repertoires, as compared to Mamu-A2*01:02 and -B*010:01, which are associated with an HLA-B27-like motif. This phenomenon may be related to the relative rarity of the amino acids comprising the corresponding motifs. For example, alleles which require the presence of relatively infrequent residues in anchor positions would be expected to have a more limited repertoire of peptides than those alleles having a less restrictive chemical specificity. The former case might be exemplified by HLA-B7, which has a narrow position 2 main anchor specificity for P, which has an amino acid frequency of about 6% in naturally occurring protein sequences (Tsuji et al. 2010). The latter case may be exemplified by alleles in the HLA A3 supertype, where K, R, Y and H, with a collective frequency in proteins of about 17%, are all tolerated at the C-terminal main anchor position. Future studies will address whether a similar variation in repertoire size is a general phenomenon, and is associated with MHC class I molecules expressed in humans and other species.
Regardless of the evolutionary ramifications of MHC polymorphism and function, our findings have important practical implications because of the potential role of Chinese-origin rhesus macaques in biomedical research. The presence of key HLA-like specificities in the Chinese rhesus population provides the scientific community valuable tools, allowing evaluation of disease pathogenesis and vaccine concepts, with consideration for broader human population coverage implications.
Supplementary Material
Various peptide ligands were used as radiolabel probes in direct binding MHC dose titration experiments to ascertain binding potential to purified MHC class I molecules. The optimal radiolabeled identified for each assay was: SHSHVGYTL for Mamu-A2*01:02, RAEDNADYL for Mamu-A7*01:03, YFAIAENESK for Mamu-B*066:01, MSAPPAEYK for Mamu-B*090:01, SDIDGDRYV for Mamu-B*087:01, and SHIDRVYTL for Mamu-B*010:01. Peptides identified as high affinity radiolabeled ligands for various other assays were utilized as negative controls (as indicated) to demonstrate assay specificity.
Acknowledgments
This research is supported by the National Institutes of Health: R01 AI070902-01A2 (AS and BRM), and R15 AI064175-01 (BRM). We would also like to thank Dr. Preston Marx from Tulane University for helping us acquire the samples and virological data for the non-human primates. Support for specimens from non-human primates at the Tulane National Primate Research Center was provided by the Office of Research Infrastructure Programs (ORIP) of the National Institutes of Health through Grant Number P51 RR00164-50.
References cited
- Allen TM, Mothe BR, Sidney J, Jing P, Dzuris JL, Liebl ME, Vogel TU, O’Connor DH, Wang X, Wussow MC, Thomson JA, Altman JD, Watkins DI, Sette A. CD8(+) lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule mamu-A*01: implications for vaccine design and testing. J Virol. 2001;75:738–49. doi: 10.1128/JVI.75.2.738-749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic acids research. 2000;28:45–8. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoni R, Sette A, Sidney J, Guidotti LG, Shapiro M, Purcell R, Chisari FV. Human class I supertypes and CTL repertoires extend to chimpanzees. Journal of immunology. 1998;161:4447–55. [PubMed] [Google Scholar]
- Campillo-Gimenez L, Laforge M, Fay M, Brussel A, Cumont MC, Monceaux V, Diop O, Levy Y, Hurtrel B, Zaunders J, Corbeil J, Elbim C, Estaquier J. Nonpathogenesis of simian immunodeficiency virus infection is associated with reduced inflammation and recruitment of plasmacytoid dendritic cells to lymph nodes, not to lack of an interferon type I response, during the acute phase. Journal of virology. 2010;84:1838–46. doi: 10.1128/JVI.01496-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GL, Lau YF, Lamirande EW, McCall AW, Subbarao K. Seasonal influenza infection and live vaccine prime for a response to the 2009 pandemic H1N1 vaccine. Proc Natl Acad Sci U S A. 108:1140–5. doi: 10.1073/pnas.1009908108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Ndhlovu ZM, Liu D, Porter LC, Fang JW, Darko S, Brockman MA, Miura T, Brumme ZL, Schneidewind A, Piechocka-Trocha A, Cesa KT, Sela J, Cung TD, Toth I, Pereyra F, Yu XG, Douek DC, Kaufmann DE, Allen TM, Walker BD. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nature immunology. 2012;13:691–700. doi: 10.1038/ni.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZW, Shen L, Miller MD, Ghim SH, Hughes AL, Letvin NL. Cytotoxic T lymphocytes do not appear to select for mutations in an immunodominant epitope of simian immunodeficiency virus gag. Journal of immunology. 1992;149:4060–6. [PubMed] [Google Scholar]
- Dzuris JL, Sidney J, Appella E, Chesnut RW, Watkins DI, Sette A. Conserved MHC class I peptide binding motif between humans and rhesus macaques. J Immunol. 2000;164:283–91. doi: 10.4049/jimmunol.164.1.283. [DOI] [PubMed] [Google Scholar]
- Earley E, Mullen C, Dunyach J, Syka JEP, Compton P, Shabanowitz J, Hunt DF. Implementation of a Glow Discharge Reagent Ion Source for the Introduction of ETD Reagent Anions into a Mass Spectrometer. 58th ASMS Conference on Mass Spectrometry.2010. [Google Scholar]
- Gardner MB, Luciw PA. Macaque models of human infectious disease. ILAR J. 2008;49:220–55. doi: 10.1093/ilar.49.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. Journal of proteome research. 2004;3:958–64. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- Giraldo-Vela JP, Rudersdorf R, Chung C, Qi Y, Wallace LT, Bimber B, Borchardt GJ, Fisk DL, Glidden CE, Loffredo JT, Piaskowski SM, Furlott JR, Morales-Martinez JP, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI. The major histocompatibility complex class II alleles Mamu-DRB1*1003 and -DRB1*0306 are enriched in a cohort of simian immunodeficiency virus-infected rhesus macaque elite controllers. Journal of virology. 2008;82:859–70. doi: 10.1128/JVI.01816-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum J, Sidney J, Chung J, Brander C, Peters B, Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–35. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulukota K, Sidney J, Sette A, DeLisi C. Two complementary methods for predicting peptides binding major histocompatibility complex molecules. J Mol Biol. 1997;267:1258–67. doi: 10.1006/jmbi.1997.0937. [DOI] [PubMed] [Google Scholar]
- Hickman-Miller HD, Bardet W, Gilb A, Luis AD, Jackson KW, Watkins DI, Hildebrand WH. Rhesus macaque MHC class I molecules present HLA-B-like peptides. J Immunol. 2005;175:367–75. doi: 10.4049/jimmunol.175.1.367. [DOI] [PubMed] [Google Scholar]
- Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, Nielsen M. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N, D’Souza C, Cheung H, Lang H, Cheuk E, Chamberlain JW. Highly conserved pattern of recognition of influenza A wild-type and variant CD8+ CTL epitopes in HLA-A2+ humans and transgenic HLA-A2+/H2 class I-deficient mice. Vaccine. 2005;23:5231–44. doi: 10.1016/j.vaccine.2005.07.032. [DOI] [PubMed] [Google Scholar]
- Joag SV, Stephens EB, Adams RJ, Foresman L, Narayan O. Pathogenesis of SIVmac infection in Chinese and Indian rhesus macaques: effects of splenectomy on virus burden. Virology. 1994;200:436–46. doi: 10.1006/viro.1994.1207. [DOI] [PubMed] [Google Scholar]
- Kaizu M, Borchardt G, Glidden C, Fisk D, Loffredo J, Watkins D, Rehrauer W. Molecular typing of major histocompatibility complex class I alleles in the Indian rhesus macaque which restrict SIV CD8+ T cell epitopes. Immunogenetics. 2007;59:693–703. doi: 10.1007/s00251-007-0233-7. [DOI] [PubMed] [Google Scholar]
- Kanthaswamy S, Capitanio JP, Dubay CJ, Ferguson B, Folks T, Ha JC, Hotchkiss CE, Johnson ZP, Katze MG, Kean LS, Kubisch HM, Lank S, Lyons LA, Miller GM, Nylander J, O’Connor DH, Palermo RE, Smith DG, Vallender EJ, Wiseman RW, Rogers J. Resources for genetic management and genomics research on non-human primates at the National Primate Research Centers (NPRCs) J Med Primatol. 2009a;38(Suppl 1):17–23. doi: 10.1111/j.1600-0684.2009.00371.x. [DOI] [PubMed] [Google Scholar]
- Kanthaswamy S, Gill L, Satkoski J, Goyal V, Malladi V, Kou A, Basuta K, Sarkisyan L, George D, Smith DG. Development of a Chinese-Indian hybrid (Chindian) rhesus macaque colony at the California National Primate Research Center by introgression. J Med Primatol. 2009b;38:86–96. doi: 10.1111/j.1600-0684.2008.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kestler H, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N, et al. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science. 1990;248:1109–12. doi: 10.1126/science.2160735. [DOI] [PubMed] [Google Scholar]
- Kondo A, Sidney J, Southwood S, del Guercio MF, Appella E, Sakamoto H, Grey HM, Celis E, Chesnut RW, Kubo RT, Sette A. Two distinct HLA-A*0101-specific submotifs illustrate alternative peptide binding modes. Immunogenetics. 1997;45:249–58. doi: 10.1007/s002510050200. [DOI] [PubMed] [Google Scholar]
- Kubo RT, Sette A, Grey HM, Appella E, Sakaguchi K, Zhu NZ, Arnott D, Sherman N, Shabanowitz J, Michel H, et al. Definition of specific peptide motifs for four major HLA-A alleles. J Immunol. 1994;152:3913–24. [PubMed] [Google Scholar]
- Kuroda MJ, Schmitz JE, Barouch DH, Craiu A, Allen TM, Sette A, Watkins DI, Forman MA, Letvin NL. Analysis of Gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I-peptide complex. J Exp Med. 1998;187:1373–81. doi: 10.1084/jem.187.9.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letvin NL, Miller MD, Shen L, Chen ZW, Yasutomi Y. Simian immunodeficiency virus-specific cytotoxic T lymphocytes in rhesus monkeys: characterization and vaccine induction. Seminars in immunology. 1993;5:215–23. doi: 10.1006/smim.1993.1025. [DOI] [PubMed] [Google Scholar]
- Li A, Wang X, Liu Y, Zhao Y, Liu B, Sui L, Zeng L, Sun Z. Preliminary observations of MHC class I A region polymorphism in three populations of Chinese-origin rhesus macaques. Immunogenetics. 2012;64:887–94. doi: 10.1007/s00251-012-0645-x. [DOI] [PubMed] [Google Scholar]
- Ling B, Veazey RS, Luckay A, Penedo C, Xu K, Lifson JD, Marx PA. SIV(mac) pathogenesis in rhesus macaques of Chinese and Indian origin compared with primary HIV infections in humans. AIDS. 2002;16:1489–96. doi: 10.1097/00002030-200207260-00005. [DOI] [PubMed] [Google Scholar]
- Loffredo JT, Bean AT, Beal DR, Leon EJ, May GE, Piaskowski SM, Furlott JR, Reed J, Musani SK, Rakasz EG, Friedrich TC, Wilson NA, Allison DB, Watkins DI. Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J Virol. 2008;82:1723–38. doi: 10.1128/JVI.02084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo JT, Burwitz BJ, Rakasz EG, Spencer SP, Stephany JJ, Vela JP, Martin SR, Reed J, Piaskowski SM, Furlott J, Weisgrau KL, Rodrigues DS, Soma T, Napoe G, Friedrich TC, Wilson NA, Kallas EG, Watkins DI. The antiviral efficacy of simian immunodeficiency virus-specific CD8+ T cells is unrelated to epitope specificity and is abrogated by viral escape. Journal of virology. 2007a;81:2624–34. doi: 10.1128/JVI.01912-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo JT, Friedrich TC, Leon EJ, Stephany JJ, Rodrigues DS, Spencer SP, Bean AT, Beal DR, Burwitz BJ, Rudersdorf RA, Wallace LT, Piaskowski SM, May GE, Sidney J, Gostick E, Wilson NA, Price DA, Kallas EG, Piontkivska H, Hughes AL, Sette A, Watkins DI. CD8+ T cells from SIV elite controller macaques recognize Mamu-B*08-bound epitopes and select for widespread viral variation. PLoS One. 2007b;2:e1152. doi: 10.1371/journal.pone.0001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, Bean AT, Beal DR, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol. 2007c;81:8827–32. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo JT, Sidney J, Bean AT, Beal DR, Bardet W, Wahl A, Hawkins OE, Piaskowski S, Wilson NA, Hildebrand WH, Watkins DI, Sette A. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J Immunol. 2009;182:7763–75. doi: 10.4049/jimmunol.0900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo JT, Sidney J, Piaskowski S, Szymanski A, Furlott J, Rudersdorf R, Reed J, Peters B, Hickman-Miller HD, Bardet W, Rehrauer WM, O’Connor DH, Wilson NA, Hildebrand WH, Sette A, Watkins DI. The high frequency Indian rhesus macaque MHC class I molecule, Mamu-B*01, does not appear to be involved in CD8+ T lymphocyte responses to SIVmac239. J Immunol. 2005;175:5986–97. doi: 10.4049/jimmunol.175.9.5986. [DOI] [PubMed] [Google Scholar]
- Loffredo JT, Sidney J, Wojewoda C, Dodds E, Reynolds MR, Napoe G, Mothe BR, O’Connor DH, Wilson NA, Watkins DI, Sette A. Identification of seventeen new simian immunodeficiency virus-derived CD8+ T cell epitopes restricted by the high frequency molecule, Mamu-A*02, and potential escape from CTL recognition. J Immunol. 2004;173:5064–76. doi: 10.4049/jimmunol.173.8.5064. [DOI] [PubMed] [Google Scholar]
- Lund O, Nielsen M, Kesmir C, Petersen AG, Lundegaard C, Worning P, Sylvester-Hvid C, Lamberth K, Roder G, Justesen S, Buus S, Brunak S. Definition of supertypes for HLA molecules using clustering of specificity matrices. Immunogenetics. 2004;55:797–810. doi: 10.1007/s00251-004-0647-4. [DOI] [PubMed] [Google Scholar]
- Maness NJ, Walsh AD, Rudersdorf RA, Erickson PA, Piaskowski SM, Wilson NA, Watkins DI. Chinese origin rhesus macaque major histocompatibility complex class I molecules promiscuously present epitopes from SIV associated with molecules of Indian origin; implications for immunodominance and viral escape. Immunogenetics. 2011;63:587–97. doi: 10.1007/s00251-011-0538-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews PC, Listgarten J, Carlson JM, Payne R, Huang KH, Frater J, Goedhals D, Steyn D, van Vuuren C, Paioni P, Jooste P, Ogwu A, Shapiro R, Mncube Z, Ndung’u T, Walker BD, Heckerman D, Goulder PJ. Co-operative additive effects between HLA alleles in control of HIV-1. PLoS One. 2012;7:e47799. doi: 10.1371/journal.pone.0047799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney DM, Erickson AL, Walker CM, Thimme R, Chisari FV, Sidney J, Sette A. Identification of five different Patr class I molecules that bind HLA supertype peptides and definition of their peptide binding motifs. J Immunol. 2000;165:4414–22. doi: 10.4049/jimmunol.165.8.4414. [DOI] [PubMed] [Google Scholar]
- McMichael AJ, Jones EY. Genetics. First-class control of HIV-1. Science. 2010;330:1488–90. doi: 10.1126/science.1200035. [DOI] [PubMed] [Google Scholar]
- Miller CJ, Alexander NJ, Sutjipto S, Lackner AA, Gettie A, Hendrickx AG, Lowenstine LJ, Jennings M, Marx PA. Genital mucosal transmission of simian immunodeficiency virus: animal model for heterosexual transmission of human immunodeficiency virus. J Virol. 1989;63:4277–84. doi: 10.1128/jvi.63.10.4277-4284.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MD, Yamamoto H, Hughes AL, Watkins DI, Letvin NL. Definition of an epitope and MHC class I molecule recognized by gag-specific cytotoxic T lymphocytes in SIVmac-infected rhesus monkeys. Journal of immunology. 1991;147:320–9. [PubMed] [Google Scholar]
- Mothe BR, Horton H, Carter DK, Allen TM, Liebl ME, Skinner P, Vogel TU, Fuenger S, Vielhuber K, Rehrauer W, Wilson N, Franchini G, Altman JD, Haase A, Picker LJ, Allison DB, Watkins DI. Dominance of CD8 responses specific for epitopes bound by a single major histocompatibility complex class I molecule during the acute phase of viral infection. Journal of virology. 2002a;76:875–84. doi: 10.1128/JVI.76.2.875-884.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothe BR, Sidney J, Dzuris JL, Liebl ME, Fuenger S, Watkins DI, Sette A. Characterization of the peptide-binding specificity of Mamu-B*17 and identification of Mamu-B*17-restricted epitopes derived from simian immunodeficiency virus proteins. Journal of immunology. 2002b;169:210–9. doi: 10.4049/jimmunol.169.1.210. [DOI] [PubMed] [Google Scholar]
- Mudd PA, Martins MA, Ericsen AJ, Tully DC, Power KA, Bean AT, Piaskowski SM, Duan L, Seese A, Gladden AD, Weisgrau KL, Furlott JR, Kim YI, Veloso de Santana MG, Rakasz E, Capuano S, 3rd, Wilson NA, Bonaldo MC, Galler R, Allison DB, Piatak M, Jr, Haase AT, Lifson JD, Allen TM, Watkins DI. Vaccine-induced CD8+ T cells control AIDS virus replication. Nature. 2012;491:129–33. doi: 10.1038/nature11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang D, Xu L, Dai Z, Shi H, Zhang G, Zheng Y, He X. Identification of major histocompatibility complex class I alleles in Chinese rhesus macaques. Acta biochimica et biophysica Sinica. 2008;40:919–27. doi: 10.1111/j.1745-7270.2008.00474.x. [DOI] [PubMed] [Google Scholar]
- Ouyang DY, Xu LH, Shi HJ, Zheng YT, He XH. Eight novel MHC class I alleles identified in Chinese-origin rhesus macaques. Tissue antigens. 2009;73:285–7. doi: 10.1111/j.1399-0039.2008.01198.x. [DOI] [PubMed] [Google Scholar]
- Pal R, Venzon D, Letvin NL, Santra S, Montefiori DC, Miller NR, Tryniszewska E, Lewis MG, VanCott TC, Hirsch V, Woodward R, Gibson A, Grace M, Dobratz E, Markham PD, Hel Z, Nacsa J, Klein M, Tartaglia J, Franchini G. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. Journal of virology. 2002;76:292–302. doi: 10.1128/JVI.76.1.292-302.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–14. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- Pinilla C, Martin R, Gran B, Appel JR, Boggiano C, Wilson DB, Houghten RA. Exploring immunological specificity using synthetic peptide combinatorial libraries. Curr Opin Immunol. 1999;11:193–202. doi: 10.1016/s0952-7915(99)80033-8. [DOI] [PubMed] [Google Scholar]
- Reed JS, Sidney J, Piaskowski SM, Glidden CE, Leon EJ, Burwitz BJ, Kolar HL, Eernisse CM, Furlott JR, Maness NJ, Walsh AD, Rudersdorf RA, Bardet W, McMurtrey CP, O’Connor DH, Hildebrand WH, Sette A, Watkins DI, Wilson NA. The role of MHC class I allele Mamu-A*07 during SIV(mac)239 infection. Immunogenetics. 2011;63:789–807. doi: 10.1007/s00251-011-0541-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Waller MJ, Stroehr P, Marsh SGE. IPD-the Immuno Polymorphism Database. Nucleic Acids Research. 2013 doi: 10.1093/nar/gki032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidewind A, Brockman MA, Sidney J, Wang YE, Chen H, Suscovich TJ, Li B, Adam RI, Allgaier RL, Mothe BR, Kuntzen T, Oniangue-Ndza C, Trocha A, Yu XG, Brander C, Sette A, Walker BD, Allen TM. Structural and functional constraints limit options for cytotoxic T-lymphocyte escape in the immunodominant HLA-B27-restricted epitope in human immunodeficiency virus type 1 capsid. J Virol. 2008;82:5594–605. doi: 10.1128/JVI.02356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A, Alexander J, Ruppert J, Snoke K, Franco A, Ishioka G, Grey HM. Antigen analogs/MHC complexes as specific T cell receptor antagonists. Annu Rev Immunol. 1994a;12:413–31. doi: 10.1146/annurev.iy.12.040194.002213. [DOI] [PubMed] [Google Scholar]
- Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50:201–12. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- Sette A, Sidney J, Bui HH, Del Guercio MF, Alexander J, Loffredo J, Watkins DI, Mothe BR. Characterization of the peptide-binding specificity of Mamu-A*11 results in the identification of SIV-derived epitopes and interspecies cross-reactivity. Immunogenetics. 2005 doi: 10.1007/s00251-004-0749-z. [DOI] [PubMed] [Google Scholar]
- Sette A, Sidney J, del Guercio MF, Southwood S, Ruppert J, Dahlberg C, Grey HM, Kubo RT. Peptide binding to the most frequent HLA-A class I alleles measured by quantitative molecular binding assays. Mol Immunol. 1994b;31:813–22. doi: 10.1016/0161-5890(94)90019-1. [DOI] [PubMed] [Google Scholar]
- Sette A, Sidney J, Southwood S, Moore C, Berry J, Dow C, Bradley K, Hoof I, Lewis MG, Hildebrand WH, McMurtrey CP, Wilson NA, Watkins DI, Mothe BR. A shared MHC supertype motif emerges by convergent evolution in macaques and mice, but is totally absent in human MHC molecules. Immunogenetics. 2012 doi: 10.1007/s00251-011-0598-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A, Vitiello A, Reherman B, Fowler P, Nayersina R, Kast WM, Melief CJ, Oseroff C, Yuan L, Ruppert J, et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J Immunol. 1994c;153:5586–92. [PubMed] [Google Scholar]
- Sette AD, Oseroff C, Sidney J, Alexander J, Chesnut RW, Kakimi K, Guidotti LG, Chisari FV. Overcoming T cell tolerance to the hepatitis B virus surface antigen in hepatitis B virus-transgenic mice. Journal of immunology. 2001;166:1389–97. doi: 10.4049/jimmunol.166.2.1389. [DOI] [PubMed] [Google Scholar]
- Sidney J, Asabe S, Peters B, Purton KA, Chung J, Pencille TJ, Purcell R, Walker CM, Chisari FV, Sette A. Detailed characterization of the peptide binding specificity of five common Patr class I MHC molecules. Immunogenetics. 2006;58:559–70. doi: 10.1007/s00251-006-0131-4. [DOI] [PubMed] [Google Scholar]
- Sidney J, Assarsson E, Moore C, Ngo S, Pinilla C, Sette A, Peters B. Quantitative peptide binding motifs for 19 human and mouse MHC class I molecules derived using positional scanning combinatorial peptide libraries. Immunome Res. 2008;4:2. doi: 10.1186/1745-7580-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, del Guercio MF, Southwood S, Engelhard VH, Appella E, Rammensee HG, Falk K, Rotzschke O, Takiguchi M, Kubo RT, et al. Several HLA alleles share overlapping peptide specificities. Journal of immunology. 1995;154:247–59. [PubMed] [Google Scholar]
- Sidney J, Dzuris JL, Newman MJ, Johnson RP, Kaur A, Amitinder K, Walker CM, Appella E, Mothe B, Watkins DI, Sette A. Definition of the Mamu A*01 peptide binding specificity: application to the identification of wild-type and optimized ligands from simian immunodeficiency virus regulatory proteins. Journal of immunology. 2000;165:6387–99. doi: 10.4049/jimmunol.165.11.6387. [DOI] [PubMed] [Google Scholar]
- Sidney J, Grey HM, Southwood S, Celis E, Wentworth PA, del Guercio MF, Kubo RT, Chesnut RW, Sette A. Definition of an HLA-A3-like supermotif demonstrates the overlapping peptide-binding repertoires of common HLA molecules. Hum Immunol. 1996;45:79–93. doi: 10.1016/0198-8859(95)00173-5. [DOI] [PubMed] [Google Scholar]
- Sidney J, Southwood S, Mann DL, Fernandez-Vina MA, Newman MJ, Sette A. Majority of peptides binding HLA-A*0201 with high affinity crossreact with other A2-supertype molecules. Hum Immunol. 2001a;62:1200–16. doi: 10.1016/s0198-8859(01)00319-6. [DOI] [PubMed] [Google Scholar]
- Sidney J, Southwood S, Oseroff C, del Guercio M-F, Sette A, Grey HM. Current Protocols in Immunology. John Wiley & Sons, Inc; 2001b. Measurement of MHC/Peptide Interactions by Gel Filtration. [DOI] [PubMed] [Google Scholar]
- Sidney J, Southwood S, Pasquetto V, Sette A. Simultaneous prediction of binding capacity for multiple molecules of the HLA B44-supertype. Journal of immunology. 2003;171:5964–5974. doi: 10.4049/jimmunol.171.11.5964. [DOI] [PubMed] [Google Scholar]
- Sidney J, Southwood S, Sette A. Classification of A1- and A24-supertype molecules by analysis of their MHC-peptide binding repertoires. Immunogenetics. 2005;57:393–408. doi: 10.1007/s00251-005-0004-2. [DOI] [PubMed] [Google Scholar]
- Solomon C, Southwood S, Hoof I, Rudersdorf R, Peters B, Sidney J, Pinilla C, Marcondes MC, Ling B, Marx P, Sette A, Mothe BR. The most common Chinese rhesus macaque MHC class I molecule shares peptide binding repertoire with the HLA-B7 supertype. Immunogenetics. 2010;62:451–64. doi: 10.1007/s00251-010-0450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwick CH, Siddiqi MF. Partial recovery and a new population estimate of rhesus monkey populations in India. Am J Primatol. 1988;16:187–197. doi: 10.1002/ajp.1350160302. [DOI] [PubMed] [Google Scholar]
- Southwood S, Solomon C, Hoof I, Rudersdorf R, Sidney J, Peters B, Wahl A, Hawkins O, Hildebrand W, Mothe BR, Sette A. Functional analysis of frequently expressed Chinese rhesus macaque MHC class I molecules Mamu-A1*02601 and Mamu-B*08301 reveals HLA-A2 and HLA-A3 supertypic specificities. Immunogenetics. 2011;63:275–90. doi: 10.1007/s00251-010-0502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji J, Nydza R, Wolcott E, Mannor E, Moran B, Hesson G, Arvidson T, Howe K, Hayes R, Ramirez M, Way M. The Frequencies of Amino Acids Encoded by Genomes that Utilize Standard and Nonstandard Genetic Codes. BIOS. 2010;81:22–31. [Google Scholar]
- Udeshi ND, Compton PD, Shabanowitz J, Hunt DF, Rose KL. Methods for analyzing peptides and proteins on a chromatographic timescale by electron-transfer dissociation mass spectrometry. Nature protocols. 2008;3:1709–17. doi: 10.1038/nprot.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Most RG, Murali-Krishna K, Whitton JL, Oseroff C, Alexander J, Southwood S, Sidney J, Chesnut RW, Sette A, Ahmed R. Identification of Db- and Kb-restricted subdominant cytotoxic T-cell responses in lymphocytic choriomeningitis virus-infected mice. Virology. 1998;240:158–67. doi: 10.1006/viro.1997.8934. [DOI] [PubMed] [Google Scholar]
- Vitiello A, Yuan L, Chesnut RW, Sidney J, Southwood S, Farness P, Jackson MR, Peterson PA, Sette A. Immunodominance analysis of CTL responses to influenza PR8 virus reveals two new dominant and subdominant Kb-restricted epitopes. J Immunol. 1996;157:5555–62. [PubMed] [Google Scholar]
- Wambua D, Henderson R, Solomon C, Hunter M, Marx P, Sette A, Mothe BR. SIV-infected Chinese-origin rhesus macaques express specific MHC class I alleles in either elite controllers or normal progressors. J Med Primatol. 2011;40:244–7. doi: 10.1111/j.1600-0684.2011.00487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman RW, Karl JA, Bimber BN, O’Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, Szekeres E, Jr, Wright C, Harkins T, O’Connor DH. Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med. 2009;15:1322–6. doi: 10.1038/nm.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia HJ, Zhang GH, Ma JP, Dai ZX, Li SY, Han JB, Zheng YT. Dendritic cell subsets dynamics and cytokine production in SIVmac239-infected Chinese rhesus macaques. Retrovirology. 2010;7:102. doi: 10.1186/1742-4690-7-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wang P, Kim Y, Haste-Andersen P, Beaver J, Bourne PE, Bui HH, Buus S, Frankild S, Greenbaum J, Lund O, Lundegaard C, Nielsen M, Ponomarenko J, Sette A, Zhu Z, Peters B. Immune epitope database analysis resource (IEDB-AR) Nucleic acids research. 2008;36:W513–8. doi: 10.1093/nar/gkn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Various peptide ligands were used as radiolabel probes in direct binding MHC dose titration experiments to ascertain binding potential to purified MHC class I molecules. The optimal radiolabeled identified for each assay was: SHSHVGYTL for Mamu-A2*01:02, RAEDNADYL for Mamu-A7*01:03, YFAIAENESK for Mamu-B*066:01, MSAPPAEYK for Mamu-B*090:01, SDIDGDRYV for Mamu-B*087:01, and SHIDRVYTL for Mamu-B*010:01. Peptides identified as high affinity radiolabeled ligands for various other assays were utilized as negative controls (as indicated) to demonstrate assay specificity.