Abstract
Bacteria contain several sophisticated macromolecular machineries responsible for translocating proteins across the cell envelope. One prominent example is the type II secretion system (T2SS), which contains a large outer membrane channel, called the secretin. These gated channels require specialized proteins, so-called pilotins, to reach and assemble in the outer membrane. Here we report the crystal structure of the pilotin GspS from the T2SS of enterohemorrhagic Escherichia coli (EHEC), an important pathogen that can cause severe disease in cases of food poisoning. In this four-helix protein, the straight helix α2, the curved helix α3 and the bent helix α4 surround the central N-terminal helix α1. The helices of GspS create a prominent groove, mainly formed by side chains of helices α1, α2 and α3. In the EHEC GspS structure this groove is occupied by extra electron density which is reminiscent of an α-helix and corresponds well with a binding site observed in a homologous pilotin. The residues forming the groove are well conserved among homologs, pointing to a key role of this groove in this class of T2SS pilotins. At the same time, T2SS pilotins in different species can be entirely different in structure, and the pilotins for secretins in non-T2SS machineries have yet again unrelated folds, despite a common function. It is striking that a common complex function, such as targeting and assembling an outer membrane multimeric channel, can be performed by proteins with entirely different folds.
Keywords: type II secretion, lipoprotein, crystal structure, EtpO, secretin, Hamburger disease
1. Introduction
Enterohemorrhagic Escherichia coli O157:H7 (EHEC) is an important pathogenic E. coli strain that spreads via contaminated food sources. The bacteria colonize intestinal epithelial cells and cause hemorrhagic colitis, and the illness caused is sometimes called Hamburger Disease (Sherman et al., 2010). EHEC is especially dangerous since it may lead to a potentially lethal disease, hemolytic uremic syndrome (Melton-Celsa et al., 2012), in particular in young children. EHEC infection and damage to the host depends on a large number of virulence factors (Farfan and Torres, 2012). A major role is played by the large pO157 plasmid which encodes the type II secretion system (T2SS), a metalloprotease StcE (secreted protease of C1-esterase inhibitor), hemolysin, a subtilisin-like serine protease and other virulence factors (Burland et al., 1998). The metalloprotease StcE, which is secreted by the T2SS, is important for early steps in colonization of epithelial cells by EHEC (Grys et al., 2005; Lathem et al., 2002; Paton and Paton, 2002; Yu et al., 2012). Another known substrate of the T2SS from EHEC is a metal binding protein YodA that is also involved in colonization process via an as yet unknown mechanism (Ho et al., 2008). Furthermore, EHEC deletion mutants of the T2SS are defective in colonization in vivo, which underscores the importance of the T2SS role in the infection process (Ho et al., 2008).
The T2SS is a sophisticated multi-protein machinery that transports folded proteins from the periplasm across the outer membrane of Gram-negative bacteria into the extracellular milieu (Douzi et al., 2012; Korotkov et al., 2012; McLaughlin et al., 2012). The T2SS spans two membranes and consists of multiple copies of at least 12 different proteins. In the cytoplasm, the secretion ATPase GspE interacts with the inner membrane platform consisting of GspL, GspM, GspF and GspC. This platform interacts with GspG, which is the most abundant subunit of a helical subassembly called the pseudopilus. The outer membrane channel is formed by the secretin GspD. Secretins are also channels for secreted proteins, fimbriae or phages in a number of other systems, including the type III secretion system (T3SS), the type IV pilin system (T4PS) and the filamentous phage assembly system (Korotkov et al., 2011).
The biogenesis of secretins in the outer membrane requires in several cases lipoprotein chaperones called pilotins (Koo et al., 2012). The related T2SS secretins PulD from Klebsiella oxytoca (KoGspDPulD) and OutD from Dickeya dadantii, formerly Erwinia chrysanthemi, (DdGspDOutD) rely, respectively, on their cognate pilotins PulS (KoGspSPulS) and OutS (DdGspSOutS) for outer membrane targeting (Hardie et al., 1996; Shevchik et al., 1997). These pilotins have an outer membrane lipoprotein-sorting signal that directs them to the outer membrane via interactions with proteins of the Lol sorting pathway (Collin et al., 2011). In addition to pilotins, some secretins require additional accessory proteins for stability (Ast et al., 2002; Gauthier et al., 2003; Schuch and Maurelli, 2001; Strozen et al., 2011).
The pilotins KoGspSPulS and DdGspSOutS have been shown to interact with the C-terminal 60 residues of their secretins, the so-called S-domain, and protect secretin monomers from proteolysis (Daefler et al., 1997; Shevchik et al., 1997). However, the absence of pilotin or deletion of S-domain does not prevent the multimerization of KoGspDPulD and DdGspDOutD, but then these multimers assemble in the inner membrane of a bacterium causing phage shock response (Guilvout et al., 2011; Guilvout et al., 2006; Shevchik and Condemine, 1998). Interestingly, KoGspDPulD can also spontaneously form multimers in liposomes in vitro (Guilvout et al., 2008).
In EHEC, the T2SS cluster on the pO157 plasmid contains the etpO gene that encodes a protein with approximately 40% amino acid sequence identity to some, but not all, other T2SS pilotins. Here we report the crystal structure of this pilotin, which we call EHEC GspS (Korotkov et al., 2012). Based on extensive extra density in a hydrophobic groove of EHEC GspS, we suggest a possible binding site of EHEC GspS for the S-domain of the EHEC secretin GspD.
2. Protein expression, purification and crystallization
The gene fragment corresponding to the residues 16–106 of EHEC GspS (EtpO) was cloned into a modified pET-28b vector (Novagen) for expression as a fusion with maltose-binding protein (MBP). The construct has an N-terminal hexahistidine tag followed by MBP, tobacco etch virus (TEV) protease cleavage site and GspS. The protein was expressed in BL21(DE3) cells (Novagen) in LB media for 4 h at 30 °C. The harvested cells were resuspended in buffer containing 20 mM HEPES pH 7.5, 300 mM NaCl and lysed using a French press. The protein was purified on a Ni-NTA column (Qiagen) followed by cleavage with TEV protease and size-exclusion chromatography on Superdex75 10/300 GL column (GE Healthcare Bio-Sciences). SeMet-labeled GspS was produced using metabolic inhibition of methionine biosynthesis (van Duyne et al., 1993) and purified using the same protocol as for native protein. Protein was flash-frozen for storage (Deng et al., 2004). The crystals of GspS were obtained by the vapor diffusion method with a crystallization solution containing 0.1 M Tris-HCl pH 8.5, 0.2 M MgCl2, 30% (w/v) PEG3350. For cryoprotection, crystals were rapidly frozen in liquid nitrogen directly from the crystallization solution.
3. Data collection, structure determination and analysis
A native dataset was collected at the beamline 8.2.1 of the Berkeley Center for Structural Biology, the Advanced Light Source, at 100 K using a wavelength of 0.9794 Å. The native crystal was exposed at a low dose in an (unsuccessful) attempt to utilize anomalous signal from sulfur for phasing because Se-Met crystals were not available at that time. A dataset from SeMet-labeled crystal was collected at the beamline 9-2 of the Stanford Synchrotron Radiation Lightsource at 100 K using a wavelength 0.97915 Å. The crystals of Se-Met substituted protein diffracted to much lower resolution and longer exposures were necessary for data collection, therefore only a single-wavelength dataset was collected due to radiation damage to the crystal. Data were indexed, integrated and scaled using HKL2000 (Otwinowski and Minor, 1997) and XDS (Kabsch, 2010).
The positions of Se sites were found using SHELXD (Sheldrick, 2008). The phasing, density modification and initial model building were carried out using SOLVE and RESOLVE (Terwilliger, 2004). The structure was completed using ARP/WARP (Langer et al., 2008) followed by manual building in Coot (Emsley et al., 2010). The structure was refined with REFMAC5 (Murshudov et al., 2011) using 1 TLS group. The stereochemical quality of the final structure was verified using Coot and Molprobity server (Chen et al., 2010). Sequence alignment was produced using ClustalW2 (Larkin et al., 2007) and rendered using Espript (Gouet et al., 2003). The omit map for the secretin peptide in the DdGspSOutS-GspDOutD complex (Gu et al., 2012) was calculated using deposited structure factors (PDB 3UYM) and PHENIX (Adams et al., 2010). Structural illustrations were generated using PyMol (Schrodinger, 2010).
4. Overall structure of GspS and putative secretin binding site
In order to obtain protein suitable for crystallization, we constructed a soluble variant of GspS from enterohemorrhagic E.coli O157:H7 that encoded residues 13–110 of mature protein and hence lacked the signal sequence and the N-terminal lipidation residue. Because this variant failed to crystallize, the construct was optimized to include residues 16–106 based on secondary structure prediction analysis (Cole et al., 2008). The resultant crystals yielded the structure of the EHEC pilotin GspS to 1.9 Å resolution. The crystals belong to space group P6122 with one molecule in the asymmetric unit. The structure was solved de novo by the single wavelength anomalous dispersion method from crystals with selenomethionine-substituted protein, yielding a structure with good refinement statistics (Table 1). Interestingly, the Se sites substructure search revealed an additional site that was associated with the disulfide bond between Cys36–Cys90. It is likely that Se from selenomethionine was shuttled to cysteine during cell growth in minimal media.
Table 1.
Data collection and refinement statistics
| Native | Se-Met | |
|---|---|---|
| Data collection | ||
| Space group | P6122 | P6122 |
| Cell dimensions | ||
| a, b, c (Å) | 73.35, 73.35, 70.73 | 73.43, 73.43, 70.39 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 47.3-1.90 (2.00-1.90)1 | 47.2-2.50 (2.64-2.50) |
| Rsym | 0.085 (0.851) | 0.111 (0.691) |
| I/σI | 24.4 (3.1) | 19.0 (2.5) |
| Completeness (%) | 99.9 (98.8) | 99.9 (99.4) |
| Redundancy | 11.5 (11.1) | 10.1 (5.1) |
| Refinement | ||
| Resolution (Å) | 47.3-1.90 | |
| No. reflections (total/free) | 9323/426 | |
| Rwork/Rfree | 0.190/0.222 | |
| No. atoms | ||
| Protein | 743 | |
| Ligand/ion | 4 | |
| Water | 61 | |
| B-factors | ||
| Protein | 38.0 | |
| Ligand/ion | 38.1 | |
| Water | 40.1 | |
| Wilson B | 30.4 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.011 | |
| Bond angles (°) | 1.102 | |
| Ramachandran distribution (%)2 | ||
| Favored | 100 | |
| Outliers | 0.0 | |
Values in parentheses are for the highest-resolution shell.
Calculated using Molprobity (Chen et al., 2010).
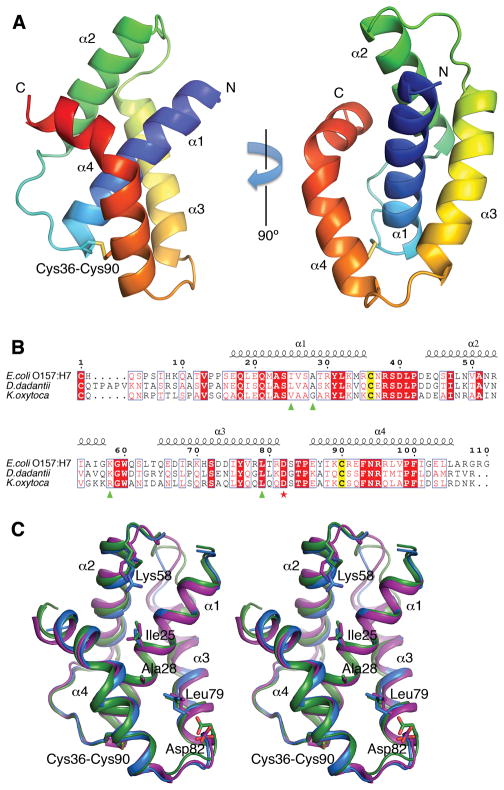
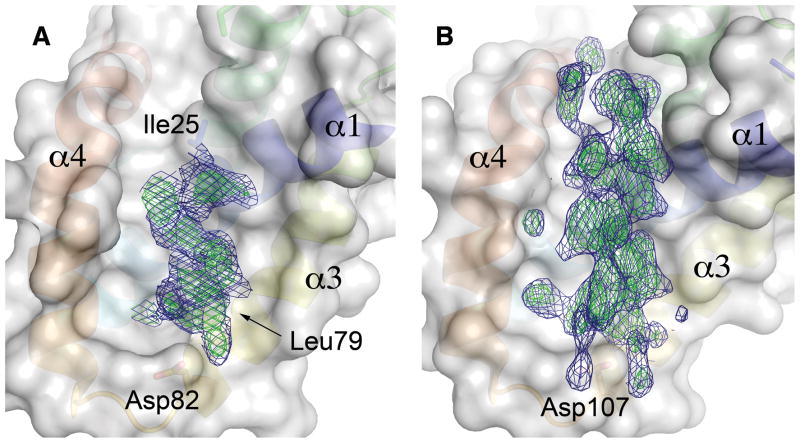
EHEC GspS is an all-helical protein containing four α-helices, with helix α1 surrounded by the other three helices (Fig. 1). Helices α1 and α2 are straight, helix α3 is curved and the C-terminal helix α4 bends at residue Leu97 where the α-helical hydrogen bond pattern is interrupted by residue Pro99. Two Cys residues, one located at the end of α1 and the other near the beginning of α4, form a disulfide bridge. A prominent feature of the structure is a deep groove formed by helices α1, α3 and α4. Intriguingly, in the final difference Fourier a distinct electron density, resembling two turns of an α helix, occupies this groove (Fig. 2A). Residues Ala28, Ile75, Leu79 and Phe93, which approach this extra density, are also hydrophobic in DdGspSOutS and KoGspSPulS (Fig. 1B). This suggests that the hydrophobic groove is the binding site for a helix from the S-domain of EHEC GspD. The extra density probably belongs to a peptide, or a mixture of peptides, that co-purified with GspS. Our efforts to identify the sequence of this peptide by mass-spectroscopy were unsuccessful due to the limited number of available crystals and possible heterogeneity of the peptide.
Fig. 1. Structure of GspS from enterohemorrhagic Escherichia coli.
(A) Overall structure of GspS in cartoon representation, colored from N-terminus (blue) to C-terminus (red). Four α-helices of GspS are labeled. The cysteine residues that form a disulfide bond are shown as sticks.
(B) Sequence alignment of EHEC GspS and homologous pilotins DdGspSOutS from D. dadantii and KoGspSPulS from K. oxytoca. Secondary structure elements are shown above the alignment. The conserved Cys residues forming the disulfide bond in GspS structures are highlighted in yellow. The amino acid residues implicated in secretin binding in the KoGspSPulS pilotin (Tosi et al., 2011) are labeled by green triangles. The capping Asp residue, involved in secretin binding in the DdGspSOutS pilotin (Gu et al., 2012), is labeled by a red star. The numbering shown is for EHEC GspS.
(C) Stereo view of EHEC GspS (purple) superimposed on pilotins KoGspSPulS from K. oxytoca (blue) (Tosi et al., 2011) and DdGspSOutS from D. dadantii (green) (Gu et al., 2012). The amino acid residues highlighted in (B) are shown in stick representation.
Fig. 2.
The putative secretin-binding site of GspS.
(A) The extra electron density in the groove on the surface of EHEC GspS. The blue mesh represents a σA-weighted 2FO-FC map contoured at the 1σ level; the green mesh represents contours at the 2.5 σ level. The amino acid residues Ile25 and Leu29 from EHEC GspS that are homologous to the residues implicated in secretin binding in KoGspSPulS (Tosi et al., 2011), as well as the secretin α-helix-capping Asp82 of EHEC.
(B) The omit maps of the 18-residue secretin peptide from the DdGspSOutS-GspDOutD structure (Gu et al., 2012) (PDB 3UYM) shown for comparison with the extra electron density in the ETEC GspS structure. The map is contoured at the same levels as in (A). Pilotin helices α1, α3 and α4 are labeled and shown in the same colors as in Fig. 1 underneath a transparent surface. The equivalent capping residue of Asp 82 in EHEC GspS (Fig 2A) is Asp107 in DdGspSOutS.
5. Structural and functional comparisons
While this manuscript was in preparation, structures of related pilotins have been reported: Klebsiella oxytoca KoGspSPulS and Dickeya dadantii DdGspSOutS (Gu et al., 2012; Tosi et al., 2011), which allows us to analyze all available structures. The EHEC GspS structure could be superimposed with KoGspSPulS with an r.m.s.d. of 0.96 Å and 43% sequence identity over 90 amino acid residues, and with DdGspSOutS with an r.m.s.d. of 1.08 Å and 39% sequence identity over 89 residues (Fig. 1C). The disulfide bridge linking α1 and α4 in EHEC GspS is conserved in DdGspSOutS and KoGspSPulS (Fig. 1).
Interestingly, substitutions in residues located in the groove of KoGspSPulS affected the binding of the KoGspDPulD S-domain in vitro, although only the Val42Asp substitution (corresponding to Ile25 of EHEC GspS, which approaches the extra density in the EHEC GspS structure (Fig. 2A)) led to abolishing T2SS secretion in vivo (Tosi et al., 2011). Moreover, in the structure of the complex between DdGspSOutS and a peptide derived from the S-domain of DdGspDOutD, the S-domain moiety is located in a concave area on the surface of DdGspSOutS, which corresponds to the hydrophobic groove of EHEC GspS (Fig. 2B) (Gu et al., 2012). Taken together, these data suggest that the T2SS pilotins KoGspSPulS, DdGspSOutS and EHEC GspS all share the same binding site for the α-helical S-domain of their cognate T2SS secretin.
Some pathogenic and non-pathogenic E. coli strains encode two clusters of the T2SS genes on the chromosome, named alpha (T2SSα) and beta (T2SSβ) (Strozen et al., 2012). Interestingly, both the T2SSα and T2SSβ clusters lack a homolog of a pilotin in contrast to the T2SS cluster of the pO157 plasmid. However, the T2SSβ cluster contains a gene yghG that encodes a small lipoprotein, which functions as a pilotin — and therefore named GspSβ or AspS (Alternate secretin pathway subunit S) — for its cognate secretin (Dunstan et al., 2013; Strozen et al., 2012). Moreover, AspS homologs have been recently identified in other bacteria, including V. cholerae (Dunstan et al., 2013). The structure of V. cholerae AspS is a novel α/β fold composed of 5 β-strands flanked by 4 α-helices (Dunstan et al., 2013). Remarkably, although the structure of AspS is unrelated to the structures of the T2SS pilotins of KoGspSPulS, DdGspSOutS and EHEC GspS family, AspS interacts with the S-domain of its cognate secretin (Dunstan et al., 2013). Therefore, the T2SS’s from different organisms, or different clusters within the same organism, appear to engage structurally unrelated pilotin chaperones to assist secretin targeting to the outer membrane.
Other secretin-containing systems, such as the T4PS and the T3SS, also contain pilotin proteins that are involved in secretin targeting and multimerization, although functional details may vary between different pilotin-secretin pairs (Koo et al., 2012). The functionally characterized T4PS pilotins include Myxococcus xanthus lipoprotein Tgl (Nudleman et al., 2006), Neisseria meningitidis PilW (Carbonnelle et al., 2006; Carbonnelle et al., 2005; Szeto et al., 2011) and Pseudomonas aeruginosa PilF (Koo et al., 2008), whereas the T3SS pilotins include Salmonella typhimurium InvH (Crago and Koronakis, 1998), Shigella flexneri MxiM (Schuch and Maurelli, 2001), Yersinia enterocolitica YscW (Burghout et al., 2004) and P. aeruginosa ExsB (Izore et al., 2011). Structural analyses of the T4PS pilotins PilW (Trindade et al., 2008) and PilF (Kim et al., 2006; Koo et al., 2008) reveal a similar arrangements of six tetratricopeptide repeat (TPR) motifs. Thus, despite multiple structural and mechanistic similarities between the T2SS and T4PS machineries (Ayers et al., 2010; Korotkov et al., 2012), PilW and PilF pilotins are unrelated in structure to the T2SS GspS and AspS pilotins. Moreover, the mode of interaction between PilW and PilF pilotins and their cognate secretins is also different, because the T4PS secretins lack the S-domain present in the T2SS secretins (Korotkov et al., 2011).
The structures of the T3SS pilotin proteins MxiM from S. flexneri (Lario et al., 2005; Okon et al., 2008) and ExsB from P. aeruginosa (Izore et al., 2011) show that these pilotins adopt two different structures: a cracked β-barrel and a β-sandwich fold. Remarkably, these T3SS pilotins are hence unrelated to (i) the T4PS pilotins PilW and PilF, and (ii) the T2SS GspS and AspS pilotins. Clearly, multiple entirely different folds have been adapted in the course of evolution to perform the same function: assisting the assembly of a multimeric protein in the bacterial outer membrane.
6. Conclusions
Since EHEC can cause severe foodborne disease, and can even be life threatening, the spread of antibiotic resistant variants of pathogenic bacteria is a source of growing concern. At the same time, the importance of commensal human microorganisms raises questions about safety of broad-spectrum anti-microbial drugs. Therefore, much attention has recently been given to the concept of devising alternative therapies based on targeting virulence factors (Cegelski et al., 2008). Increasing our understanding of T2SS secretin biogenesis in EHEC, such as identifying the critical hydrophobic groove EHEC pilotin, might assist in the development of compounds which bind tightly to this site. Such compounds would prevent the interaction of the pilotin with the S-domain of the secretin, leading to mis-targeting of the secretin multimer, and inhibition of secretion of potentially lethal virulence factors. Hence, such compounds might be a starting point for developing novel therapies against the diseases caused by EHEC.
Acknowledgments
We thank Stewart Turley for assistance during data collection and Steve Moseley for providing E.coli O157:H7 DNA. We thank the staff of the Berkeley Center for Structural Biology and the Stanford Synchrotron Radiation Lightsource for support during data collection. The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. The Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393) and the National Center for Research Resources (P41RR001209). This study was supported by National Institutes of Health Grants AI34501 (to WGJH). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS, NCRR or NIH.
Footnotes
7. Accession numbers
The structure factors and coordinates have been deposited in the Protein Data Bank under accession number 3SOL.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast VM, Schoenhofen IC, Langen GR, Stratilo CW, Chamberlain MD, et al. Expression of the ExeAB complex of Aeromonas hydrophila is required for the localization and assembly of the ExeD secretion port multimer. Mol Microbiol. 2002;44:217–231. doi: 10.1046/j.1365-2958.2002.02870.x. [DOI] [PubMed] [Google Scholar]
- Ayers M, Howell PL, Burrows LL. Architecture of the type II secretion and type IV pilus machineries. Future Microbiol. 2010;5:1203–1218. doi: 10.2217/fmb.10.76. [DOI] [PubMed] [Google Scholar]
- Burghout P, Beckers F, de Wit E, van Boxtel R, Cornelis GR, et al. Role of the pilot protein YscW in the biogenesis of the YscC secretin in Yersinia enterocolitica. J Bacteriol. 2004;186:5366–5375. doi: 10.1128/JB.186.16.5366-5375.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burland V, Shao Y, Perna NT, Plunkett G, Sofia HJ, et al. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 1998;26:4196–4204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnelle E, Helaine S, Nassif X, Pelicic V. A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol Microbiol. 2006;61:1510–1522. doi: 10.1111/j.1365-2958.2006.05341.x. [DOI] [PubMed] [Google Scholar]
- Carbonnelle E, Helaine S, Prouvensier L, Nassif X, Pelicic V. Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occurring after pilus assembly, essential for fibre stability and function. Mol Microbiol. 2005;55:54–64. doi: 10.1111/j.1365-2958.2004.04364.x. [DOI] [PubMed] [Google Scholar]
- Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin S, Guilvout I, Nickerson NN, Pugsley AP. Sorting of an integral outer membrane protein via the lipoprotein-specific Lol pathway and a dedicated lipoprotein pilotin. Mol Microbiol. 2011;80:655–665. doi: 10.1111/j.1365-2958.2011.07596.x. [DOI] [PubMed] [Google Scholar]
- Crago AM, Koronakis V. Salmonella InvG forms a ring-like multimer that requires the InvH lipoprotein for outer membrane localization. Mol Microbiol. 1998;30:47–56. doi: 10.1046/j.1365-2958.1998.01036.x. [DOI] [PubMed] [Google Scholar]
- Daefler S, Guilvout I, Hardie KR, Pugsley AP, Russel M. The C-terminal domain of the secretin PulD contains the binding site for its cognate chaperone, PulS, and confers PulS dependence on pIVf1 function. Mol Microbiol. 1997;24:465–475. doi: 10.1046/j.1365-2958.1997.3531727.x. [DOI] [PubMed] [Google Scholar]
- Deng J, Davies DR, Wisedchaisri G, Wu M, Hol WGJ, et al. An improved protocol for rapid freezing of protein samples for long-term storage. Acta Crystallogr D Biol Crystallogr. 2004;60:203–204. doi: 10.1107/s0907444903024491. [DOI] [PubMed] [Google Scholar]
- Douzi B, Filloux A, Voulhoux R. On the path to uncover the bacterial type II secretion system. Philos Trans R Soc Lond B Biol Sci. 2012;367:1059–1072. doi: 10.1098/rstb.2011.0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunstan RA, Heinz E, Wijeyewickrema LC, Pike RN, Purcell AW, et al. Assembly of the type II secretion system such as found in Vibrio cholerae depends on the novel pilotin AspS. PLoS Pathog. 2013;9:e1003117. doi: 10.1371/journal.ppat.1003117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farfan MJ, Torres AG. Molecular mechanisms that mediate colonization of Shiga toxin-producing Escherichia coli strains. Infect Immun. 2012;80:903–913. doi: 10.1128/IAI.05907-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier A, Puente JL, Finlay BB. Secretin of the enteropathogenic Escherichia coli type III secretion system requires components of the type III apparatus for assembly and localization. Infect Immun. 2003;71:3310–3319. doi: 10.1128/IAI.71.6.3310-3319.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grys TE, Siegel MB, Lathem WW, Welch RA. The StcE protease contributes to intimate adherence of enterohemorrhagic Escherichia coli O157:H7 to host cells. Infect Immun. 2005;73:1295–1303. doi: 10.1128/IAI.73.3.1295-1303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Rehman S, Wang X, Shevchik VE, Pickersgill RW. Structural and functional insights into the pilotin-secretin complex of the type II secretion system. PLoS Pathog. 2012;8:e1002531. doi: 10.1371/journal.ppat.1002531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilvout I, Nickerson NN, Chami M, Pugsley AP. Multimerization-defective variants of dodecameric secretin PulD. Res Microbiol. 2011;162:180–190. doi: 10.1016/j.resmic.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Guilvout I, Chami M, Engel A, Pugsley AP, Bayan N. Bacterial outer membrane secretin PulD assembles and inserts into the inner membrane in the absence of its pilotin. EMBO J. 2006;25:5241–5249. doi: 10.1038/sj.emboj.7601402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilvout I, Chami M, Berrier C, Ghazi A, Engel A, et al. In vitro multimerization and membrane insertion of bacterial outer membrane secretin PulD. J Mol Biol. 2008;382:13–23. doi: 10.1016/j.jmb.2008.06.055. [DOI] [PubMed] [Google Scholar]
- Hardie KR, Lory S, Pugsley AP. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 1996;15:978–988. [PMC free article] [PubMed] [Google Scholar]
- Ho TD, Davis BM, Ritchie JM, Waldor MK. Type 2 secretion promotes enterohemorrhagic Escherichia coli adherence and intestinal colonization. Infect Immun. 2008;76:1858–1865. doi: 10.1128/IAI.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izore T, Perdu C, Job V, Attree I, Faudry E, et al. Structural characterization and membrane localization of ExsB from the type III secretion system (T3SS) of Pseudomonas aeruginosa. J Mol Biol. 2011;413:236–246. doi: 10.1016/j.jmb.2011.07.043. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Oh J, Han D, Kim EE, Lee B, et al. Crystal structure of PilF: functional implication in the type 4 pilus biogenesis in Pseudomonas aeruginosa. Biochem Biophys Res Commun. 2006;340:1028–1038. doi: 10.1016/j.bbrc.2005.12.108. [DOI] [PubMed] [Google Scholar]
- Koo J, Burrows LL, Howell PL. Decoding the roles of pilotins and accessory proteins in secretin escort services. FEMS Microbiol Lett. 2012;328:1–12. doi: 10.1111/j.1574-6968.2011.02464.x. [DOI] [PubMed] [Google Scholar]
- Koo J, Tammam S, Ku SY, Sampaleanu LM, Burrows LL, et al. PilF is an outer membrane lipoprotein required for multimerization and localization of the Pseudomonas aeruginosa type IV pilus secretin. J Bacteriol. 2008;190:6961–6969. doi: 10.1128/JB.00996-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkov KV, Gonen T, Hol WGJ. Secretins: dynamic channels for protein transport across membranes. Trends Biochem Sci. 2011;36:433–443. doi: 10.1016/j.tibs.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkov KV, Sandkvist M, Hol WG. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol. 2012;10:336–351. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lario PI, Pfuetzner RA, Frey EA, Creagh L, Haynes C, et al. Structure and biochemical analysis of a secretin pilot protein. EMBO J. 2005;24:1111–1121. doi: 10.1038/sj.emboj.7600610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lathem WW, Grys TE, Witowski SE, Torres AG, Kaper JB, et al. StcE, a metalloprotease secreted by Escherichia coli O157:H7, specifically cleaves C1 esterase inhibitor. Mol Microbiol. 2002;45:277–288. doi: 10.1046/j.1365-2958.2002.02997.x. [DOI] [PubMed] [Google Scholar]
- McLaughlin LS, Haft RJ, Forest KT. Structural insights into the Type II secretion nanomachine. Curr Opin Struct Biol. 2012;22:208–216. doi: 10.1016/j.sbi.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton-Celsa A, Mohawk K, Teel L, O’Brien A. Pathogenesis of Shiga-toxin producing Escherichia coli. Curr Top Microbiol Immunol. 2012;357:67–103. doi: 10.1007/82_2011_176. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudleman E, Wall D, Kaiser D. Polar assembly of the type IV pilus secretin in Myxococcus xanthus. Mol Microbiol. 2006;60:16–29. doi: 10.1111/j.1365-2958.2006.05095.x. [DOI] [PubMed] [Google Scholar]
- Okon M, Moraes TF, Lario PI, Creagh AL, Haynes CA, et al. Structural characterization of the type-III pilot-secretin complex from Shigella flexneri. Structure. 2008;16:1544–1554. doi: 10.1016/j.str.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Paton AW, Paton JC. Reactivity of convalescent-phase hemolytic-uremic syndrome patient sera with the megaplasmid-encoded TagA protein of Shiga toxigenic Escherichia coli O157. J Clin Microbiol. 2002;40:1395–1399. doi: 10.1128/JCM.40.4.1395-1399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. [Google Scholar]
- Schuch R, Maurelli AT. MxiM and MxiJ, base elements of the Mxi-Spa type III secretion system of Shigella, interact with and stabilize the MxiD secretin in the cell envelope. J Bacteriol. 2001;183:6991–6998. doi: 10.1128/JB.183.24.6991-6998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Sherman PM, Ossa JC, Wine E. Bacterial infections: new and emerging enteric pathogens. Curr Opin Gastroenterol. 2010;26:1–4. doi: 10.1097/MOG.0b013e328333d73b. [DOI] [PubMed] [Google Scholar]
- Shevchik VE, Condemine G. Functional characterization of the Erwinia chrysanthemi OutS protein, an element of a type II secretion system. Microbiology. 1998;144:3219–3228. doi: 10.1099/00221287-144-11-3219. [DOI] [PubMed] [Google Scholar]
- Shevchik VE, Robert-Baudouy J, Condemine G. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 1997;16:3007–3016. doi: 10.1093/emboj/16.11.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strozen TG, Li G, Howard SP. YghG (GspSβ) is a novel pilot protein required for localization of the GspSβ type II secretion system secretin of enterotoxigenic Escherichia coli. Infect Immun. 2012;80:2608–2622. doi: 10.1128/IAI.06394-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strozen TG, Stanley H, Gu Y, Boyd J, Bagdasarian M, et al. Involvement of the GspAB complex in assembly of the type II secretion system secretin of Aeromonas and Vibrio species. J Bacteriol. 2011;193:2322–2331. doi: 10.1128/JB.01413-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto TH, Dessen A, Pelicic V. Structure/function analysis of Neisseria meningitidis PilW, a conserved protein that plays multiple roles in type IV pilus biology. Infect Immun. 2011;79:3028–3035. doi: 10.1128/IAI.05313-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- Tosi T, Nickerson NN, Mollica L, Jensen MR, Blackledge M, et al. Pilotin-secretin recognition in the type II secretion system of Klebsiella oxytoca. Mol Microbiol. 2011;82:1422–1432. doi: 10.1111/j.1365-2958.2011.07896.x. [DOI] [PubMed] [Google Scholar]
- Trindade MB, Job V, Contreras-Martel C, Pelicic V, Dessen A. Structure of a widely conserved type IV pilus biogenesis factor that affects the stability of secretin multimers. J Mol Biol. 2008;378:1031–1039. doi: 10.1016/j.jmb.2008.03.028. [DOI] [PubMed] [Google Scholar]
- van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- Yu AC, Worrall LJ, Strynadka NC. Structural insight into the bacterial mucinase StcE essential to adhesion and immune evasion during enterohemorrhagic E. coli infection. Structure. 2012;20:707–717. doi: 10.1016/j.str.2012.02.015. [DOI] [PubMed] [Google Scholar]