Abstract
Caveolin-1 is an essential component of membrane caveolae. It is an important regulator of cellular processes such as signal transduction and endocytosis. We report here, for the first time, that caveolin-1 is a target of the K-RAS oncogene in colon carcinogenesis. Caveolin-1 is induced in colon cancer cells and in human colon tumor samples, in response to K-RAS activating mutations. An activated K-RAS oncogene transcriptionally induces caveolin-1 expression in human colon cancer cells and this effect is not restricted to the type of activating K-RAS mutation. Inhibition of the P-I3 Kinase-AKT pathway, but not the ERK MAPK pathway, both important K-RAS effectors, leads to a decrease in caveolin-1 expression indicating that the AKT pathway is involved in caveolin-1 expression in response to an activated K-RAS. Increased AKT signaling induces caveolin-1 expression by increasing the activity of the transcription factor, Sp1. Interestingly; caveolin-1 depletion alters K-RAS-dependent signaling by decreasing Grb2-SOS activity. Consistent with these finding, caveolin-1-depleted cells shows decreased migration in vitro. However, caveolin-1 over-expression by itself does not increase migration whereas an activated Src can increase migration in a caveolin-1-dependent manner. This increased migration is highly dependent on the RhoA GTPase, indicating that an activated K-RAS modulates migration in part via caveolin-1 induction, and increasing RhoA activity via phospho-caveolin-1. Our findings indicate that K-RAS regulates both caveolin-1 expression and other factors affecting caveolin-1 functions in colon cancer-derived cell migration.
Keywords: Colon cancer, K-RAS, Caveolin-1, invasion and migration, AKT
Introduction
Caveolin-1, a component of membrane caveolae, is a multi-functional protein, governing signal transduction, membrane transport, and lipid homeostasis 1. The role of caveolin-1 in cancer development and progression has been a subject of controversy among biologists. Initial studies in murine fibroblast (NIH-3T3) cells showed that loss of caveolin-1 was sufficient to induce anchorage-independent growth and tumor formation in SCID mice 2. Similar studies showed that expression of oncogenes like K-RAS, H-RAS, V-ABL and SRC were sufficient to reduce caveolin-1 expression in NIH-3T3 cells 3–4. Engelman et al showed that the CAV1 gene can be mapped to the D7S522 locus (chromosome 7q31.1), a site commonly deleted in human cancers 5. These findings strengthened the hypothesis that caveolin-1 functions as a potential tumor suppressor and its loss aids in tumorigenesis. With respect to its function during cancer development, the tumor suppressive activities of caveolin-1 have been attributed to its ability to bind to signaling molecules via its scaffolding domain, and negatively regulate their activity. Indeed, re-expression of caveolin-1 in transformed murine fibroblasts has been shown to be sufficient to down-regulate signaling via the Ras-Raf-Erk pathway 6. Consistent with these findings, caveolin-1 is down-regulated in several cancers such as breast and ovarian 7.
However, the other domains present in caveolin-1 can nullify its tumor suppressive functions. In human tumors, caveolin-1 seems to play a tumor-promoting role in certain types of cancers. In prostate cancer, caveolin-1 can maintain activated AKT by inhibiting serine/threonine phosphatases PP1 and PP2A 8. Caveolin-1 has the ability to be secreted by prostate cancer cells after phosphorylation at residue Ser80, and secreted caveolin-1 can act as an autocrine growth factor 9. During the later stages of cancer, transformed cells become resistant to standard chemotherapeutic agents and acquire the multi-drug resistance (MDR) phenotype. This phenomenon is associated with an increase in expression of P-glycoprotein (P-gp). P-gp has been shown to be localized in caveolae of MDR-cells, implicating these membrane micro-domains in conferring the MDR phenotype 10. In line with these observations, an increased expression of caveolin-1 has been reported to be associated with increased metastasis in prostate cancer. Thus caveolin-1 can have tumorigenic as well as tumor-suppressive properties.
With regards to the colon, certain groups have reported that caveolin-1 is down-regulated in colon cancer tissue, as compared to normal colon tissue 11. Other studies have revealed that caveolin-1 is over-expressed in adenocarcinoma of the colon 12–13. Thus, there is still a major conflict regarding caveolin-1 expression during colon cancer progression.
We have previously demonstrated that caveolin-1 is induced by the APC tumor suppressor gene 14. In this study, we have shown that caveolin-1 is a transcriptional target of the K-RAS oncogene. Acquisition of K-RAS mutations is a late event in colon cancer progression 15. K-RAS is commonly mutated at codon 12 or 13, or in more rare instances, codon 61; 16–17. Interestingly, caveolin-1 increases K-RAS activity through increased SOS activation and migration through the activation of the RhoA-ROCK pathway. Studies regarding caveolin-1 expression in human colon tumor samples have not accounted for mutations in the tumor samples. Our findings demonstrate the upregulation of caveolin-1 in colon tumor cells and tissue samples, harboring K-RAS mutations and provide a possible mechanism by which the K-RAS/Caveolin-1 pathway can aid in colon cancer progression.
Materials and Methods
Cell Culture
The HCT116 cells (with a G13D mutation in one of the K-RAS alleles) was obtained from American Type Culture Collection (ATCC) and maintained in DMEM medium supplemented with 10% FBS (Fetal Bovine Serum) and 1% Penicillin-Streptomycin. The Hkh2 cells, which are a clone of HCT116 cells wherein the activated K-RAS oncogene has been disrupted by homologous recombination, was a kind gift from Drs. Shirasawa and Sasazuki 18 and maintained in DMEM supplemented with 10% FBS, 1% Penicillin-Streptomycin and 600 μg/ml G418. The Caco2 colon cancer cells, transfected with pcDNA3.0 empty vector (Caco/Neo#3) or an activated K-RAS (G12V) expression vector (clones Kras#6 and Kras#26), were developed in our laboratory and have been previously characterized 19. The HCT116-Mock and Caveolin-1 antisense cells were a kind gift of Drs. Cadvallo-Medved and Sloane 20. The Caco2-Mock and Caco2-caveolin-1 cells have been previously described 21. All cells were grown at 37 °C in a humidified incubator with 5% carbon dioxide.
Reagents and antibodies
All chemicals and reagents were of the highest grade. LY294002 (PI-3-Kinase inhibitor) was obtained from Calbiochem, San Diego, CA. Dimethyl sulfoxide (DMSO) were purchased from Sigma, St. Louis, MO. G418 sulfate was purchased from CellGro. Lipofectamine 2000, Hygromycin B and all media were from Invitrogen, Carlsbad, CA. A list of all antibodies used in this study is mentioned in Supplementary Table 1. All secondary antibodies were purchased from Santa Cruz Biotechnology, Santa Cruz, CA. Pan-Src kinase inhibitor PP2 was purchased from EMD Chemicals, Gibbstown, NJ
cDNA Microarray Study
cDNA microarray analysis was performed as previously described 22. Briefly, HCT116 and Caco2 isogenic cells, cells were plated for 48 hours after which total cellular RNA was extracted using the Qiagen RNEASY Kit (Catalog Number: 74104) obtained from Qiagen, Valencia, CA, using manufacturer’s instructions. The RNA was quantified and used for cDNA microarray analysis.
Tissue immunohistochemistry
Colon cancer tissues were obtained from patients with metastatic colorectal cancer (Stage IV colorectal cancer). All the patients were treated at the Oncology Unit of Ospedale Santa Chiara in Pisa, Italy and expressed their written informed consent to molecular analyses. All tissues were obtained at colonoscopy in accordance with the Institutional Review Board regulations of the Department of Oncology, University of Pisa. Formalin-fixed, paraffin-embedded blocks of colorectal cancer specimens were analyzed for the presence of K-RAS mutation. Caveolin-1 expression was analyzed in samples with WT and mutant K-RAS using anti-caveolin-1 antibody from Cell Signaling Technologies, using standard immunohistochemistry (IHC) procedure. Immunoreactivity was determined as: Absent (no or weak staining in 10% or fewer cells), Moderate (moderate staining in 10%–25% of cells), Strong (strong staining in greater than 25% cells). K-RAS mutational status of the tumor samples used in the study are shown in Supplementary Table 2.
Western Blotting and Immunoprecipitation
Cells (2 × 106 cells/100 mm plate) were plated for 48 hours. They were lysed in radio-immunoprecipitation assay (RIPA) buffer with protease inhibitors (10 μg/ml Aprotinin, 10 μg/ml phenyl-methyl-sulfony chloride (PMSF) and 50 μM sodium orthovanadate). Samples were kept on ice for 30 minutes, followed by centrifugation at 14,000 rpm for 10 minutes. Supernatants were collected and protein concentration was determined with the Bio-Rad DC protein assay (Bio-Rad, Hercules, CA). Fifty micrograms of protein were resolved on a 12.5% SDS-PAGE gel and transferred overnight to a Hybond-C nitrocellulose membrane, at 4 °C. The next day, the membrane was blocked in Blotto A [5% non-fat dry milk in Tris-buffered saline with 0,05% Tween-20 (TBST)] for 1 hour at room temperature. Membranes were probed with antibodies in blocking buffer for 2 hours at room temperature, or overnight at 4 °C, as described in table 3. Alternately, primary antibodies were diluted in 5% bovine serum albumin (BSA) in TBST. After washing with TBST three times, the membrane was probed with horse-radish peroxidase (HRP)-conjugated secondary antibody, washed and protein was detected with an enhanced chemiluminescence detection reagent (Amersham). All blots were then stripped with Pierce Restore Western Blot Stripping Buffer (Pierce, Rockford, IL) as described by the manufacturer and reprobed with appropriate protein loading controls. For immunoprecipitation, whole cell extracts of indicated cell lines were precleared with normal rabbit serum and Protein A/GPlus-Agarose (Santa Cruz Biotechnology, Inc.) for 1 hour at 4°C. CAV-1 was immunoprecipitated from precleared lysates with polyclonal CAV-1 antibody (Santa Cruz Biotechnology, Inc.) or Sp1 antibody (Cell Signaling, Danvers, MA) overnight at 4°C, in the presence of protein A/GPlus-Agarose. Immunoprecipitated beads were washed four times, and then boiled in SDS sample buffer, separated by SDS-PAGE, and immunoblotted with antibodies as indicated in Figure Legends.
Chromatin Immuniprecipitation (ChIP) Assay
Cells (2 × 105 cells/100 mm plate) were plated and next day, transfected with respective plasmids. Medium was changed 24 hours after transfection. Cells were harvested 28 hours after transfection and ChIP assay was carried out using the ChIP assay kit (Catalog No: 53006) from Active Motif, Carlsbad, CA. The antibody used for ChIP analysis of Sp1 was from Cell Signaling, Danvers, MA.
Q-RT-PCR
For total cellular RNA extraction, cells (2.5 × 106) were plated in 100 mm plates. Cells were harvested by trypsinization after 48 hours of plating. Total cellular RNA was extracted using the Qiagen RNAEASY Kit (Catalog Number: 74104) obtained from Qiagen, Valencia, CA, using manufacturer’s instructions. Reverse transcription was performed on 1 μg of total RNA with random primers and the Reverse Transcription System kit (Promega Corp., Madison, WI). The cDNA template (100 ng) obtained from the above reaction was amplified using specific primers and puReTaq™Ready-To-Go™PCR Beads (Amersham Biosciences Corp., Piscataway, NJ). For caveolin-1 RT-PCR analysis, usually 200 ng of cDNA (for the HCT116 cells) and 500 ng of cDNA (for the Caco2 derived cells) were used as template. The PCR products were then resolved on 2% agarose gels and photographed using the Kodak 1D Imaging software. PCR primers used, along with their Tm and amplicon sizes for various genes were: Caveolin-1 Forward 5′ TCA ACC GCG ACC CTA AAC ACC 3′; Caveolin-1 Reverse 5′ TGA AAT AGC TCA GAA GAG ACA T 3′. Caveolin-1 Tm=60°C, amplicon size is 561 bp. GAPDH Forward 5′ TGGTATCGTGG-AAGGACTCATGAC3′, GAPDH Reverse 5′ AGTCCAGTGAGCTTCCCGTTCAGC3′. GAPDH Tm=59°C, amplicon size is 198 bp All PCR primers were synthesized through Invitrogen Custom Primer Synthesis.
Plasmids and Transfection
The human caveolin-1 promoter region, corresponding to −737 to −37 in the promoter region, cloned into the PGL2 vector was a kind gift from Dr. Vijay Shah 23. The constitutively active (MYR) AKT and dominant negative (DN) AKT plasmids were obtained from Dr. M. J. Quon, National Institutes of Health, Bethesda, MD 24. The Renilla-TK plasmid was purchased from Promega, (Madison, WI) and used as a transfection efficiency control in all promoter-reporter transfection experiments. For transfection with promoter-reporter constructs, 5 × 105 cells were plated per well, in 6-well plates. The next day, cells were transfected with Lipofectamine 2000 transfection reagent, as described by the manufacturer. Typically, cells were transfected with 2 μg of promoter and 0.01 μg of Renilla-TK vector. For studies with DN-AKT and MYR-AKT, 1.5 μg of MYR/DN AKT was transfected with 2 μg of promoter and 0.01 μg of Renilla-TK vector. The next day, the cells were lysed in Passive Lysis Buffer from the Dual Luciferase Assay kit, Promega, Madison, WI and dual luciferase activity was measured, as described by the manufacturer.
The caveolin-1 expression vector (pCINeo-Caveolin-1) was a kind of Dr. Eric Smart, University of Kentucky 25. Caveolin-1 Y14F was generated from the parent plasmid using the QuickChange Site-directed Mutaganesis Kit from Stratagene, La Jolla, CA. The Src plasmids were a kind gift from Dr. Sara Courtneidge, from University of California, San Diego 26. The RhoA constructs (RhoA T19N – Plasmid Number: 15901 and RhoA Q63L – Plasmid Number: 15900) were obtained from Addgene, Cambridge, MA.
Caveolin-1 and Sp1 siRNA experiments
HCT116 and Caco-2 isogenic cell lines were transfected with caveolin-1 siRNA purchased from Dharmacon, and used for migration assays. Dharmacon siGENOME SMARTpool anti-CAV-1 siRNA, anti-Sp1 siRNA and ON-TARGET plus siCONTROL™ negative control siRNA (catalog numbers L-003467-00, L-026959-00 and D-001820-01, respectively) were prepared as per manufacturer’s instructions. Briefly, the lyophilized sequences were resuspended in the manufacturer’s siRNA Suspension Buffer to a concentration of 20 μM. The suspension was incubated at 90°C for 1 minute, then at 37°C for 60 minutes to disrupt aggregates formed during lyophilization. All siRNA sequences were used at a final concentration of 75nM during cell transfection. Sixteen to eighteen hours preceding transfection, 2.0×105 cells/well were plated in 6-well plates in DMEM with 10% fetal bovine serum and no antibiotics. Just prior to transfection, cells were rinsed 3x with saline. Cells were then incubated in Opti-MEM with siRNA and Lipofectamine2000 (Invitrogen) (12 μl/well) for 6 hours. After the 6 hours, an equal volume of Opti-MEM plus 10% fetal bovine serum was added to each well. All experiments were performed 48 hours after transfection.
Migration and Invasion Assays
The cell migration assays were performed with BIOCOAT® control cell culture inserts (BD Biosciences Discovery Labware, Bedford, MA). Inserts were coated with 2 μg/well of laminin (Sigma-Aldrich, Life Science Research, St. Louis, MO). Cell suspensions were prepared at a concentration of 5.0x105 cells/ml in serum-free medium. Two hundred microliters of the cell suspensions were plated in the inner chamber of the coated insert in a 24-well tissue plate. Five hundred microliters of complete growth medium was added to the outer chamber of the insert. Twenty hours after plating, the cells that migrated through the surface of the membrane were stained with 100 μl/well of staining solution (0.5% crystal violet, 20% methanol, 80% water) for 1 minute. The stained cells were dissolved in 200 μl 0.1 M citric acid in a 96 well plate. The plate was placed on a high speed titre plate shaker for 5 minutes. One hundred and fifty microliters of the supernatant was then transferred to a new well and read at 560 nm 51 on an EL800 Universal Microplate Reader (Bio-Tek Instruments, Inc.,Winooski, VT). Migration experiments were carried out in sextuplet. The Matrigel cell invasion assays were performed similar to the migration assays on prepared 24-well plate sized Matrigel inserts (BD Biosciences Discovery Labware, Bedford, MA).
Lipid raft fractionation
Lipid raft fractionation was carried out by using a detergent-free, alkaline lysis method as described previously 27. Briefly, HCT116 or Caco2 cells (7 × 106cells/150 mm plate) were plated for 48 hours. Each plate was then lysed with 2 ml of 500 mM sodium carbonate (pH-11.0). The lysate was sonicated for three 20-s bursts using a SONIC Vibra Cell sonicator. The lysate was then adjusted to 45% sucrose by mixing with equal volumes of 90% sucrose prepared in Mes-buffered saline (MBS - 25 mM Mes, pH 6.5, 0.15 M NaCl), and placed at the bottom of an ultracentrifuge tube. A 5–35% discontinuous sucrose gradient was formed above (4 ml of 5% sucrose/4 ml of 35% sucrose; both in MBS containing 250 mM sodium carbonate) and centrifuged at 39,000 rpm for 16 hours in an SW40-Ti rotor (Beckman Instruments, Palo Alto, CA). A light-scattering band at the 5–35% sucrose interface was observed. This fraction contains caveolin-1/lipid raft proteins. Twelve 1 ml fractions were collected from top to bottom of the tube. For detection of caveolin-1 in the fractions, equal volume from each fraction were loaded on a 12.5 % SDS-PAGE gel and visualised as described in Western Blotting.
Ras and RhoA Activity Assay
Activated RAS and RhoA levels were measured using the RAS activation assay kit (Catalog No: 17-218) and the RhoA activation assay kit (Catalog No: 17-294) from Upstate, Millipore, Billerica, MA. For all these assays, cells for grown for 48 hours under regular tissue culture conditions and processed as per the manufacturer’s instructions. However, since activity of these GTPases are highly dependent on the ECM used for the assay 28–29, we have deliberately measured basal levels of the active GTPases on non-ECM coated plates.
Statistical Analysis
All statistical analysis was carried out using Microsoft Excel Software and the paired student’s t test was used to establish significance. In all figures shown, *p<0.05. Evaluation of caveolin-1 staining by immunohistochemistry was done using Fisher’s Exact test.
Results
An activated K-RAS oncogene induces caveolin-1 expression in human colon tumor -derived cells
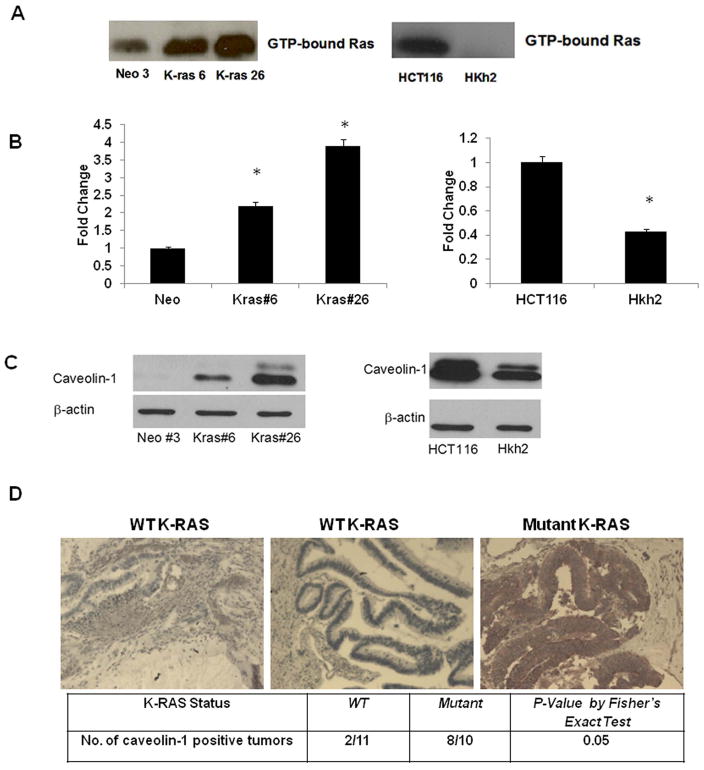
The Caco2 cells expressing an activated K-RAS oncogene have been previously characterized in our laboratory 19. Here we performed a RAS activity assay to confirm that the K-RAS transfected clones have higher GTP-bound (active) K-RAS protein. As shown in figure 1A (left panel), the K-RAS clones have significantly increased levels of GTP-bound K-RAS. In a cDNA microarray study carried out in our laboratory, caveolin-1 was found to be a transcriptional target of the activated K-RAS oncogene (Supplementary Table 3). Validation of the micro array results was then performed with quantitative RT-PCR and immunoblotting. Caveolin-1 mRNA and protein levels were significantly increased in the K-RAS expressing clones, as compared to the vector control cells (Figure 1B, left panel and 1C, left panel). The cell culture experiments indicated that an activating mutation in the K-RAS oncogene induces caveolin-1 expression. We confirmed whether the effect of activated K-RAS observed in the Caco2 cell system was specific to a certain codon in the K-RAS gene. In order to test this, another isogenic cell line system was employed. The HCT116 cells are a colon adenocarcinoma cell line, which possess a mutation in the K-RAS gene at codon 13 (G13D) 18. The Hkh2 cells are derived from the HCT116 cells, wherein the activated K-RAS oncogene has been disrupted by homologous recombination 18. Again, a RAS activity assay revealed that loss of the oncogene leads to a decrease in GTP-bound RAS in Hkh2 cells (Figure 1A, right panel). A cDNA microarray analysis using these two cell lines showed that the Hkh2 cells had lower caveolin-1 mRNA levels, as compared to HCT116 cells (Table 1). These results were validated at mRNA and protein levels (Figure 1B and 1C, right panels). Caveolin-1 is expressed as a longer (caveolin-1α) and a shorter isoform (caveolin-1β) that are translated from methionine 1 and 32 respectively leading two two bands on a Western blot 30. These studies confirm that a mutant K-RAS induces caveolin-1 expression in cell culture and in human tumor samples and that caveolin-1 overexpression is observed in the presence of both K-RASG12V and K-RASG13D mutations.
Figure 1. An activated K-RAS oncogene influences caveolin-1 expression in the colon cancer.
1A. -RAS activation in mutant K-RAS expressing Caco/Kras clones (left panel) and in HCT116 and Hkh2 cells (right panel) grown to sub-confluency for 48 hours. RAS activity assay was carried out, as described in Materials and Methods. Levels of GTP-bound (active) RAS is shown. Each figure is representative of two independent experiments
1B. Quantitative RT PCR analysis of CAV-1 mRNA levels in Caco/Kras clones (left panel) and in HCT116-Hkh2 cells (right panel) grown to sub-confluency for 48 hours. Total RNA was extracted and processed for PCR analysis as described in Materials and Methods. Left panel-Each figure is representative of three independent experiments (* = p <0.05 by t-test).
1C. Western blot analysis of caveolin-1 protein levels in Caco/Kras clones (left panel) and HCT116-Hkh2 cells (right panel) grown to sub-confluency for 72 hours and lysed in RIPA buffer. Lysates were resolved on a 12.5% SDS-PAGE gel and probed for caveolin-1 (21–24 kDa). Data shown are representative of three independent experiments. Note the double bands on the Western blot corresponding to a longer (caveolin-1α) and a shorter isoform (caveolin-1β).
1D. Caveolin-1 expression in human colon tumors samples with WT (N=10) and activated K-RAS (N=10) by IHC. Caveolin-1 immunoreactivity was determined as: Absent (no or weak staining in 10% or fewer cells), Moderate (moderate staining in 10%-25% of cells), Strong (strong staining in greater than 25% cells). P<0.05 by Fisher’s exact test. Representative stained samples with WT and activated K-RAS are shown.
We extended our analysis to human colon tumor samples and analyzed the expression of caveolin-1 in 10 colon tumor samples harboring WT and 10 samples harboring mutant K-RAS (K-RAS mutation status of samples is shown in Supplementary Table 2). The tumor samples were from patients with late stage disease (Stage IV colorectal cancer). We focused our analysis on samples from Stage IV patients since K-RAS mutations occur late in the disease progression and are often associated with metastatic disease 31. Interestingly, we found that 8 out of 10 tumor samples with mutant K-RAS had high levels of caveolin-1 expression as against 2 sample out of 11 in which the WT allele was present (p <0.05 by Fisher’s Exact test) (Figure 1D). As seen in Figure 1D, the sample with WT K-RAS shows few caveolin-1 positive cells. On the other hand, the tumor samples with mutant K-RAS showed intense staining of caveolin-1. To confirm whether the increased caveolin-1 expression we observed in the colon tumor samples corresponded to increased levels of caveolin-1 mRNA, we analyzed caveolin-1 mRNA levels in tumors samples harboring WT and mutated K-RAS alleles and found that caveolin-1 mRNA was significantly increased (p <0.05 by paired T-test) in samples with K-RAS mutations (Supplementary Figure 1).
K-RAS-dependent caveolin-1 upregulation occurs via the AKT pathway
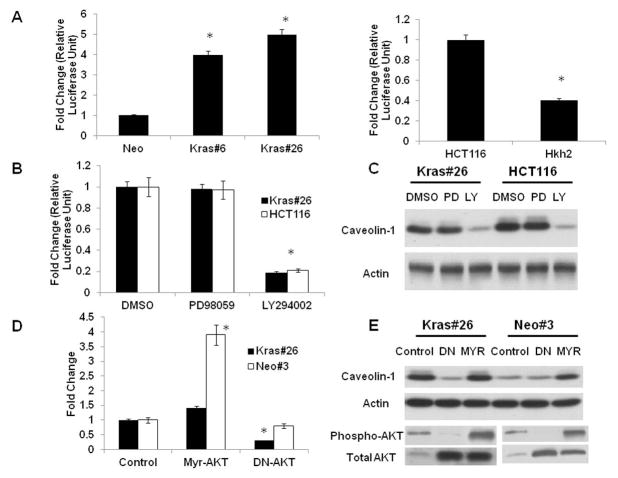
In order to assess whether the effect observed was because of increased transcription versus other mechanisms, the Caco2 isogenic clones were transfected with a caveolin-1 promoter reporter construct. As shown in Figure 2A, an activated K-RAS oncogene increases caveolin-1 promoter reporter activity in the Caco2 isogenic cell system. Similarly, we wanted to assess whether the decrease in caveolin-1 expression in the Hkh2 cells was a result of decreased transcription. The Hkh2 cells showed a dramatic reduction in caveolin-1 promoter reporter activity as compared to the HCT116 cells (Figure 2A). Activation of the RAS oncogene can result in upregulation of several signaling pathways in transformed cells, which in turn can have different outcomes. In order to assess the role of these pathways in K-RAS mediated caveolin-1 up-regulation in Caco-Kras26 and HCT116 cells, we used PD98059 to inhibit the Raf/Mek/Erk pathway and LY294002 to inhibit the P-I3Kinase/AKT pathway. We then determined caveolin-1 promoter-reporter activity, as well as the levels of caveolin-1 mRNA (Supplementary Figure 2A) and protein levels in PD98059 and LY294002 treated cells. While PD98059 did not affect caveolin-1 promoter-reporter activity, treatment with LY294002 diminished promoter reporter activity (Figure 2B), indicating that caveolin-1 is regulated by the P-I3 kinase pathway at the level of transcription. As shown in Figure 2C, there is a decrease in caveolin-1 protein levels in LY294002-treated cell lines indicating that the PI-3 Kinase/AKT signaling pathway is the main K-RAS effector pathway for caveolin-1 upregulation in colon cancer cells expressing an activated K-RAS oncogene. To further confirm the pharmacological findings, we transfected the Caco-Kras26 cells with a dominant negative (DN) AKT construct. This construct has a mutation in the ATP-binding site, K179A, and acts in a dominant inhibitory manner 24. As shown in Figure 2D, there is a significant reduction in caveolin-1 mRNA (left panel) and protein (right panel) levels in the presence of the dominant negative AKT construct in the CacoKras#26 cells. Similarly, transfection of the Caco-Neo3 cells with a constitutively active (MYR) AKT, which produces a high level of AKT activity 24 leads to the induction of caveolin-1 mRNA and protein levels (Figure 2D, left and right panels)). Similar results were obtained in the HCT116 and Hkh2 cells (Supplementary Figure 2B), thus confirming that caveolin-1 is transcriptionally induced by the AKT pathway.
Figure 2. An activated K-RAS activated caveolin-1 transcription via the AKT pathway.
2A. Caveolin-1 promoter activity in Caco-2 and HCT116 isogenic systems.
Caco/Kras clones and HCT116-Hkh2 (0.5 × 106 cells/well in 6-plate) were grown for 24 hours and transfected with a caveolin-1 promoter reporter. Forty eight hours after transfection, cells were lysed and luciferase activity measured as described in Materials and Methods. Results are the average of three independent transfections. *P<0.05 by t-test.
2B. Effect of MAPK and P-I3 kinase inhibitors on caveolin-1 promoter activity in Caco-2 and HCT116 isogenic systems.
CacoKras#26 and HCT116 cells (0.5 × 106 cells/well in 6-plate) were grown for 24 hours and transfected with a caveolin-1 promoter reporter. Twenty four hours after transfection, cells were treated with inhibitors (50 μM of PD98059/Ly294002) for an additional 24 hours, lysed and luciferase activity measured as described in Materials and Methods. Results are the average of three independent transfections. *P<0.05 by t-test.
2C. Western blot analysis of caveolin-1 protein level in CacoKras#26 and HCT116 cells after treatment with ERK and P-I3 kinase inhibitors. Cells were grown in DMSO (control) or PD98059/LY294002 (50 μM each) for 48 hours. Cells were lysed in RIPA buffer and lysates were resolved on a 12.5% SDS-PAGE and probed for caveolin-1. Blot is representative of three independent experiments.
2D and 2E. Caveolin-1 transcript and protein levels are influenced by AKT activity.
Caco/Kras clones (0.5 × 106 cells/well in 6-plate) were grown for 24 hours and transfected with a dominant negative (DN) or an activated (Myr) AKT construct. The next day, fresh medium was added. After 48 hours, RNA and protein were extracted and caveolin-1 levels were determined by quantitative RT-PCR (2D) and Western blotting (2E) as described in Materials and Methods. Western blot depicting expression of phospho-AKT and total AKT protein are shown in the lower panel of Figure 2E. Figure is representative of three independent experiments *P<0.05 by t-test.
AKT increases caveolin-1 transcription through Sp1
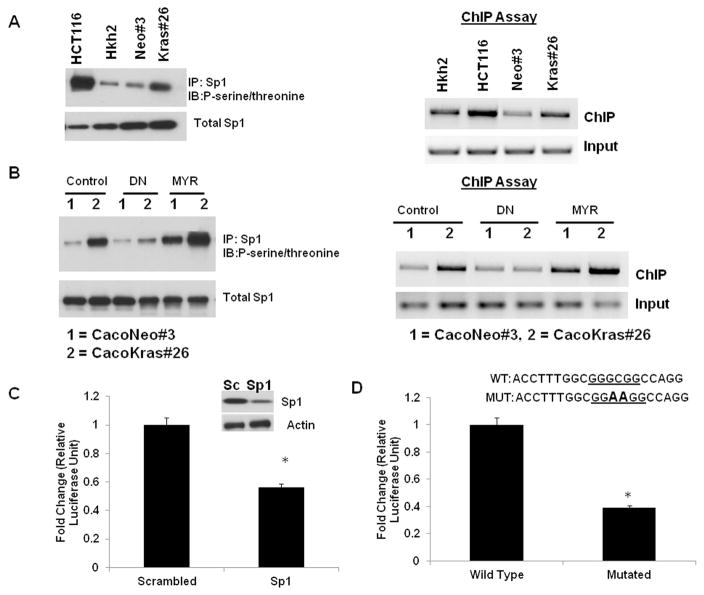
In order to elucidate the mechanism by which AKT up-regulates caveolin-1, we sought to determine which transcription factors are activated in response to AKT signaling. One of the key transcription factors whose DNA binding activity is increased by AKT-dependent phosphorylation is Sp1 32–33. The human caveolin-1 promoter (AF019742) has a consensus Sp1 binding site at the −148 position from the transcriptional start site 34. To assess the role of Sp1 in AKT-dependent caveolin-1 up-regulation, we first determined the levels of phospho-Sp1 in the two independent isogenic cell line systems. As seen in Figure 3A – left panel, both the HCT116 cells and the Caco-Kras 26 cells have higher ratios of phospho-Sp1 to total Sp1 protein levels, as compared to their wild-type K-RAS counterparts respectively. To determine whether increased phospho-Sp1 corresponds to increased binding to the caveolin-1 promoter site, we determined the extent of Sp1 binding using Chromatin Immuno-Precipitation (ChIP) assay with caveolin-1 promoter-specific primers. As seen in Figure 3A (right panel), there is increased Sp1 binding to the caveolin-1 promoter region in the HCT116 and the Caco-Kras26 cells. While these findings suggested that AKT might activate Sp1, to conclusively determine the contribution of the AKT-Sp1 pathway, we established whether activation of the AKT pathway in the Caco-Neo cells or inhibition of the AKT pathway in the K-RAS expressing clone can alter caveolin-1 expression. Introduction of a constitutively active AKT (Myr-AKT) increases both phospho-Sp1 levels (Figure 3B-left panel) and Sp1 binding to the caveolin-1 promoter (Figure 3B-right panel), in the Caco-Neo cells. Conversely, inhibition of the AKT pathway in the Caco-Kras cells decreases levels of phosphorylated Sp1 and promoter-bound Sp1, indicating that AKT activity is sufficient to increase Sp1-DNA binding to the caveolin-1 promoter. In line with these findings, use of Sp1 siRNA or mutation of the Sp1 binding site in the caveolin-1 promoter, is sufficient to decrease caveolin-1 promoter-driven luciferase activity (Figure 3C left and right panels), confirming that AKT-dependent Sp1 phosphorylation is a key event in K-RAS-dependent up-regulation of caveolin-1.
Figure 3. AKT induces caveolin-1 expression via Sp1.
3A. Left Panel: The levels of total, phosphorylated (left panel-IP and Western blot) and caveolin-1 promoter-bound (right panel-Chip assay) Sp1 protein in HCT116 and Caco-2 isogenic systems. HCT116/Hkh2 cells and CacoNeo/Kras#26 were grown for 48 hours and lysed in RIPA buffer. Sp1 protein was immunoprecipitated from the lysates using an anti-Sp1 antibody. Immunoprecipitates were resolved on a 10% SDS-PAGE gel and immunoblotted for anti-phospho-serine/threonine antibody. Right Panel: For ChIP experiments, chromatin prepared from 48-hour cultures of HCT116/Hkh2 cells and CacoNeo/Kras#26 was immunoprecipitated with anti-Sp1 antibody. Caveolin-1 promoter-specific primers were used to amplify the precipitated DNA as described in Materials and Methods. Figure is representative of two independent experiments
3B. Effect of constitutively active (MYR) and dominant negative (DN) AKT on the total, phosphorylated (left panel – IP Western) and bound (right panel-Chip assay) Sp1 protein in Caco-2 isogenic system.
Left Panel: CacoNeo/Kras#26 were transfected with the indicated constructs, grown for 48 hours and lysed in RIPA buffer. Sp1 protein was immunoprecipitated from the lysates using an anti-Sp1 antibody. Immunoprecipitates were resolved on a 10% SDS-PAGE gel and immunoblotted for anti-phospho-serine/threonine antibody. Right Panel: For ChIP experiments, chromatin prepared from 48-hour cultures of HCT116/Hkh2 cells and CacoNeo/Kras#26 was immunoprecipitated with anti-Sp1 antibody. Caveolin-1 promoter-specific primers were used to amplify the precipitated DNA as described in Materials and Methods.
3C and 3D. Suppression of caveolin-1 promoter activity in the presence of Sp1 siRNA (3C) or mutation in Sp1 binding site (3D).
3C - CacoNeo/Kras#26 were transfected with scrambled (Sc) or Sp1 siRNA and the caveolin-1 promoter-reporter construct. Forty eight hours after transfection, cells were assayed for promoter-reporter activity *P<0.05 by t-test. Figure is representative of two independent experiments
3D -CacoKras#26 were transfected with caveolin-1 promoter-reporter constructs containing either wild-type (WT Sp1) and mutated Sp1 (mut Sp1) sites. Forty eight hours after transfection, cells were assayed for promoter-reporter activity. *P<0.05 by t-test. Figure is representative of two independent experiments
Caveolin-1 regulates K-RAS activity and is required for K-RAS signaling of migration in colon cancer cells
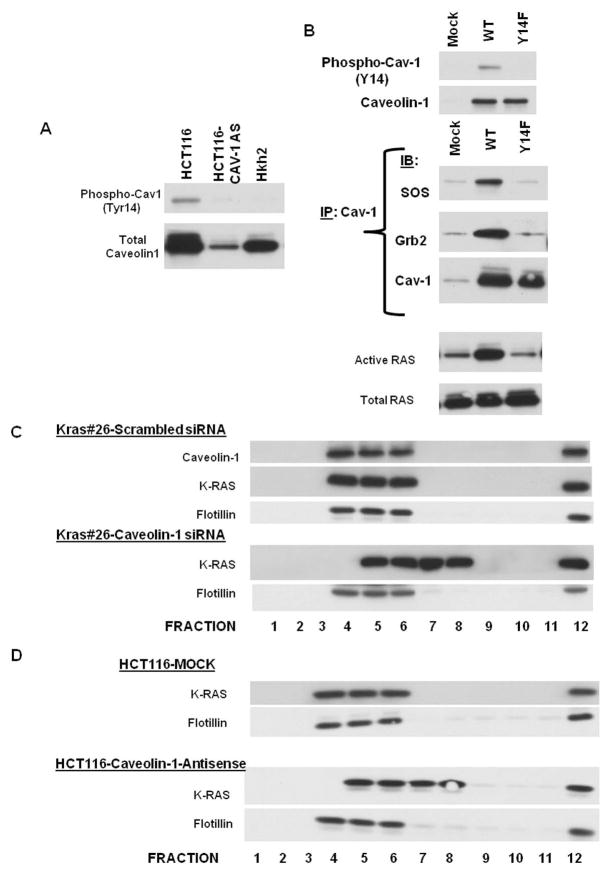
Caveolin-1 expression is required to maintain correct sub-cellular localization of H-RAS and loss of caveolin-1 disrupts H-RAS – membrane localization in mouse embryo fibroblasts isolated from mice with knockout of caveolin-1 gene 35. Similarly, we wanted to determine whether caveolin-1 expression is required for the activity of K-RAS in colon cancer cells. Caveolin-1 is tyrosine phosphorylated at tyrosine residue 14 and can serve as an adaptor for signaling molecules with SH2 binding domains such as Shc and Grb2 36–37. As seen in Figure 4A, phospho-caveolin-1 levels are elevated in the HCT116 cells, as compared to the HCT116-Cav-1-AS cells, and the Hkh2 cells, suggesting that phospho-caveolin-1 levels might be a regulator of K-RAS activation. In order to determine whether phospho-caveolin-1 expression sustains RAS signaling, we introduced in to HCT116-Cav-1 AS cells either a full-length caveolin-1 expression construct (WT) or a mutant form of caveolin-1 that is deficient in tyrosine phophorylation at residue 14 (Caveolin-1 Y14F) and assayed for the association of caveolin-1 with the two signaling molecules Grb2 and SOS1. Grb2 is an adaptor protein with an SH2 domain that binds to tyrosine-phosphorylated proteins and recruits SOS, the RAS GEDF that is responsible for increasing the activity of RAS proteins. As seen in Figure 4B, while introduction of a full-length caveolin-1 protein was sufficient to form a complex of Grb2 and SOS, indicating the recruitment of SOS1 to the membrane, this complex was not seen in the cells expressing caveolin-1 Y14F. Introduction of caveolin-1 in Caco2 cells (that do not express caveolin-1 and have a WT K-RAS) leads to a similar activation of Grb2 and SOS1 (data not shown). In line with these findings RAS activity was restored in the cells expressing WT and not mutant caveolin-1 (Figure 4B). These studies strongly suggest that phosphorylated caveolin-1 sustains K-RAS signaling through increased activation of SOS1. Thus, even in the presence of WT K-RAS caveolin-1 expression can lead to sustained SOS1 and Grb2-dependent signal transduction.
Figure 4. Caveolin-1 is required for K-RAS function and localization.
4A. Intracellular levels of phospho-caveolin-1 in HCT116 isogenic system
4B. Phosphorylation of caveoilin-1 is necessary for caveolin-1 interactions with K-RAS signaling molecules Grb2 and SOS1.
HCT116-Caveolin-1-anti-sense cells were transfected with WT or mutant (Y14F) caveolin-1 and lysed in RIPA buffer 48 hours after transfection. Lysates were immunoprecipitated with indicated proteins and immune complexes were resolved on SDS-PAGE gels and probed for indicated proteins.
4C. K-RAS protein localization in caveolin-1 depleted Caco-2 isogenic system.
CacoKras#26 were transfected with scrambled or caveolin-1 siRNA 24 hour after subculture and fractionated into lipid rafts 48 hours after transfection as described in 61. Caveolin-1, K-RAS and flotilling-1 protein levels were determined by Western blotting. Figure is representative of three independent experiments
4D. K-RAS localization in HCT116 mock and HCT116 CAV-1 AS isogenic system.
HCT116-Mock and Caveolin-1 anti-sense cells were fractionated into lipid rafts.
K-RAS and flotilling-1 protein levels were determined by Western blotting. Figure is representative of three independent experiments
Since K-RAS has been shown to associate with caveolin-1, we also assessed whether caveolin-1 alters K-RAS localization. We knocked out caveolin-1 expression in the K-Ras#26 cells via transient transfection of caveolin-1 siRNA and determined the distribution of caveolin-1 protein in lipid raft by sucrose density gradient fractionation. As seen in Figure 4C, K-Ras#26 cells treated with scrambled siRNA show a regular distribution of caveolin-1 protein in fractions 4, 5 and 6 in the sucrose density gradient, following lipid raft fractionation. In these cells, K-RAS is primarily distributed in fractions 4, 5 and 6 also with flotillin-1 (a protein commonly localized to lipid rafts 38). Interestingly, K-RAS distribution was altered in the K-ras expressing cells with reduced caveolin-1 expression and is seen in fractions 5, 6, 7 and 8. We also determined the distribution of K-RAS in HCT116-Mock (control) and HCT116-Cav-AS cells (HCT116 cells stably expressing a caveolin-1 antisense construct and showing reduced levels of caveolin-1 20) and found that K-RAS distribution is altered in lipid rafts following caveolin-1 depletion in HCT116 cells (Figure 4D). Thus, caveolin-1 expression is required for maintaining distribution of the K-RAS protein to lipid raft microdomains. This effect is specific to K-RAS since the distribution of flotillin-1, another lipid raft-associated protein, does not get altered in caveolin-1 depleted cells.
Caveolin-1 protein modulates the levels of active RAS (GTP-bound) following its depletion in the K-Ras#26 cells or in HCT116-Cav-1-AS cells without changes in total RAS protein levels (Supplementary Figure 3). In line with these findings, we also found that phosphorylated Erk1/2 and Akt levels are substantially decreased in the lines with reduced caveolin-1 expression, indicating that loss of caveolin-1 expression disrupts K-RAS-mediated intracellular signaling. (Supplementary Figure 3). It should be mentioned that we did not detect any evidence of interaction between K-RAS and caveolin-1 in co-immunoprecipitation experiments.
K-RAS increases migration of colon cancer cells in a caveolin-1 and Src dependent manner
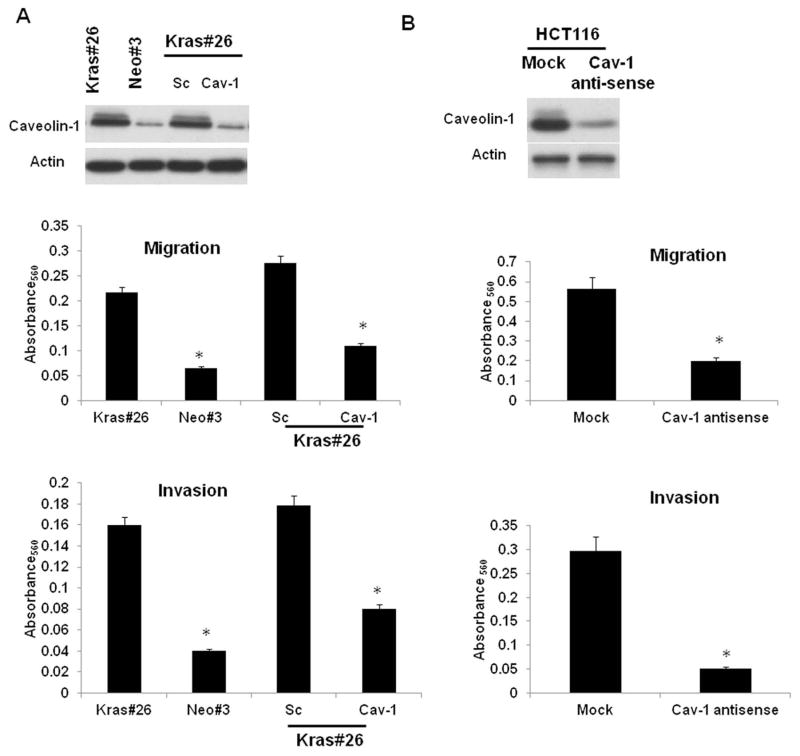
Acquisition of K-RAS mutations is a relatively late event in colorectal carcinogenesis and is associated with poor prognosis and increased metastatic disease. Cancer cell migration is an important aspect of cancer-associated metastasis. We and other groups have demonstrated that caveolin-1 is required for the regulated secretion of extracellular proteases associated with cell migration 20, 39. In order to determine whether caveolin-1 is required for K-RAS-dependent migration, we determined the effect of caveolin-1 depletion on migration of Caco2 cells expressing mutant K-RAS and HCT116 cells. As seen in Figure 5A, the Kras#26 cells migrate significantly higher than the Caco-Neo#3 (control) cells in an in vitro transwell migration assay. Depletion of caveolin-1 in Caco-Kras cells using siRNA dramatically reduces their caveolin-1 expression and migration, almost similar to that of control cells. Similarly, knockdown of caveolin-1 expression in HCT116 cells reduced their migration and migration (Figure 5B). These studies indicate that caveolin-1 is an essential regulator of cancer cell migration.
Figure 5. Caveolin-1 is required for K-RAS-dependent migration.
5A. Caveolin-1 levels in CacoNeo#3, Kras#26 and CacoKras#26 transfected with scrambled or caveolin-1 siRNA. Expression of caveolin-1 was determined by Western blotting. Migration and invasion assay of cells was measured as described in Materials and Methods and in Henkhaus et al.39. Figure is representative of three independent experiments. * = P<0.05 by t-test
5B. Caveolin-1 levels in HCT116-Mock/Caveolin-1 anti-sense cells. Down-regulation of caveolin-1 expression in the anti-sense cells was confirmed by Western blotting and migration and invasion assay of cells was measured as described in Materials and Methods and in Henkhaus et al.39. Figure is representative of three independent experiments. * = P<0.05 by t-test
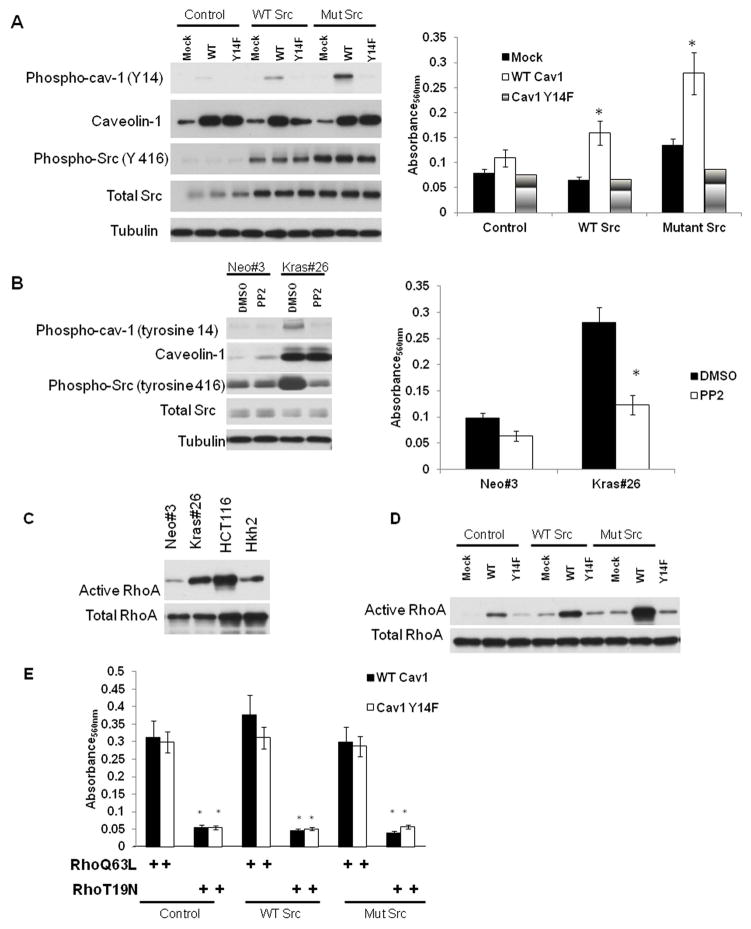
In order to determine whether caveolin-1 by itself can affect migration, we determined the rate of migration of Caco2 cells expressing an empty vector, or an expression vector with WT or mutant (Y14F) caveolin-1. As seen in Figure 6A, caveolin-1 is increased in cells transfected with a caveolin-1 expression construct. Interestingly, caveolin-1 expression by itself is not sufficient to increase migration of Caco2 cells (Figure 6A). We had previously demonstrated that activated K-RAS increases caveolin-1 phosphorylation in a Src-dependent manner 40. Src, a cytoplasmic tyrosine kinase, is localized in caveolae and can phosphorylate caveolin-1 at tyrosine-14 7, 41. Several recent reports have demonstrated that phosphorylated caveolin-1 can increase migration in cancer cells 42–43. To assess whether tyrosine-14 phosphorylation of caveolin-1 by Src can increase migration, we transfected the Caco2-Mock and Caco2-Cav-1 with an activated Src plasmid. As seen in Figure 6A, introduction of an activated Src induces tyrosine-14 phosphorylation of caveolin-1 with a concurrent increase in migration of Caco2-Cav-1 cells. However, presence of activated Src protein is unable to increase migration in Caco2 cells expressing caveolin-1 Y14F. These data suggest that phosphorylated caveolin-1 can increase migration in colon cancer cells in a Src-dependent manner. To conclusively determine whether an activated K-RAS increases migration of colon cancer cells by phoshorylating caveolin-1 via Src, we inhibited Src activity in Caco-Kras cells using the Pan-Src kinase inhibitor, PP2. Treatment with 25 μM PP2 leads to a dramatic reduction in phospho-caveolin-1 tyrosine-14 and phospho-Src (Figure 6B). Reduction in caveolin-1 phosphorylation is accompanied by a decrease in migration of the Caco-Kras cells (Figure 6B) indicating that Src plays a critical role in K-RAS dependent migration through caveolin-1 phosphorylation.
Figure 6. Src regulates K-RAS-dependent migration through Src/phospho-caveolin-1/RhoA pathway.
6A. Caco2 cells were transfected with either an empty vector, caveolin-1 or caveolin-1 Y14F, or expression constructs for wild-type or constitutively active Src. Levels of phosphorylated caveolin-1 (tyrosine 14), total caveolin-1, active Src (tyrosine phosphorylated at residue 416) and total Src protein of caveolin-1 was determined by Western blotting. Migration of cells was measured as described in 39.
6B. Caco-Neo#3 and Kras#26 cells were treated with 25 μM the pan-Src inhibitor for 24 hours. Levels of phosphorylated caveolin-1 (tyrosine 14), total caveolin-1, active Src (tyrosine phosphorylated at residue 416) and total Src protein of caveolin-1 was determined by Western blotting. Migration of cells was measured as described in 39. Figure is representative of three independent experiments. * = P<0.05 by t-test
6C and 6D. RhoA activity (GTP-bound-RhoA levels) was determined using the RhoA activity assay kit from Upstate Biotechnology, using manufacturers’ instructions. Active RhoA levels and total Rhoa A levels of the indicated cell lines are shown in the figure. 6C: RhoA activity in cells in HCT116/Hkh2 and Caco-Neo#3/Kras#26 cells, 6D: RhoA activity in Caco2 cells transfected with (a) either an empty vector, caveolin-1 or caveolin-1 Y14F and (b) expression constructs for wild-type or constitutively active Src, Figure is representative of two independent experiments.
6E. Caco2 cells were transfected with (a) either an empty vector, caveolin-1 or caveolin-1 Y14F (b) expression constructs for wild-type or constitutively active Src and (c) a constitutively active form (Q63L) or a dominant negative form (T19N) of RhoA. Migration of cells was measured as described in 39. Each experiment was performed in triplicate at least twice. Results of a representative experiment are shown.
Tyrosine phosphorylated caveolin-1 increases migration through activation of the RhoA GTPase
Our data suggest that an activated K-RAS uses two distinct mechanisms to increase caveolin-1-dependent migration: 1) at the transcriptional level, it regulates caveolin-1 expression and 2) at the post-translational level, it activates Src-dependent phosphorylation of caveolin-1 (Figure 6A and B). To determine the mechanism by which phospho-caveolin-1 increases migration, we examined the activation state of various pro-migratory pathways. The Rho family of GTPases is important players in cancer cell migration and has been shown to increase migration through increased Src activity. Interestingly, RhoA activity is increased in cells expressing an activated RAS 44, and caveolin-1 has been shown to be necessary for maintaining RhoA activity 45. We first determined the levels of active RhoA levels in the cells expressing active K-RAS. As seen in Figure 6C, active RhoA levels are increased in cells expressing an active K-RAS gene. Consistent with previously published reports 44, we found no change in active Cdc42 and Rac1 levels in cells expressing mutant K-RAS protein. Since caveolin-1 phosphorylation status correlates with active RhoA levels, we then determined whether caveolin-1 expression was sufficient to increase RhoA activity. As seen in Figure 6D, constitutive expression of caveolin-1 is not sufficient to increase the activity of RhoA GTPase. However, concurrent expression of an active form of Src that increase tyrosine phosphorylation of caveolin-1 (Figure 6A) increases the levels of active RhoA. This increase is dependent on phospho-caveolin-1 since a phosphorylation-deficient form of caveolin-1 is unable to increase RhoA activity, even in the presence of a constitutively active form of the Src kinase. These findings suggest that RhoA activity might be a critical event in caveolin-1-dependent migration. To assess the contribution of active RhoA in caveolin-1-dependent migration, we introduced in the Caco2 cells either a constitutively active RhoA mutant (RhoQ63L) or a dominant negative form of the protein (RhoT19N). As seen in Figure 6E, presence of the active form of RhoA is sufficient to induce migration in the cells expressing mutant caveolin-1 Y14F. More importantly, expression of RhoT19N blocks migration induced in caveolin-1-expressing cells in the presence of active Src. These results prove that RhoA activation by phosphorylated caveolin-1 is a crucial event in migration induced by an activated K-RAS protein.
Discussion
In the present study, we have shown that caveolin-1 is a transcriptional target of the K-RAS oncogene in late stage colon cancer. This upregulation is independent of the type of activating mutation present in the gene, since both G12V and G13D mutations are sufficient to upregulate caveolin-1 expression in human colon tumor-derived cells and in human colon tumor tissue harboring activating K-RAS mutations. An activated K-RAS can mediate signal transduction via multiple pathways, leading to various cancer phenotypes like decreased apoptosis and increased proliferation and migration. Using a combination of pharmacological inhibitors and genetics, we show that the P-I3 Kinase-AKT pathway is involved in K-RAS mediated caveolin-1 expression in these colon cancer cell lines. The P-I3 Kinase/AKT pathway is an important signal transduction pathway which is involved in cell survival and apoptosis resistance. We finally demonstrate that caveolin-1 is required for K-RAS-dependent signal transduction and migration. We propose a model for K-RAS-mediated caveolin-1 expression and activity (Supplementary Figure 4). An activated K-RAS oncogene increases Sp1 transactivation in an AKT-dependent manner. This in turn increases caveolin-1 transcription. Caveolin-1 protein then localizes to the membrane and is phosphorylated by Src. Caveolin-1 phosphorylated at tyrosine residue 14 then recruits signaling molecules such as Grb2 and SOS1 to sustain signal transduction and RhoA-dependent migration. In fact, we have found that depletion of caveolin-1 in colon cancer cells harboring an activated K-RAS oncogene decreases GTP-bound (active) RAS suggesting that caveolin-1 expression is necessary to sustain RAS-dependent signaling. These studies suggest that the K-RAS-caveolin-1 pathway might elicit a positive feedback loop to aid tumor cell migration and metastasis especially in the late stages of colorectal cancer.
The role of RAS signaling in caveolin-1 expression is highly debated. In studies carried out by Engelman et al, the RAS oncogene was shown to downregulate caveolin-1 expression in NIH-3T3 cells 3. However, in a study carried out by Sasai et al in human dermal fibroblasts, an activated K-RAS oncogene was not sufficient to downregulate caveolin-1 expression levels 46. Thus, transformation by oncogenes in murine cell culture systems is differentially regulated as compared to human cell culture systems 47. Also, specific and differential pathways are activated in murine and human cells, in response to an activated RAS oncogene 47. Thus, K-RAS-dependent upregulation of caveolin-1 in human colon cancer cells is a unique finding and highlights the differential regulation of gene expression by K-RAS in murine and human carcinogenesis. The involvement of the P-I3 Kinase pathway in caveolin-1 upregulation has been demonstrated previously. This pathway has been implicated in progestin-mediated caveolin-1 expression in human breast cancer cells, and this is necessary for breast cancer proliferation 48. Our study defines the role of AKT activation through K-RAS, in the expression of caveolin-1 through the transcription factor Sp1.
We have previously shown that caveolin-1 is transcriptionally regulated by the APC tumor suppressor 49. The APC tumor suppressor is mutated in as many as 85% of diagnosed colorectal cancer cases, and is often considered to be an initiating mutation 50. The K-RAS oncogene in mutated in 35% cases of sporadic colorectal cancer, and occurs at the later stages of cancer development 51–52. Thus, caveolin-1 expression might be differentially regulated at different stages of colorectal cancer progression. In the initial stages, following an APC mutation, there is downregulation of caveolin-1 14. However, acquisition of a K-RAS mutation might then be a potential mechanism to upregulate caveolin-1. It is interesting to note that caveolin-1 expression is elevated in late stage colon adenocarcinoma where K-RAS is often activated through mutations 12–13. Thus, caveolin-1 up-regulation might occur at late stages of colorectal carcinogenesis. 50, 53.
Our studies also identify caveolin-1 as a novel regulator of K-RAS-dependent signaling and cancer cell migration. Caveolin-1 has been shown to be necessary for the activation of the P-I3 Kinase/AKT pathway 54. In prostate cells, caveolin-1 can confer a survival advantage by maintaining an activated AKT through negative regulation of the AKT phosphatases, PP1 and PP2A 8. Additionally, caveolin-1 is phosphorylated at residue Tyr14 by several oncogenes, and this can help recruitment of GRB7 through its SH2 domain and lead to sustained signaling41. Thus, while caveolin-1 can negatively regulate signal transduction 55, tyrosine phosphorylation of caveolin-1 converts it into a highly efficient scaffold for proteins with SH2 domain thereby increasing mitogenic signaling. Interestingly, we have found that colon cancer cells harboring an activated K-RAS mutation are sensitive to caveolin-1 levels since depletion of caveolin-1 decreases active K-RAS levels and mitogenic signaling. This could be explained by the redistribution of K-RAS protein from lipid rafts in cells that have been depleted of caveolin-1. While these studies highlight a novel role for caveolin-1 in K-RAS-dependent mitogenic signaling, it is noteworthy to point out that work by Roy and colleagues demonstrates that K-RAS is unaffected by the expression of a dominant negative mutant of caveolin-1 in BHK cells 56 Their study shows that lipid-raft associated H-RAS is more susceptible to a caveolin-1 mutant than K-RAS in BHK cells. However, work from Kranenburg et al demonstrate that in COS cells, K-RAS localizes to caveolae and that its activity it highly dependent on this localization 57 with disruption of caveolar structures leading to altered K-RAS activity. Thus, the effect of caveolin-1 on K-RAS might be cell-type specific and sensitive to the cholesterol content of the cells. In colon cancer cells, depletion of caveolin-1 decreases free cholesterol (our unpublished observation). Thus, K-RAS activity in these cells might be sensitive to caveolin-1 depletion because of altered cholesterol metabolism.
Caveolin-1 was originally identified as a substrate of the Src tyrosine kinase. Recent reports have demonstrated that phosphorylated caveolin-1 has been associated with increased tumor cell migration 42, 58. Phosphorylated caveolin-1 increases cell migration via the modulation of the RHO/Rock pathway. Thus, acquisition of a K-RAS mutation, followed by consequent upregulation of caveolin-1 and Src-dependent caveolin-1 phosphorylation might aid in cancer progression by increasing the metastatic potential of cancer cells. Furthermore, caveolin-1 has been shown to be necessary for the localization and secretion of the tumor proteases, cathepsin B and kallikrein 6, both of which are necessary for migration 20, 39, 59–60.
Our data suggest that caveolin-1 is a conditional “oncogene” in colon cancer whose activity is dependent on mutant K-RAS. Our data suggests that activation of AKT and Src regulate caveolion-1 expression and post-translational modification. In fact, phospho-AKT and phosphor-Src levels are elevated in the tumors samples harboring K-RAS mutations. Through regulation by the K-RAS oncogene at late stages of colorectal carcinogenesis, caveolin-1 might play pleiotropic roles in cancer development, depending on the stage of cancer.
Supplementary Material
Novelty and Impact Statement.
In this manuscript, we present our novel finding about the regulation of the expression of the membrane protein, caveolin-1, by mutant K-RAS. The expression of caveolin-1 is highly debated in colorectal cancer. Here, we demonstrate that caveolin-1 is induced in response to mutant K-RAS through AKT activation in colon cancer in vitro and in vivo, and is required for K-RAS signaling. Our data suggest a positive feedback loop between K-RAS-dependent- caveolin-1 expression and signaling.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health, CA095060 (EWG, PI) and CA123065 (NAI and EWG, Co-PI’s). The authors wish to thank David E. Stringer, Dr. H. Yerushalmi, and Karen Kachel for assistance with experimental set-up as well as Dr. Shirasawa et al. for the kind gift of the Hkh2 cell line. We are grateful to Dr. Vijay Shah for providing us with CAV-1 promoter-reporter plasmid and Dr. M.J. Quon for MYR AKT and DN AKT plasmids.
References
- 1.Parton RG. Caveolae and caveolins. Curr Opin Cell Biol. 1996;8:542–8. doi: 10.1016/s0955-0674(96)80033-0. [DOI] [PubMed] [Google Scholar]
- 2.Galbiati F, Volonte D, Engelman JA, Watanabe G, Burk R, Pestell RG, Lisanti MP. Targeted downregulation of caveolin-1 is sufficient to drive cell transformation and hyperactivate the p42/44 MAP kinase cascade. Embo J. 1998;17:6633–48. doi: 10.1093/emboj/17.22.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engelman JA, Zhang XL, Razani B, Pestell RG, Lisanti MP. p42/44 MAP kinase-dependent and -independent signaling pathways regulate caveolin-1 gene expression. Activation of Ras-MAP kinase and protein kinase a signaling cascades transcriptionally down-regulates caveolin-1 promoter activity. J Biol Chem. 1999;274:32333–41. doi: 10.1074/jbc.274.45.32333. [DOI] [PubMed] [Google Scholar]
- 4.Koleske AJ, Baltimore D, Lisanti MP. Reduction of caveolin and caveolae in oncogenically transformed cells. Proc Natl Acad Sci U S A. 1995;92:1381–5. doi: 10.1073/pnas.92.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engelman JA, Zhang XL, Lisanti MP. Genes encoding human caveolin-1 and -2 are co-localized to the D7S522 locus (7q31. 1), a known fragile site (FRA7G) that is frequently deleted in human cancers. FEBS Lett. 1998;436:403–10. doi: 10.1016/s0014-5793(98)01134-x. [DOI] [PubMed] [Google Scholar]
- 6.Engelman JA, Chu C, Lin A, Jo H, Ikezu T, Okamoto T, Kohtz DS, Lisanti MP. Caveolin-mediated regulation of signaling along the p42/44 MAP kinase cascade in vivo. A role for the caveolin-scaffolding domain. FEBS Lett. 1998;428:205–11. doi: 10.1016/s0014-5793(98)00470-0. [DOI] [PubMed] [Google Scholar]
- 7.Cohen AW, Hnasko R, Schubert W, Lisanti MP. Role of caveolae and caveolins in health and disease. Physiol Rev. 2004;84:1341–79. doi: 10.1152/physrev.00046.2003. [DOI] [PubMed] [Google Scholar]
- 8.Li L, Ren CH, Tahir SA, Ren C, Thompson TC. Caveolin-1 maintains activated Akt in prostate cancer cells through scaffolding domain binding site interactions with and inhibition of serine/threonine protein phosphatases PP1 and PP2A. Mol Cell Biol. 2003;23:9389–404. doi: 10.1128/MCB.23.24.9389-9404.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tahir SA, Yang G, Ebara S, Timme TL, Satoh T, Li L, Goltsov A, Ittmann M, Morrisett JD, Thompson TC. Secreted caveolin-1 stimulates cell survival/clonal growth and contributes to metastasis in androgen-insensitive prostate cancer. Cancer Res. 2001;61:3882–5. [PubMed] [Google Scholar]
- 10.Lavie Y, Liscovitch M. Changes in lipid and protein constituents of rafts and caveolae in multidrug resistant cancer cells and their functional consequences. Glycoconj J. 2000;17:253–9. doi: 10.1023/a:1026553626537. [DOI] [PubMed] [Google Scholar]
- 11.Bender FC, Reymond MA, Bron C, Quest AF. Caveolin-1 levels are down-regulated in human colon tumors, and ectopic expression of caveolin-1 in colon carcinoma cell lines reduces cell tumorigenicity. Cancer Res. 2000;60:5870–8. [PubMed] [Google Scholar]
- 12.Patlolla JM, Swamy MV, Raju J, Rao CV. Overexpression of caveolin-1 in experimental colon adenocarcinomas and human colon cancer cell lines. Oncol Rep. 2004;11:957–63. [PubMed] [Google Scholar]
- 13.Fine SW, Lisanti MP, Galbiati F, Li M. Elevated expression of caveolin-1 in adenocarcinoma of the colon. Am J Clin Pathol. 2001;115:719–24. doi: 10.1309/YL54-CCU7-4V0P-FDUT. [DOI] [PubMed] [Google Scholar]
- 14.Roy UK, Henkhaus RS, Ignatenko NA, Mora J, Fultz KE, Gerner EW. Wild-type APC regulates caveolin-1 expression in human colon adenocarcinoma cell lines via FOXO1a and C-myc. Mol Carcinog. 2008;47:947–55. doi: 10.1002/mc.20451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 16.Shaw P, Tardy S, Benito E, Obrador A, Costa J. Occurrence of Ki-ras and p53 mutations in primary colorectal tumors. Oncogene. 1991;6:2121–8. [PubMed] [Google Scholar]
- 17.Yanez L, Groffen J, Valenzuela DM. c-K-ras mutations in human carcinomas occur preferentially in codon 12. Oncogene. 1987;1:315–8. [PubMed] [Google Scholar]
- 18.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–8. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 19.Ignatenko NA, Zhang H, Watts GS, Skovan BA, Stringer DE, Gerner EW. The chemopreventive agent alpha-difluoromethylornithine blocks Ki-ras-dependent tumor formation and specific gene expression in Caco-2 cells. Mol Carcinog. 2004;39:221–33. doi: 10.1002/mc.20008. [DOI] [PubMed] [Google Scholar]
- 20.Cavallo-Medved D, Mai J, Dosescu J, Sameni M, Sloane BF. Caveolin-1 mediates the expression and localization of cathepsin B, pro-urokinase plasminogen activator and their cell-surface receptors in human colorectal carcinoma cells. J Cell Sci. 2005;118:1493–503. doi: 10.1242/jcs.02278. [DOI] [PubMed] [Google Scholar]
- 21.Vogel U, Sandvig K, van Deurs B. Expression of caveolin-1 and polarized formation of invaginated caveolae in Caco-2 and MDCK II cells. J Cell Sci. 1998;111 ( Pt 6):825–32. doi: 10.1242/jcs.111.6.825. [DOI] [PubMed] [Google Scholar]
- 22.Ignatenko NA, Yerushalmi HF, Watts GS, Futscher BW, Stringer DE, Marton LJ, Gerner EW. Pharmacogenomics of the polyamine analog 3,8,13,18-tetraaza-10,11-[(E)-1,2-cyclopropyl]eicosane tetrahydrochloride, CGC-11093, in the colon adenocarcinoma cell line HCT1161. Technol Cancer Res Treat. 2006;5:553–64. doi: 10.1177/153303460600500602. [DOI] [PubMed] [Google Scholar]
- 23.Cao S, Fernandez-Zapico ME, Jin D, Puri V, Cook TA, Lerman LO, Zhu XY, Urrutia R, Shah V. KLF11-mediated repression antagonizes Sp1/sterol-responsive element-binding protein-induced transcriptional activation of caveolin-1 in response to cholesterol signaling. J Biol Chem. 2005;280:1901–10. doi: 10.1074/jbc.M407941200. [DOI] [PubMed] [Google Scholar]
- 24.Cong LN, Chen H, Li Y, Zhou L, McGibbon MA, Taylor SI, Quon MJ. Physiological role of Akt in insulin-stimulated translocation of GLUT4 in transfected rat adipose cells. Mol Endocrinol. 1997;11:1881–90. doi: 10.1210/mend.11.13.0027. [DOI] [PubMed] [Google Scholar]
- 25.Matveev S, Uittenbogaard A, van Der Westhuyzen D, Smart EJ. Caveolin-1 negatively regulates SR-BI mediated selective uptake of high-density lipoprotein-derived cholesteryl ester. Eur J Biochem. 2001;268:5609–16. doi: 10.1046/j.1432-1033.2001.02496.x. [DOI] [PubMed] [Google Scholar]
- 26.Blake RA, Broome MA, Liu X, Wu J, Gishizky M, Sun L, Courtneidge SA. SU6656, a selective src family kinase inhibitor, used to probe growth factor signaling. Mol Cell Biol. 2000;20:9018–27. doi: 10.1128/mcb.20.23.9018-9027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song KS, Li S, Okamoto T, Quilliam LA, Sargiacomo M, Lisanti MP. Co-purification and direct interaction of Ras with caveolin, an integral membrane protein of caveolae microdomains. Detergent-free purification of caveolae microdomains. J Biol Chem. 1996;271:9690–7. doi: 10.1074/jbc.271.16.9690. [DOI] [PubMed] [Google Scholar]
- 28.Jones GE, Ridley AJ, Zicha D. Rho GTPases and cell migration: measurement of macrophage chemotaxis. Methods Enzymol. 2000;325:449–62. doi: 10.1016/s0076-6879(00)25465-7. [DOI] [PubMed] [Google Scholar]
- 29.Ridley A. Rho GTPases. Integrating integrin signaling. J Cell Biol. 2000;150:F107–9. doi: 10.1083/jcb.150.4.f107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujimoto T, Kogo H, Nomura R, Une T. Isoforms of caveolin-1 and caveolar structure. J Cell Sci. 2000;113(Pt 19):3509–17. doi: 10.1242/jcs.113.19.3509. [DOI] [PubMed] [Google Scholar]
- 31.Etienne-Grimaldi MC, Formento JL, Francoual M, Francois E, Formento P, Renee N, Laurent-Puig P, Chazal M, Benchimol D, Delpero JR, Letoublon C, Pezet D, et al. K-Ras mutations and treatment outcome in colorectal cancer patients receiving exclusive fluoropyrimidine therapy. Clin Cancer Res. 2008;14:4830–5. doi: 10.1158/1078-0432.CCR-07-4906. [DOI] [PubMed] [Google Scholar]
- 32.Pore N, Liu S, Shu HK, Li B, Haas-Kogan D, Stokoe D, Milanini-Mongiat J, Pages G, O’Rourke DM, Bernhard E, Maity A. Sp1 is involved in Akt-mediated induction of VEGF expression through an HIF-1-independent mechanism. Mol Biol Cell. 2004;15:4841–53. doi: 10.1091/mbc.E04-05-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bae IH, Park MJ, Yoon SH, Kang SW, Lee SS, Choi KM, Um HD. Bcl-w promotes gastric cancer cell invasion by inducing matrix metalloproteinase-2 expression via phosphoinositide 3-kinase, Akt, and Sp1. Cancer Res. 2006;66:4991–5. doi: 10.1158/0008-5472.CAN-05-4254. [DOI] [PubMed] [Google Scholar]
- 34.Bist A, Fielding PE, Fielding CJ. Two sterol regulatory element-like sequences mediate up-regulation of caveolin gene transcription in response to low density lipoprotein free cholesterol. Proc Natl Acad Sci U S A. 1997;94:10693–8. doi: 10.1073/pnas.94.20.10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baran J, Mundy DI, Vasanji A, Parat MO. Altered localization of H-Ras in caveolin-1-null cells is palmitoylation-independent. J Cell Commun Signal. 2007;1:195–204. doi: 10.1007/s12079-008-0017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wary KK, Mariotti A, Zurzolo C, Giancotti FG. A requirement for caveolin-1 and associated kinase Fyn in integrin signaling and anchorage-dependent cell growth. Cell. 1998;94:625–34. doi: 10.1016/s0092-8674(00)81604-9. [DOI] [PubMed] [Google Scholar]
- 37.Biedi C, Panetta D, Segat D, Cordera R, Maggi D. Specificity of insulin-like growth factor I and insulin on Shc phosphorylation and Grb2 recruitment in caveolae. Endocrinology. 2003;144:5497–503. doi: 10.1210/en.2003-0417. [DOI] [PubMed] [Google Scholar]
- 38.Dermine JF, Duclos S, Garin J, St-Louis F, Rea S, Parton RG, Desjardins M. Flotillin-1-enriched lipid raft domains accumulate on maturing phagosomes. J Biol Chem. 2001;276:18507–12. doi: 10.1074/jbc.M101113200. [DOI] [PubMed] [Google Scholar]
- 39.Henkhaus RS, Gerner EW, Ignatenko NA. Kallikrein 6 is a mediator of K-RAS-dependent migration of colon carcinoma cells. Biol Chem. 2008;389:757–64. doi: 10.1515/BC.2008.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy UK, Rial NS, Kachel KL, Gerner EW. Activated K-RAS increases polyamine uptake in human colon cancer cells through modulation of caveolar endocytosis. Mol Carcinog. 2008;47:538–53. doi: 10.1002/mc.20414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee H, Volonte D, Galbiati F, Iyengar P, Lublin DM, Bregman DB, Wilson MT, Campos-Gonzalez R, Bouzahzah B, Pestell RG, Scherer PE, Lisanti MP. Constitutive and growth factor-regulated phosphorylation of caveolin-1 occurs at the same site (Tyr-14) in vivo: identification of a c-Src/Cav-1/Grb7 signaling cassette. Mol Endocrinol. 2000;14:1750–75. doi: 10.1210/mend.14.11.0553. [DOI] [PubMed] [Google Scholar]
- 42.Joshi B, Strugnell SS, Goetz JG, Kojic LD, Cox ME, Griffith OL, Chan SK, Jones SJ, Leung SP, Masoudi H, Leung S, Wiseman SM, et al. Phosphorylated caveolin-1 regulates Rho/ROCK-dependent focal adhesion dynamics and tumor cell migration and invasion. Cancer Res. 2008;68:8210–20. doi: 10.1158/0008-5472.CAN-08-0343. [DOI] [PubMed] [Google Scholar]
- 43.Thomas S, Overdevest JB, Nitz MD, Williams PD, Owens CR, Sanchez-Carbayo M, Frierson HF, Schwartz MA, Theodorescu D. Src and caveolin-1 reciprocally regulate metastasis via a common downstream signaling pathway in bladder cancer. Cancer Res. 2011;71:832–41. doi: 10.1158/0008-5472.CAN-10-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sahai E, Olson MF, Marshall CJ. Cross-talk between Ras and Rho signalling pathways in transformation favours proliferation and increased motility. EMBO J. 2001;20:755–66. doi: 10.1093/emboj/20.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grande-Garcia A, Echarri A, de Rooij J, Alderson NB, Waterman-Storer CM, Valdivielso JM, del Pozo MA. Caveolin-1 regulates cell polarization and directional migration through Src kinase and Rho GTPases. J Cell Biol. 2007;177:683–94. doi: 10.1083/jcb.200701006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sasai K, Kakumoto K, Hanafusa H, Akagi T. The Ras-MAPK pathway downregulates Caveolin-1 in rodent fibroblast but not in human fibroblasts: implications in the resistance to oncogene-mediated transformation. Oncogene. 2007;26:449–55. doi: 10.1038/sj.onc.1209792. [DOI] [PubMed] [Google Scholar]
- 47.Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–83. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 48.Salatino M, Beguelin W, Peters MG, Carnevale R, Proietti CJ, Galigniana MD, Vedoy CG, Schillaci R, Charreau EH, Sogayar MC, Elizalde PV. Progestin-induced caveolin-1 expression mediates breast cancer cell proliferation. Oncogene. 2006;25:7723–39. doi: 10.1038/sj.onc.1209757. [DOI] [PubMed] [Google Scholar]
- 49.Roy UK, Rial NS, Kachel KL, Gerner EW. Activated K-RAS increases polyamine uptake in human colon cancer cells through modulation of caveolar endocytosis. Mol Carcinog. 2008 doi: 10.1002/mc.20414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–90. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 51.Castagnola P, Giaretti W. Mutant KRAS, chromosomal instability and prognosis in colorectal cancer. Biochim Biophys Acta. 2005;1756:115–25. doi: 10.1016/j.bbcan.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 52.Smakman N, Borel Rinkes IH, Voest EE, Kranenburg O. Control of colorectal metastasis formation by K-Ras. Biochim Biophys Acta. 2005;1756:103–14. doi: 10.1016/j.bbcan.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 53.Maier TJ, Janssen A, Schmidt R, Geisslinger G, Grosch S. Targeting the beta-catenin/APC pathway: a novel mechanism to explain the cyclooxygenase-2-independent anticarcinogenic effects of celecoxib in human colon carcinoma cells. Faseb J. 2005;19:1353–5. doi: 10.1096/fj.04-3274fje. [DOI] [PubMed] [Google Scholar]
- 54.Sedding DG, Hermsen J, Seay U, Eickelberg O, Kummer W, Schwencke C, Strasser RH, Tillmanns H, Braun-Dullaeus RC. Caveolin-1 facilitates mechanosensitive protein kinase B (Akt) signaling in vitro and in vivo. Circ Res. 2005;96:635–42. doi: 10.1161/01.RES.0000160610.61306.0f. [DOI] [PubMed] [Google Scholar]
- 55.Kurzchalia TV, Parton RG. Membrane microdomains and caveolae. Curr Opin Cell Biol. 1999;11:424–31. doi: 10.1016/s0955-0674(99)80061-1. [DOI] [PubMed] [Google Scholar]
- 56.Roy S, Luetterforst R, Harding A, Apolloni A, Etheridge M, Stang E, Rolls B, Hancock JF, Parton RG. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nat Cell Biol. 1999;1:98–105. doi: 10.1038/10067. [DOI] [PubMed] [Google Scholar]
- 57.Kranenburg O, Verlaan I, Moolenaar WH. Regulating c-Ras function. cholesterol depletion affects caveolin association, GTP loading, and signaling. Curr Biol. 2001;11:1880–4. doi: 10.1016/s0960-9822(01)00582-6. [DOI] [PubMed] [Google Scholar]
- 58.Luanpitpong S, Talbott SJ, Rojanasakul Y, Nimmannit U, Pongrakhananon V, Wang L, Chanvorachote P. Regulation of lung cancer cell migration and invasion by reactive oxygen species and caveolin-1. J Biol Chem. 2010;285:38832–40. doi: 10.1074/jbc.M110.124958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cavallo-Medved D, Dosescu J, Linebaugh BE, Sameni M, Rudy D, Sloane BF. Mutant K-ras regulates cathepsin B localization on the surface of human colorectal carcinoma cells. Neoplasia. 2003;5:507–19. doi: 10.1016/s1476-5586(03)80035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goetz JG, Minguet S, Navarro-Lerida I, Lazcano JJ, Samaniego R, Calvo E, Tello M, Osteso-Ibanez T, Pellinen T, Echarri A, Cerezo A, Klein-Szanto AJ, et al. Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Cell. 2011;146:148–63. doi: 10.1016/j.cell.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Henkhaus RS, Roy UK, Cavallo-Medved D, Sloane BF, Gerner EW, Ignatenko NA. Caveolin-1-mediated expression and secretion of kallikrein 6 in colon cancer cells. Neoplasia. 2008;10:140–8. doi: 10.1593/neo.07817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.