Abstract
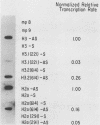
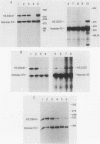
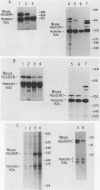
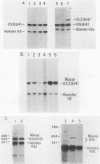
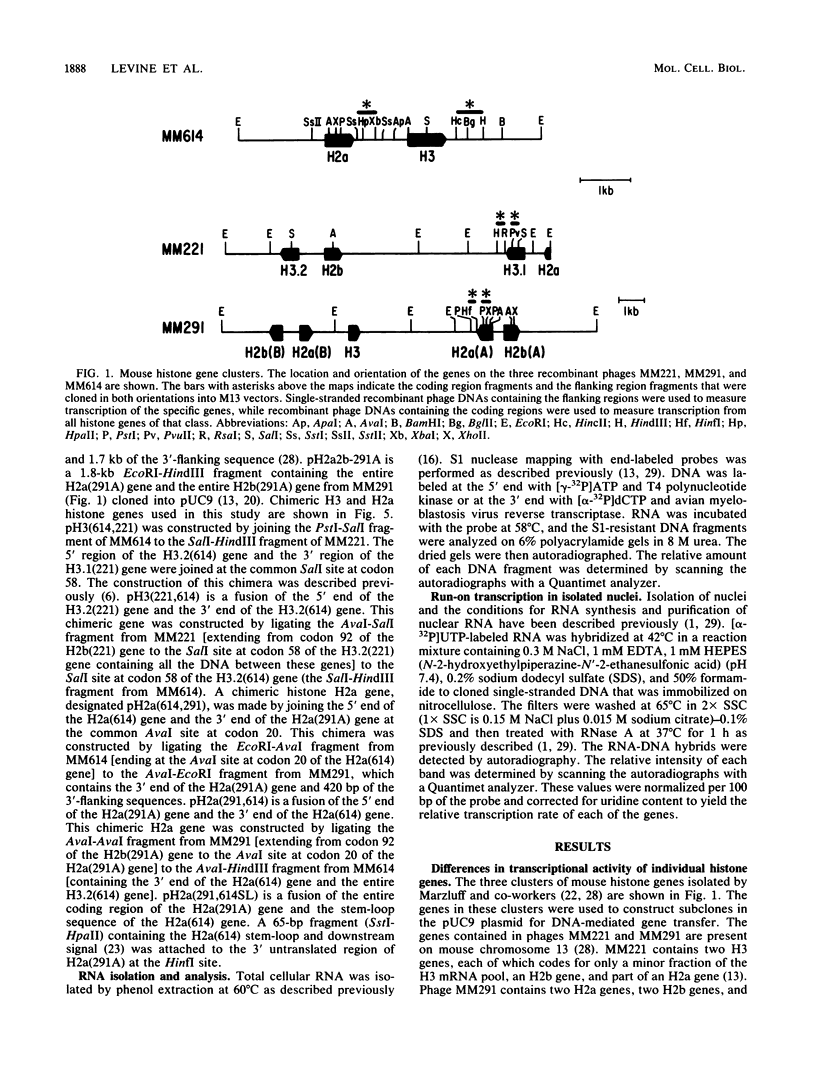
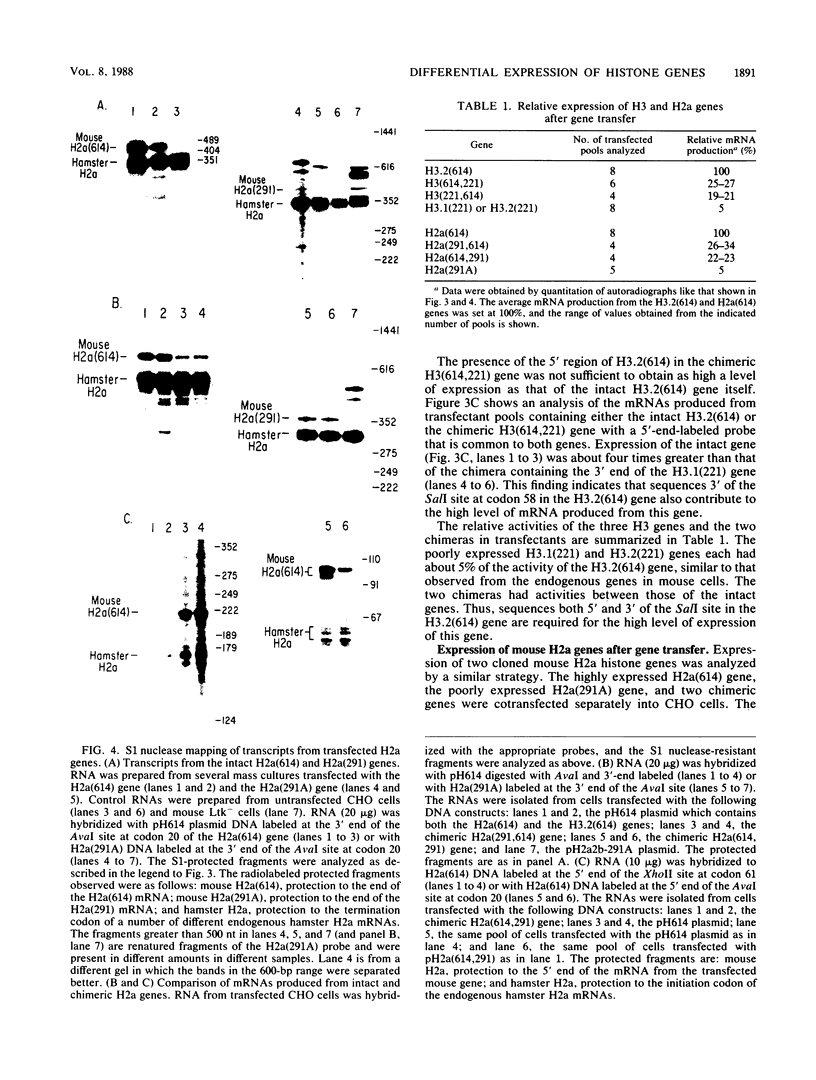
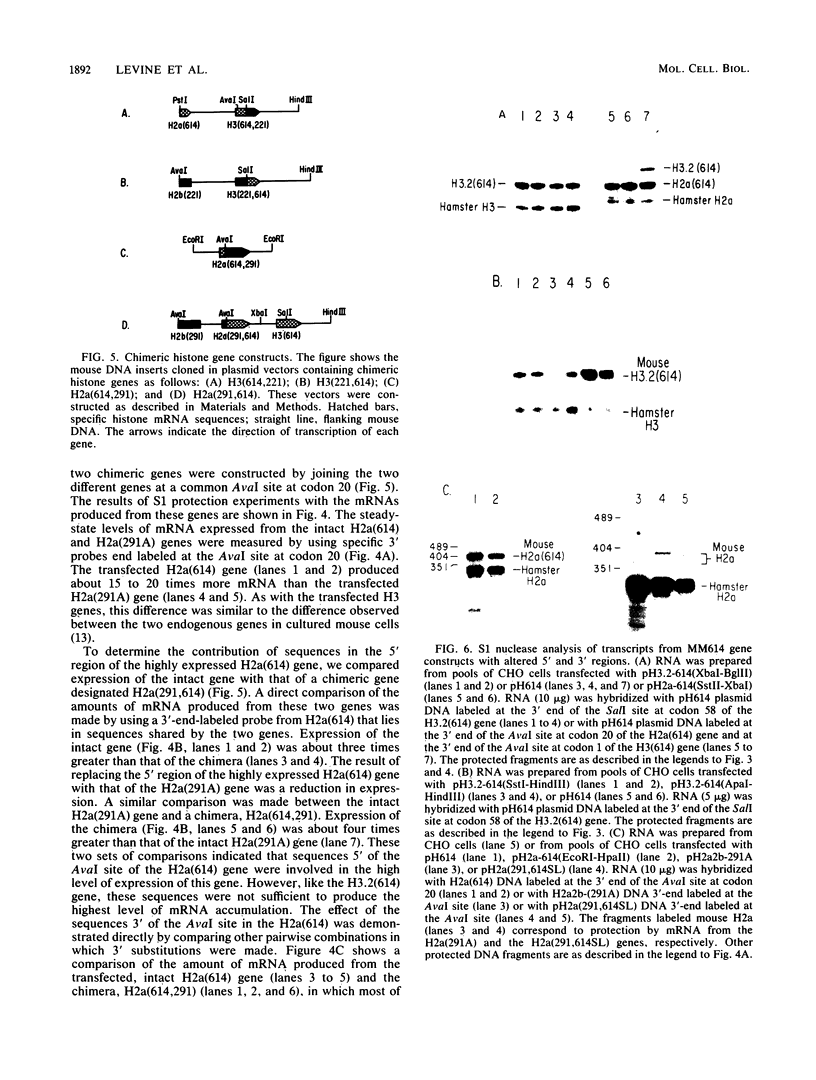
Histone proteins are encoded by a multigene family. The H3.2(614) and H2a(614) genes are present as single copies which are expressed at high levels, accounting for 30 to 40% of the H3 and H2a mRNAs, respectively, in different types of mouse cells. The other genes which have been isolated each contribute only a very small amount to the total type-specific mRNA pool. We demonstrate here that the differences in the level of expression of these genes are partly due to differences in their transcription rates. To investigate the sequences responsible for these differences in expression among the members of each family, we carried out DNA-mediated gene transfer experiments with both intact and chimeric histone genes. The 5' region of a highly expressed gene [H3.2(614) or H2a(614)] was attached to the 3' region of a histone gene which was expressed at low levels (H3-221 or H2a-291) and vice versa. The results show that sequences in both the 5' and 3' regions of the H3.2(614) and H2a(614) genes contribute to their high level of mRNA production by two independent mechanisms. The effect of the 3' sequences on mRNA accumulation has been narrowed to a 65-base-pair region including the 3'-terminal palindrome and downstream signal implicated in mRNA processing.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alterman R. B., Ganguly S., Schulze D. H., Marzluff W. F., Schildkraut C. L., Skoultchi A. I. Cell cycle regulation of mouse H3 histone mRNA metabolism. Mol Cell Biol. 1984 Jan;4(1):123–132. doi: 10.1128/mcb.4.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alterman R. B., Sprecher C., Graves R., Marzluff W. F., Skoultchi A. I. Regulated expression of a chimeric histone gene introduced into mouse fibroblasts. Mol Cell Biol. 1985 Sep;5(9):2316–2324. doi: 10.1128/mcb.5.9.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Brown D. T., Wellman S. E., Sittman D. B. Changes in the levels of three different classes of histone mRNA during murine erythroleukemia cell differentiation. Mol Cell Biol. 1985 Nov;5(11):2879–2886. doi: 10.1128/mcb.5.11.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaney W. G., Howard D. R., Pollard J. W., Sallustio S., Stanley P. High-frequency transfection of CHO cells using polybrene. Somat Cell Mol Genet. 1986 May;12(3):237–244. doi: 10.1007/BF01570782. [DOI] [PubMed] [Google Scholar]
- Chodchoy N., Levine B. J., Sprecher C., Skoultchi A. I., Marzluff W. F. Expression of mouse histone genes: transcription into 3' intergenic DNA and cryptic processing sites downstream from the 3' end of the H3 gene. Mol Cell Biol. 1987 Mar;7(3):1039–1047. doi: 10.1128/mcb.7.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea R. J., Coles L. S., Lesnikowski C., Tabe L., Wells J. R. Chromosomal organization of chicken histone genes: preferred associations and inverted duplications. Mol Cell Biol. 1985 Nov;5(11):3108–3115. doi: 10.1128/mcb.5.11.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLisle A. J., Graves R. A., Marzluff W. F., Johnson L. F. Regulation of histone mRNA production and stability in serum-stimulated mouse 3T6 fibroblasts. Mol Cell Biol. 1983 Nov;3(11):1920–1929. doi: 10.1128/mcb.3.11.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Gick O., Krämer A., Keller W., Birnstiel M. L. Generation of histone mRNA 3' ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986 Jun;5(6):1319–1326. doi: 10.1002/j.1460-2075.1986.tb04362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves R. A., Marzluff W. F., Giebelhaus D. H., Schultz G. A. Quantitative and qualitative changes in histone gene expression during early mouse embryo development. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5685–5689. doi: 10.1073/pnas.82.17.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves R. A., Marzluff W. F. Rapid reversible changes in the rate of histone gene transcription and histone mRNA levels in mouse myeloma cells. Mol Cell Biol. 1984 Feb;4(2):351–357. doi: 10.1128/mcb.4.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves R. A., Wellman S. E., Chiu I. M., Marzluff W. F. Differential expression of two clusters of mouse histone genes. J Mol Biol. 1985 May 25;183(2):179–194. doi: 10.1016/0022-2836(85)90211-6. [DOI] [PubMed] [Google Scholar]
- Heintz N., Sive H. L., Roeder R. G. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol Cell Biol. 1983 Apr;3(4):539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Hsiung N., Roginski R. S., Henthorn P., Smithies O., Kucherlapati R., Skoultchi A. I. Introduction and expression of a fetal human globin gene in mouse fibroblasts. Mol Cell Biol. 1982 Apr;2(4):401–411. doi: 10.1128/mcb.2.4.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob E. Histone-gene reiteration in the genome of mouse. Eur J Biochem. 1976 May 17;65(1):275–284. doi: 10.1111/j.1432-1033.1976.tb10415.x. [DOI] [PubMed] [Google Scholar]
- Levine B. J., Chodchoy N., Marzluff W. F., Skoultchi A. I. Coupling of replication type histone mRNA levels to DNA synthesis requires the stem-loop sequence at the 3' end of the mRNA. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6189–6193. doi: 10.1073/pnas.84.17.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher B., Schümperli D. RNA 3' processing regulates histone mRNA levels in a mammalian cell cycle mutant. A processing factor becomes limiting in G1-arrested cells. EMBO J. 1987 Jun;6(6):1721–1726. doi: 10.1002/j.1460-2075.1987.tb02423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey N. B., Marzluff W. F. The stem-loop structure at the 3' end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol. 1987 Dec;7(12):4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumb M., Stein J., Stein G. Coordinate regulation of multiple histone mRNAs during the cell cycle in HeLa cells. Nucleic Acids Res. 1983 Apr 25;11(8):2391–2410. doi: 10.1093/nar/11.8.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D. H., Parker C. S. The 3' end of drosophila histone H3 mRNA is produced by a processing activity in vitro. Cell. 1984 Sep;38(2):423–429. doi: 10.1016/0092-8674(84)90497-5. [DOI] [PubMed] [Google Scholar]
- Schümperli D. Cell-cycle regulation of histone gene expression. Cell. 1986 May 23;45(4):471–472. doi: 10.1016/0092-8674(86)90277-1. [DOI] [PubMed] [Google Scholar]
- Sierra F., Lichtler A., Marashi F., Rickles R., Van Dyke T., Clark S., Wells J., Stein G., Stein J. Organization of human histone genes. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1795–1799. doi: 10.1073/pnas.79.6.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F. Histone mRNA concentrations are regulated at the level of transcription and mRNA degradation. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1849–1853. doi: 10.1073/pnas.80.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F. Structure of a cluster of mouse histone genes. Nucleic Acids Res. 1983 Oct 11;11(19):6679–6697. doi: 10.1093/nar/11.19.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Stauber C., Lüscher B., Eckner R., Lötscher E., Schümperli D. A signal regulating mouse histone H4 mRNA levels in a mammalian cell cycle mutant and sequences controlling RNA 3' processing are both contained within the same 80-bp fragment. EMBO J. 1986 Dec 1;5(12):3297–3303. doi: 10.1002/j.1460-2075.1986.tb04643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. D., Wellman S. E., Marzluff W. F. Sequences of four mouse histone H3 genes: implications for evolution of mouse histone genes. J Mol Evol. 1986;23(3):242–249. doi: 10.1007/BF02115580. [DOI] [PubMed] [Google Scholar]