Abstract
Background
Synaptic dysfunction has been shown to be involved in the pathogenesis of autism. We hypothesized that the protocadherin α gene cluster (PCDHA), which is involved in synaptic specificity and in serotonergic innervation of the brain, could be a suitable candidate gene for autism.
Methods
We examined 14 PCDHA single nucleotide polymorphisms (SNPs) for genetic association with autism in DNA samples of 3211 individuals (841 families, including 574 multiplex families) obtained from the Autism Genetic Resource Exchange.
Results
Five SNPs (rs251379, rs1119032, rs17119271, rs155806 and rs17119346) showed significant associations with autism. The strongest association (p < 0.001) was observed for rs1119032 (z score of risk allele G = 3.415) in multiplex families; SNP associations withstand multiple testing correction in multiplex families (p = 0.041). Haplotypes involving rs1119032 showed very strong associations with autism, withstanding multiple testing corrections. In quantitative transmission disequilibrium testing of multiplex families, the G allele of rs1119032 showed a significant association (p = 0.033) with scores on the Autism Diagnostic Interview–Revised (ADI-R)_D (early developmental abnormalities). We also found a significant difference in the distribution of ADI-R_A (social interaction) scores between the A/A, A/G and G/G genotypes of rs17119346 (p = 0.002).
Limitations
Our results should be replicated in an independent population and/or in samples of different racial backgrounds.
Conclusion
Our study provides strong genetic evidence of PCDHA as a potential candidate gene for autism.
Introduction
Autism is a complex neurodevelopmental disorder characterized by social and communicative deficits and ritualistic-repetitive behaviours that are detectable in early childhood. It is one of the most heritable (> 90% heritability) genetic disorders.
Altered synaptic development and plasticity due to dys-regulated synaptic protein synthesis has been proposed to be causative of autistic phenotypes.1 During development and throughout life, billions of synapses in the mammalian central nervous system are continuously reconfigured, both structurally and functionally — a process called synaptic plasticity, which forms the basis for learning and memory.2 For this process to be well defined, the neurons involved should have their own uniquely identifiable chemical characteristics to be recognized by a specific synaptic partner. This much-needed synaptic specificity is achieved through a combination of diverse and unique molecular cues in the pre- and postsynaptic neurons.3,4
The protocadherins (PCDH), which comprise the largest family in the cadherin superfamily of calcium-dependent cell adhesion molecules, gain importance in this context.5,6 The PCDH genes, organized into α, β and γ clusters, are predominantly expressed in the brain.7
The PCDH α (PCDHA) proteins in particular have been proposed to participate in specific synaptic connections based on their localization to synaptic junctions.5 The PCDHA gene (5q31.3) cluster is composed of 15 large, variable region exons (V) organized in a tandem array, followed by a constant region composed of 3 exons shared by all the splice variants in the cluster6 (Appendix 1, Fig. S1, available at cma.ca/jpn). Individual PCDHA mRNAs are assembled from one of the V exons and the 3 constant exons in a process involving differential promoter activation and alternative splicing.6,8,9 Esumi and colleagues10 reported monoallelic yet combinatorial expression of the V exons of PCDHA in single neurons. This type of regulation could account for the expression of a wide variety of different combinations of PCDHA transcripts in individual neurons, thereby establishing specific neuron identity and synaptic specificity. It would be interesting to examine the role of PCDHA in the pathogenesis of autism, since impaired neuronal connectivity11,12 involving altered synapses13 has been found to lead to cognitive deficits.
Furthermore, PCDHA proteins have also been found to be important for the serotonergic projections to appropriately innervate the target brain areas.14 This is another interesting aspect of PCDHA that makes it a potential candidate gene for autism, since a growing body of evidence links autism to abnormalities in serotonin function.15–17
Previously, structural variations of PCDHs, such as PCDH918 and PCDH10,19 have been implicated in autism. Recently, a de novo gene disruption (deletion leading to frame shift) in PCDHA13 was reported in autism.20
Given the role of PCDHA in the aforementioned processes, we hypothesized that PCDHA is a suitable candidate gene for autism. In the present study, we analyzed the genetic association of PCDHA with autism in a large cohort of 841 families with affected children.
Methods
Our study protocol was approved by the Ethics Committee of Hamamatsu University School of Medicine, Japan.
Samples
We obtained the DNA samples used in this study from Autism Genetic Resource Exchange (AGRE; www.agre.org).21 They have obtained informed consent, allowing the distribution of DNA samples to approved researchers, from all individuals listed in their pedigree catalogue. We used DNA from 841 families (3211 individuals in total) in the present association study. This includes 1467 patients (1178 male, 289 female) with autism. Among the 841 families, 574 were multiplex, meaning that more than 1 child had autism.
Pedigree information for each individual, along with the diagnoses based on Autism Diagnostic Interview-Revised (ADI-R),22 were available on the AGRE website. The ADI-R scores were available for 1523 individuals with autism. We excluded families with nonidiopathic autism flags (e.g., fragile-X, abnormal brain imaging results, dysmorphic features, birth trauma) recorded for any family members.
Single nucleotide polymorphism selection
The genomic structure of PCDHA (140145500..140375000 in chromosome 5) was based on the University of California, Santa Cruz, February 2009 assembly of the human genome (http://genome.ucsc.edu/).
We selected the SNPs (MAF > 0.1) for the association study from the International HapMap Project (www.hapmap.org) database of white populations. Using the Tagger in Haploview version 4.1 (www.broad.mit.edu/mpg/haploview), we selected 14 SNPs by pairwise tagging with an r2 threshold of 0.8 (Appendix 1, Table S1). Except for rs3733709, a missense mutation located in the coding region of PCHDA2 and PCDHA3, all the SNPs were located in the introns.
Genotyping
We used Assay-on-Demand SNP genotyping products (Applied Biosystems) to score genotypes based on the TaqMan assay method.23 Genotypes were determined in the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems) and analyzed using SDS version 2.0 software (Applied Biosystems).
Statistical analysis
All statistical analyses were performed separately for the whole set of 841 families (hereafter referred to as all families) and for the 574 multiplex families. Power analysis was conducted using Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/dtdt.html). We used FBAT version 2.0.3 (http://biosun1.harvard.edu/~fbat/fbat.htm) to examine the genetic association of SNPs with autism in a family-based association test under an additive model; the FBAT–MM option was used for multimarker testing.
Pairwise linkage disequilibrium (LD) among the SNPs, based on D’ (LD coefficient) values,24 was estimated using Haploview. We defined LD blocks in PCDHA using the Haploview confidence interval algorithm.25 Using FBAT (HBAT option) and Haploview, the association of haplotypes within the LD blocks was analyzed. The –p option (100 000 permutations) of HBAT was used to compute the global test for haplotypes via Monte-Carlo simulations.
Using the Quantitative Transmission Disequilibrium Test (QTDT) version 2.5.1 (www.sph.umich.edu/csg/abecasis/QTDT/index.html), we further examined the association between quantitative ADI-R scores (ADI-R_A, ADI-R_BV, ADI-R_BNV, ADI-R_C, ADI-R_D) and SNPs selected from FBAT analysis (all p < 0.05). We used the Bonferroni method to adjust for multiple testing.
In the autism samples, we performed 1-way analysis of variance (ANOVA) followed by post hoc pairwise comparison with Bonferroni correction to examine the variability in the distribution of ADI-R phenotypic data across the homozygous and heterozygous genotypes of the SNPs selected from FBAT analysis (p < 0.05).
Results
Single SNP association analysis
Table 1 shows the results of FBAT analysis of PCDHA SNPs. In all families, 4 SNPs showed significant associations with autism, rs251379 (p = 0.009, z score for risk allele T = 2.608), rs1119032 (p = 0.014, z score for risk allele G = 2.454), rs155806 (p = 0.029, z score for risk allele C = 2.177) and rs17119346 (p = 0.017, z score for risk allele G = 2.382). However, the SNP associations did not withstand multiple testing correction.
Table 1.
FBAT analysis of PCDHA with autism in all 841 families (all) and in the 574 multiplex families
| No. informative families | Frequency | z score† | p value‡ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||
| Marker | Location | Allele* | All | Multiplex | All | Mutiplex | All | Mutiplex | All | Mutiplex |
| rs3733709 | 140161832 | A | 300 | 211 | 0.890 | 0.888 | 0.376 | 0.919 | 0.71 | 0.36 |
| G | 0.110 | 0.112 | −0.376 | −0.919 | ||||||
| rs3756337 | 140166622 | G | 607 | 423 | 0.525 | 0.531 | 0.945 | 0.288 | 0.34 | 0.77 |
| C | 0.475 | 0.469 | −0.945 | −0.288 | ||||||
| rs3806843 | 140192722 | G | 606 | 421 | 0.540 | 0.532 | −1.384 | −0.086 | 0.17 | 0.93 |
| A | 0.460 | 0.468 | 1.384 | 0.086 | ||||||
| rs628871 | 140211436 | A | 416 | 292 | 0.753 | 0.744 | −0.203 | −0.298 | 0.84 | 0.77 |
| G | 0.247 | 0.256 | 0.203 | 0.298 | ||||||
| rs251379 | 140239106 | T | 585 | 402 | 0.651 | 0.651 | 2.608 | 1.977 | 0.009 | 0.048 |
| C | 0.349 | 0.349 | −2.608 | −1.977 | ||||||
| rs1119032 | 140262615 | A | 343 | 235 | 0.860 | 0.862 | −2.454 | −3.415 | 0.014 | < 0.001 |
| G | 0.140 | 0.138 | 2.454 | 3.415 | ||||||
| rs155363 | 140281727 | C | 600 | 419 | 0.536 | 0.531 | −1.050 | 0.144 | 0.29 | 0.89 |
| T | 0.464 | 0.469 | 1.050 | −0.144 | ||||||
| rs17119271 | 140289890 | T | 129 | 81 | 0.953 | 0.959 | −1.527 | −2.495 | 0.13 | 0.013 |
| C | 0.047 | 0.041 | 1.527 | 2.495 | ||||||
| rs155806 | 140330024 | T | 286 | 191 | 0.889 | 0.894 | −2.177 | −2.057 | 0.029 | 0.039 |
| C | 0.111 | 0.106 | 2.177 | 2.057 | ||||||
| rs17119328 | 140331257 | A | 169 | 110 | 0.939 | 0.944 | −1.326 | −1.617 | 0.18 | 0.11 |
| C | 0.061 | 0.056 | 1.326 | 1.617 | ||||||
| rs11743888 | 140334091 | G | 167 | 106 | 0.941 | 0.948 | −1.292 | −1.261 | 0.20 | 0.21 |
| A | 0.059 | 0.052 | 1.292 | 1.261 | ||||||
| rs31874 | 140349502 | T | 477 | 334 | 0.745 | 0.733 | −0.337 | −0.504 | 0.74 | 0.61 |
| C | 0.255 | 0.267 | 0.337 | 0.504 | ||||||
| rs31872 | 140352406 | T | 569 | 394 | 0.640 | 0.632 | −0.768 | −1.088 | 0.44 | 0.28 |
| C | 0.360 | 0.368 | 0.768 | 1.088 | ||||||
| rs17119346 | 140354018 | G | 562 | 383 | 0.680 | 0.677 | 2.382 | 1.857 | 0.017 | 0.06 |
| A | 0.320 | 0.323 | −2.382 | −1.857 | ||||||
| p value after multimarker testing (FBAT-MM) | 0.107 | 0.041 | ||||||||
Major allele is listed first.
Positive score indicates the risk allele; negative score indicates the protective allele.
p < 0.05, additive model.
We further examined SNP associations with autism in families with male and female autistic individuals separately. The SNPs rs251379 (p = 0.031, z score for risk allele T = 2.160) and rs17119346 (p = 0.026, z score for risk allele G = 2.223) showed nominal association with autism in families with male autistic patients. No significant association was observed in families with female autistic patients.
Considering only the multiplex families, 4 SNPs showed significant associations with autism: rs251379 (p = 0.048, z score for risk allele T = 1.977), rs1119032 (p < 0.001, z score for risk allele G = 3.415), rs17119271 (p = 0.013, z score for risk allele C = 2.495) and rs155806 (p = 0.039, z score for risk allele C = 2.057). The SNP associations remained significant after multiple testing correction (p = 0.041).
We further examined SNP associations with autism in multiplex families with male and female autistic patients separately. The SNPs rs1119032 (p = 0.004, z score for risk allele G = 2.851) and rs17119271 (p = 0.028, z score for risk allele C = 2.197) showed nominal association with autism in families with male autistic patients. No significant association was observed in families with female autistic patients.
Power analysis showed that the overall sample size of 841 families provided 91% power to detect an odds ratio of 1.5 for an allele frequency of 0.1 at an α of 0.05. The sample size of families with affected male patients (n = 784) provided 89% power to detect an association under similar criteria. However, the sample size of families with affected female patients (n = 254) was underpowered (< 80%) to detect an association.
Linkage disequlibrium structure of PCDHA
The Haploview confidence interval algorithm identified 3 LD blocks in PCDHA: the first block consisted of SNPs rs3733709 and rs3756337; the second consisted of rs1119032 and rs155363; and the third consisted of rs17119328, rs11743888, rs31874, rs31872 and rs17119346. The LD structure was similar when all families were considered (Appendix 1, Fig. S2) and when only multiplex families were considered.
Haplotype association analysis
The results of the haplotype association analysis are shown in Table 2. In all families, the haplotypes AC (p = 0.002, z score = −2.851) and GC (p = 0.008, z score = 2.506) in block 2, and the haplotype AGTTA (p = 0.036, z score = −2.029) in block 3 showed significant associations with autism. The p value after global testing was significant for block 2 (p = 0.006), but not for block 3.
Table 2.
Haplotype association analysis of PCDHA with autism in all 841 families (all), including 574 multiplex families
| Frequency | z score* | p value† | |||||
|---|---|---|---|---|---|---|---|
|
|
|
|
|||||
| Block | Haplotype | All | Mutiplex | All | Mutiplex | All | Mutiplex |
| Block 1 | AC | 0.478 | 0.469 | 0.34 | 0.75 | ||
| (SNPs 01, 02) | AG | 0.411 | 0.418 | 0.24 | 0.36 | ||
| GG | 0.111 | 0.112 | 0.75 | 0.37 | |||
| Block 2 | AT | 0.461 | 0.469 | 0.23 | 0.95 | ||
| (SNPs 06, 07) | AC | 0.399 | 0.395 | −2.851 | −2.259 | 0.002 | 0.014 |
| GC | 0.140 | 0.137 | 2.506 | 3.425 | 0.008 | < 0.001 | |
| Block 3 | AGTTA | 0.323 | 0.322 | −2.029 | 0.036 | 0.08 | |
| (SNPs 10–14) | AGTTG | 0.256 | 0.265 | 0.35 | 0.45 | ||
| AGCCG | 0.251 | 0.250 | 0.64 | 0.97 | |||
| AGTCG | 0.105 | 0.102 | 0.57 | 0.56 | |||
| CATTG | 0.052 | 0.048 | 0.12 | 0.11 | |||
| Global haplotype test (100 000 permutations)‡ | 0.006 | 0.004 | |||||
SNP = single nucleotide polymorphism.
z scores of only the significantly associated haplotypes are shown.
Global test for haplotypes in block 2.
In multiplex families only, the haplotypes AC (p = 0.014, z score = −2.259) and GC (p < 0.001, z score = 3.425) of block 2 showed significant associations with autism. The p value remained significant (p = 0.004) after global testing.
Taken together, the AC and GC haplotypes of block 2 may be considered as protective and risk haplotypes, respectively.
Quantitative Transmission Disequilibrium Test analysis
In the QTDT analysis of multiplex families, the G allele of rs1119032 showed a significant association with ADI-R_D (developmental abnormalities before 36 months) scores (p = 0.033, F = 4.578). However, the association did not withstand multiple testing correction. In all families, the G allele of rs1119032 was found to have a tendency for association with the ADIR_D score (p = 0.05, F = 3.688). None of the other SNPs were associated with ADI-R scores.
One-way ANOVA (genotype v. ADI_R)
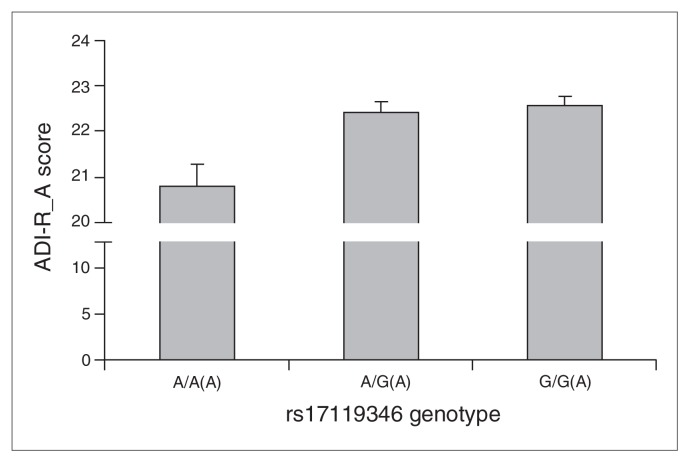
Considering only the autistic individuals in all families, we observed a significant difference in the distribution of ADI-R_A (social interaction) scores between the A/A, A/G and G/G genotypes of rs17119346 (p = 0.002; Fig. 1). Individuals with the A/A genotype had lower ADI-R_A scores than individuals with A/G and G/G genotypes. After post hoc pairwise comparison with Bonferroni correction, A/A versus A/G and A/A versus G/G differences in ADI-R_A scores were significant at p < 0.01.
Fig. 1.
Comparison of the distribution of Autism Diagnostic Interview–Revised (ADI-R)_A scores of autistic individuals across the A/A, A/G and G/G genotypes of rs17119346. A significant variation was observed in the distribution of ADI-R_A scores among the 3 groups (1-way analysis of variance, p = 0.002). Comparisons of A/A versus A/G and A/A versus G/G genotypes were significant at p < 0.01 following post hoc pairwise comparison with Bonferroni correction.
Discussion
We report a strong association between the PCDHA gene cluster and autism. The G allele of SNP rs1119032 was found to be a risk allele in both single SNP association and haplotype association tests. The z scores were found to be 2.454 and 3.415 in the single SNP association analysis of all families and of only multiplex families, respectively. In haplotype analysis, the z scores for the haplotype involving the G allele were found to be 2.506 and 3.425 for all families and for only multiplex families, respectively.
Anney and colleagues26 recently conducted a genome-wide association study (GWAS) of 1558 families from the Autism Genome Project consortium, genotyped at more than 1 million SNPs. In addition to the data published online, further information on SNPs in the region spanned by PCDHA was provided to us by the authors through personal communication (Prof. Bernie Devlin, Department of Psychiatry, University of Pittsburgh School of Medicine: personal communication, February–March 2012). We could thus compare these results with that from our study. Among the 5 SNPs for which we have reported significant association in this study, 2 were also found to be nominally associated in the GWAS: rs1119032 (GWAS p values, 0.004 for strict diagnosis and 0.04 for spectrum diagnosis), the SNP for which we obtained most significant association in multiplex families, and rs251379 (GWAS p values, 0.02 for strict diagnosis and 0.005 for spectrum diagnosis), the SNP showing the most significant association in all families. Furthermore, 8 other PCDHA SNPs were found to be nominally associated (p < 0.01) with spectrum diagnosis in the GWAS: rs11750201 (p = 0.005), rs13182228 (p = 0.005), rs155800 (p = 0.006), rs155817 (p = 0.006), rs251378 (p = 0.008), rs265311 (p = 0.008), rs59479 (p = 0.004) and rs6874218 (p = 0.003). All these SNPs are located in introns and may not have a functional significance.
In the present study, the strongest allelic (p < 0.001) and haplotypic associations for rs1119032 was observed in multiplex families. Multiplex sibships tend to be enriched in frequency for a disease susceptibility allele, and thus create a stronger contrast with unrelated controls, as well as with parents and unaffected siblings.27 In a genetically complex, heterogeneous, polygenic disorder like autism, the common genetic predisposing factors are therefore likely to be enriched in multiplex families; in simplex families, a fraction of the cases are likely to be caused by rare chromosomal rearrangements or other sporadic events.28
The SNP associations in multiplex families and the haplotype associations in all families and multiplex families withstood multiple testing correction. In several of the previous candidate gene studies of autism, the SNP associations were not found to be significant after adjusting for multiple markers.29–31 In spite of the large sample size and large number of markers in the present study, our SNP and haplotype associations were found to withstand multiple testing correction.
In QTDT analysis, the G allele of rs1119032 showed a significant association with ADI-R_D scores, which reflect developmental abnormalities observed in patients with autism before the age of 36 months. Delayed speech, unusual socioemotional reactions and poor attention to and exploration of the environment are among the first clinically noticeable behavioural symptoms of autism during the second and third years of life.32,33 Since autism is a pervasive developmental disorder, neuroanatomical, neurochemical and neurobiological abnormalities should precede the appearance of clinical symptoms. It has been reported that PCDHA functions during neurogenesis in early development owing to its role in neuronal circuit maturation.10,34 A new microdeletion syndrome of 5q31.3, the locus of PCDHA, has been reported in patients with severe developmental delays.35 Therefore, the association of the risk allele of rs1119032 with ADI-R_D is an interesting observation. It should be noted that in the GWAS by Anney and colleagues,26 rs119035 was associated most significantly with strict autism, but not with other autism-spectrum disorders.
In addition to rs1119032, we also observed significant associations of 4 more SNPs (rs251379, rs17119271, rs155806 and rs17119346) with autism. Since all the associated SNPs are located in introns, we used SNP Function Prediction (FuncPred; http://snpinfo.niehs.nih.gov/snpfunc.htm) and FastSNP (http://fastsnp.ibms.sinica.edu.tw/pages/input_SNPListAnalysis.jsp) to predict their functions. The SNPs were analyzed for putative roles in splicing regulation, transcription factor binding site, miRNA binding site and for other regulatory roles. A transcription factor binding site was predicted for rs251379.
To further expand our search for possible functional polymorphisms, we also included in FuncPred and FastSNP analyses other PCDHA SNPs in the LD bins of significantly associated SNPs: 3 SNPs in the LD bin of rs251379 were located in the coding region. Among these, rs251354 is a non-synonymous SNP (Leu/Val) with a benign effect; the other 2 SNPs (rs155807 and rs155820) are synonymous SNPs. Two SNPs (rs12153295 and rs11744560) in the LD bin of rs1119032, located in the coding region, are also synonymous SNPs. Transcription factor binding sites were predicted in several of the SNPs in the LD bins of rs251379, rs1119032, rs17119271 and rs155806. We predicted miRNA binding sites for 1 SNP each in the LD bins of rs251379 and rs17119271.
Autistic patients with the A/A genotype of rs17119346 were found to have lower ADI-R_A scores than patients with A/G and G/G genotypes. The difference in ADI-R_A scores of patients with A/A versus A/G and A/A versus G/G genotypes was significant after Bonferroni correction. The ADI-R_A scores are indicative of social interaction. Impaired social interaction is a hallmark feature of autism.
Autism-spectrum disorders are typically detected and diagnosed during the second or third years of life. Their onset and detection corresponds with a period of robust synapse formation and maturation in humans. The complex genomic organization of the PCDHA, along with the allelic and combinatorial expression of their distinct V exons, contributes in specifying neuronal cell identities.10 This enormous diversity of neurons is essential to generate complex neuronal networks. Loss-of-function mice have revealed that Pcdha plays pivotal roles in neuronal survival,36,37 synaptic connectivity,38 axonal convergence,39 and learning and memory.40
It has been found that PCDHA is essential for the serotonergic projections to appropriately innervate the target brain regions during early development.14 In PCDHA knockout mutant mice, abnormal distribution of serotonergic fibres was observed in various target regions, along with abnormal levels of serotonin in the hippocampus.14 Abberant serotonergic innervations have been implicated in several behavioural abnormalities related to autism.41–43
Recently, a de novo deletion in PCDHA13 has been observed in autism.20 Several other members of the cadherin superfamily, such as CDH8,44CDH9,45CDH10,45CDH18,18PCDH918 and PCDH10,19 have been implicated in autism. The PCDHA gene is also a candidate locus for schizophrenia and bipolar disorder.46,47
After analyzing the families with male and female autistic patients separately, we observed significant associations only for male patients in our analysis of all families and of only multiplex families. However, the lack of association in female patients could be explained by the small number of families with affected female patients.
Limitations
Our study involved mainly white samples. The results need to be replicated in an independent population and/or in samples of different racial backgrounds.
Conclusion
Our study provides strong genetic evidence of PCDHA as a potential candidate gene for autism. More research is warranted into the molecular mechanisms through which PCDHA is involved in the pathogenesis of autism-spectrum disorders.
Acknowledgements
We gratefully acknowledge the resources provided by the Autism Genetic Resource Exchange (AGRE) Consortium and the participating AGRE families. The AGRE is a program of Autism Speaks and is supported, in part, by grant 1U24MH081810 from the National Institute of Mental Health to Clara M. Lajonchere (PI). This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (23591700 to A. Ayyappan and 23390288 to K. Nakamura). We thank Tae Takahashi for technical assistance.
Footnotes
Competing interests: As above for A. Ayyappan and K. Nakamura; none declared for all others.
Contributors: A. Ayyappan, I. Thanseem, K. Nakamura, T. Sugiyama, M. Tsujii, T. Yoshikawa and N. Mori designed the study. K. Yamada, Y. Iwayama, T. Toyota, Y. Iwata and K. Suzuki acquired the data; A. Ayyappan, I. Thanseem, K. Nakamura, K. Yamada, Y. Iwayama and T. Toyota analyzed it. A. Ayyappan, I. Thanseem and K. Nakamura wrote the article. All authors reviewed the article and approved its publication.
References
- 1.Kelleher RJ, III, Bear MF. The autistic neuron: Troubled translation? Cell. 2008;135:401–6. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 2.Munno DW, Syed NI. Synaptogenesis in the CNS: an odyssey from wiring together to firing together. J Physiol. 2003;552:1–11. doi: 10.1113/jphysiol.2003.045062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sperry RW. Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc Natl Acad Sci U S A. 1963;50:703–10. doi: 10.1073/pnas.50.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jontes JD, Phillips GR. Selective stabilization and synaptic specificity: a new cell-biological model. Trends Neurosci. 2006;29:186–91. doi: 10.1016/j.tins.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Kohmura N, Senzaki K, Hamada S, et al. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron. 1998;20:1137–51. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 6.Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–90. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 7.Frank M, Kemler R. Protocadherins. Curr Opin Cell Biol. 2002;14:557–62. doi: 10.1016/s0955-0674(02)00365-4. [DOI] [PubMed] [Google Scholar]
- 8.Tasic B, Nabholz CE, Baldwin KK, et al. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol Cell. 2002;10:21–33. doi: 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- 9.Ribich S, Tasic B, Maniatis T. Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proc Natl Acad Sci U S A. 2006;103:19719–24. doi: 10.1073/pnas.0609445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esumi S, Kakazu N, Taguchi Y, et al. Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat Genet. 2005;37:171–6. doi: 10.1038/ng1500. [DOI] [PubMed] [Google Scholar]
- 11.Belmonte MK, Allen G, Beckel-Mitchener A, et al. Autism and abnormal development of brain connectivity. J Neurosci. 2004;24:9228–31. doi: 10.1523/JNEUROSCI.3340-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jou RJ, Jackowski AP, Papademetris X, et al. Diffusion tensor imaging in autism spectrum disorders: preliminary evidence of abnormal neural connectivity. Aust N Z J Psychiatry. 2011;45:153–62. doi: 10.3109/00048674.2010.534069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoghbi HY. Postnatal neurodevelopmental disorders: Meeting at the synapse? Science. 2003;302:826–30. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]
- 14.Katori S, Hamada S, Noguchi Y, et al. Protocadherin-alpha family is required for serotonergic projections to appropriately innervate target brain areas. J Neurosci. 2009;29:9137–47. doi: 10.1523/JNEUROSCI.5478-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook EH, Leventhal BL. The serotonin system in autism. Curr Opin Pediatr. 1996;8:348–54. doi: 10.1097/00008480-199608000-00008. [DOI] [PubMed] [Google Scholar]
- 16.Chugani DC. Role of altered brain serotonin mechanisms in autism. Mol Psychiatry. 2002;7(Suppl 2):S16–7. doi: 10.1038/sj.mp.4001167. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura K, Sekine Y, Ouchi Y, et al. Brain serotonin and dopamine transporter bindings in adults with high-functioning autism. Arch Gen Psychiatry. 2010;67:59–68. doi: 10.1001/archgenpsychiatry.2009.137. [DOI] [PubMed] [Google Scholar]
- 18.Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–88. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrow EM, Yoo SY, Flavell SW, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–23. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iossifov I, Ronemus M, Levy D, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–99. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geschwind DH, Sowinski J, Lord C, et al. The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am J Hum Genet. 2001;69:463–6. doi: 10.1086/321292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–85. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- 23.Ranade K, Chang MS, Ting CT, et al. High-throughput genotyping with single nucleotide polymorphisms. Genome Res. 2001;11:1262–8. doi: 10.1101/gr.157801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewontin RC. On measures of gametic disequilibrium. Genetics. 1988;120:849–52. doi: 10.1093/genetics/120.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 26.Anney R, Klei L, Pinto D, et al. A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet. 2010;19:4072–82. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Risch N. Implications of multilocus inheritance for gene-disease association studies. Theor Popul Biol. 2001;60:215–20. doi: 10.1006/tpbi.2001.1538. [DOI] [PubMed] [Google Scholar]
- 28.Campbell DB, Sutcliffe JS, Ebert PJ, et al. A genetic variant that disrupts MET transcription is associated with autism. Proc Natl Acad Sci U S A. 2006;103:16834–9. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lerer E, Levi S, Salomon S, et al. Association between the oxytocin receptor (OXTR) gene and autism: relationship to Vineland Adaptive Behavior Scales and cognition. Mol Psychiatry. 2008;13:980–8. doi: 10.1038/sj.mp.4002087. [DOI] [PubMed] [Google Scholar]
- 30.Ma DQ, Rabionet R, Konidari I, et al. Association and gene-gene interaction of SLC6A4 and ITGB3 in autism. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:477–83. doi: 10.1002/ajmg.b.31003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sousa I, Clark TG, Holt R, et al. Polymorphisms in leucine-rich repeat genes are associated with autism spectrum disorder susceptibility in populations of European ancestry. Mol Autism. 2010;1:7. doi: 10.1186/2040-2392-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahlgren SO, Gillberg C. Symptoms in the first two years of life. A preliminary population study of infantile autism. Eur Arch Psychiatry Neurol Sci. 1989;238:169–74. doi: 10.1007/BF00451006. [DOI] [PubMed] [Google Scholar]
- 33.De Giacomo A, Fombonne E. Parental recognition of developmental abnormalities in autism. Eur Child Adolesc Psychiatry. 1998;7:131–6. doi: 10.1007/s007870050058. [DOI] [PubMed] [Google Scholar]
- 34.Morishita H, Murata Y, Esumi S, et al. CNR/Pcdhalpha family in subplate neurons, and developing cortical connectivity. Neuroreport. 2004;15:2595–9. doi: 10.1097/00001756-200412030-00007. [DOI] [PubMed] [Google Scholar]
- 35.Shimojima K, Isidor B, Le Caignec C, et al. A new microdeletion syndrome of 5q31.3 characterized by severe developmental delays, distinctive facial features, and delayed myelination. Am J Med Genet A. 2011;155A:732–6. doi: 10.1002/ajmg.a.33891. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Weiner JA, Levi S, et al. Gamma protocadherins are required for survival of spinal interneurons. Neuron. 2002;36:843–54. doi: 10.1016/s0896-6273(02)01090-5. [DOI] [PubMed] [Google Scholar]
- 37.Emond MR, Jontes JD. Inhibition of protocadherin-alpha function results in neuronal death in the developing zebrafish. Dev Biol. 2008;321:175–87. doi: 10.1016/j.ydbio.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 38.Weiner JA, Wang X, Tapia JC, et al. Gamma protocadherins are required for synaptic development in the spinal cord. Proc Natl Acad Sci U S A. 2005;102:8–14. doi: 10.1073/pnas.0407931101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hasegawa S, Hamada S, Kumode Y, et al. The protocadherin-alpha family is involved in axonal coalescence of olfactory sensory neurons into glomeruli of the olfactory bulb in mouse. Mol Cell Neurosci. 2008;38:66–79. doi: 10.1016/j.mcn.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 40.Fukuda E, Hamada S, Hasegawa S, et al. Down-regulation of protocadherin-alpha A isoforms in mice changes contextual fear conditioning and spatial working memory. Eur J Neurosci. 2008;28:1362–76. doi: 10.1111/j.1460-9568.2008.06428.x. [DOI] [PubMed] [Google Scholar]
- 41.Baumgarten HG, Grozdanovic Z. Psychopharmacology of central serotonergic systems. Pharmacopsychiatry. 1995;28(Suppl 2):73–9. doi: 10.1055/s-2007-979623. [DOI] [PubMed] [Google Scholar]
- 42.Chugani DC, Muzik O, Behen M, et al. Developmental changes in brain serotonin synthesis capacity in autistic and nonautistic children. Ann Neurol. 1999;45:287–95. doi: 10.1002/1531-8249(199903)45:3<287::aid-ana3>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 43.Gould GG, Hensler JG, Burke TF, et al. Density and function of central serotonin (5-HT) transporters, 5-HT1A and 5-HT2A receptors, and effects of their targeting on BTBR T+tf/J mouse social behavior. J Neurochem. 2011;116:291–303. doi: 10.1111/j.1471-4159.2010.07104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pagnamenta AT, Wing K, Sadighi Akha E, et al. A 15q13.3 microdeletion segregating with autism. Eur J Hum Genet. 2009;17:687–92. doi: 10.1038/ejhg.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang K, Zhang H, Ma D, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–33. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwab SG, Eckstein GN, Hallmayer J, et al. Evidence suggestive of a locus on chromosome 5q31 contributing to susceptibility for schizophrenia in German and Israeli families by multipoint affected sib-pair linkage analysis. Mol Psychiatry. 1997;2:156–60. doi: 10.1038/sj.mp.4000263. [DOI] [PubMed] [Google Scholar]
- 47.Pedrosa E, Stefanescu R, Margolis B, et al. Analysis of protocadherin alpha gene enhancer polymorphism in bipolar disorder and schizophrenia. Schizophr Res. 2008;102:210–9. doi: 10.1016/j.schres.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]