Abstract
Context:
Men and women differ in body fat distribution and adipose tissue metabolism as well as in obesity comorbidities and their response to obesity treatment.
Objective:
The objective of the study was a search for sex differences in adipose tissue function.
Design and Setting:
This was an exploratory study performed at a university hospital.
Participants and Main Outcome Measures:
Resting metabolic rate (RMR), body composition, and sc adipose tissue genome-wide expression were measured in the SOS Sib Pair study (n = 732).
Results:
The relative contribution of fat mass to RMR and the metabolic rate per kilogram adipose tissue was higher in women than in men (P value for sex by fat mass interaction = .0019). Women had increased expression of genes involved in mitochondrial function, here referred to as a mitochondrial gene signature. Analysis of liver, muscle, and blood showed that the pronounced mitochondrial gene signature in women was specific for adipose tissue. Brown adipocytes are dense in mitochondria, and the expression of the brown adipocyte marker uncoupling protein 1 was 5-fold higher in women compared with men in the SOS Sib Pair Study (P = 7.43 × 10−7), and this was confirmed in a cross-sectional, population-based study (n = 83, 6-fold higher in women, P = .00256).
Conclusions:
The increased expression of the brown adipocyte marker uncoupling protein 1 in women indicates that the higher relative contribution of the fat mass to RMR in women is in part explained by an increased number of brown adipocytes.
It is well established that obesity and its associated complications are increasing in both men and women, but there are clear sex differences in fat distribution and metabolism and in the clinical manifestations of obesity. Women have a greater proportion of body fat than men, with larger amounts of sc adipose tissue (1). By contrast, men are more prone to developing visceral obesity (2), which is associated with increased risk of obesity comorbidities (3, 4). In addition, women have a higher fat metabolism under periods of prolonged exertion (5), and the lipolytic capacity of abdominal sc fat cells is higher in obese women than in obese men (6).
Adipose tissue is a complex organ containing not only white adipocytes but also a large number of other cell types, such as immune cells and preadipocytes. The study of adipose tissue biology is central to the understanding of obesity and its comorbidities and to unravel the clinical differences between obese men and women. Sex differences in adipose tissue gene expression have been reported for proteins that have key metabolic functions, such as hormone-sensitive lipase (7), leptin (8), and adiponectin (9). Differences in gene expression in adipose tissue between women and men may represent regulatory changes in all cell types in the adipose tissue but may also be due to an altered cellular composition between the sexes.
Two central functions of adipose tissue is energy storage and energy release. However, the overall metabolic activity in adipose tissue is lower than in most other organs (10), and the contribution of adipose tissue to whole body metabolism has not been investigated in detail. Our aim was therefore to investigate the relative contribution of adipose tissue to resting metabolic rate in women and men.
Materials and Methods
Subjects
Study subjects received written and oral information before giving written informed consent. The Regional Ethics Committee in Gothenburg approved the studies.
The SOS Sib Pair study (n = 732) includes 154 nuclear families with body mass index (BMI) discordant sibling pairs (BMI difference ≥ 10 kg/m2) (Supplemental Table 1, published on The Endocrine Society's Journals Online web site at http://jcem.endojournals.org). The subjects were extensively phenotyped (11), including measurements of resting metabolic rate (RMR) in a ventilated hood and body composition by the total body potassium method (12). Body composition and RMR data were available for 405 women and 257 men. Subcutaneous adipose tissue gene expression was measured by microarray in 262 women and 113 men from the SOS Sib Pair offspring cohort (Gene Expression Omnibus database no. GSE27916) (Supplemental Table 1). Gene expression was also analyzed by real-time PCR in a subset of the cohort (30 women, 28 men) (Supplemental Table 1).
The Mölndal Metabolic study, a cross-sectional, randomly selected, population-based study assessing body composition and metabolic rate, includes 50 women and 50 men (13). RNA from sc adipose tissue was available from 39 women and 44 men (Supplemental Table 1).
Microarray profiles from perithyroid adipose tissue containing islets of brown adipose tissue (BAT) and paired sc adipose tissue profiles from patients (7 women and 2 men; aged 21–76 y) undergoing surgery in the thyroid area were available (GSE27657) (14).
The very low-calorie diet (VLCD) study included 6 women and 18 men (BMI of 37.6 ± 4.9 kg/m2; age of 25–61 y) (Supplemental Table 1). The subjects were given a VLCD of 450 kcal per day containing all essential nutrients for 16 weeks as previously described (13). All patients had scheduled visits at the Obesity Unit at Sahlgrenska University Hospital at weeks 0 (baseline) 8, and 16. Body weight was registered at each visit, and patients were given support and counseling to enhance compliance. After 16 weeks, average weight loss was 27.7 ± 1.8 kg. Abdominal sc adipose tissue biopsies were obtained and used for microarray analysis of gene expression (GSE35710).
Expression analysis
Total RNA was prepared as previously described (11, 14). Gene expression was analyzed using the Human Genome U133 plus 2.0 or U133A DNA microarray (Affymetrix, Santa Clara, CA) according to standard Affymetrix protocols. The raw expression signals were preprocessed according to the Probe Logarithmic Intensity Error (Affymetrix). The robust quantile method (15) was applied to get normalized expression values.
Additional expression profiles from adipose tissue and whole blood (GSE7965) were from Emilsson et al (Icelandic Cohort) (16). Expression profiles from skeletal muscle (GSE9676) and liver (GSE9588) were from Welle et al (17) and Schadt et al (18), respectively. Data were preprocessed as described above. For data sets with no sex information, the normalized gene expression values of genes on the X and Y chromosome were used to determine the sex by linear discrimination analysis based on Eigen vectors.
Integrated analysis of the microarray data and Gene Ontology (GO) information was performed by mapping all Q values of differential gene expression on GO networks and then using the reporter algorithm (19, 20). The results were reported as enrichment P values. Analysis of empirical cumulative distribution of fold changes was performed with Kolmogorov-Smirnov statistics (KS test).
In the Mölndal Metabolic study, uncoupling protein 1 (UCP1) real-time PCR was performed as previously described (13) with the exception that the δ-delta cycle threshold method was used for data analysis. In the subset from the SOS Sib Pair cohort, cDNA was preamplified using the TaqMan PreAmp master mix kit (Applied Biosystems, Carlsbad, CA), and UCP1 mRNA was measured by real-time PCR (13). Oxphos-CR genes were analyzed using TaqMan LDA cards (Applied Biosystems).
Statistical analysis and dimension reduction
Moderated Student's t test (21) was used to determine differences in microarray gene expression analysis and then the calculated P values were transformed to Q values by correcting for multiple testing using the method of Benjamini and Hochberg (22). With a cutoff value of 0.01, singular value decomposition analysis based on left Eigen vectors called Eigen arrays (23) were performed to capture sex differences. To eliminate undue influence of metabolic and body composition confounders on the analysis of the cumulative distribution of the fold changes, each transcript within the mitochondrial gene signature and Oxphos-CR genes was adjusted using a multiple regression procedure, which included fat mass, waist circumference, and homeostasis model assessment-insulin resistance (HOMA-IR) as covariates. A stepwise selection was used and covariates contributing to the model at the P < .1 significance level were retained. The residuals, i.e., the adjusted gene expression levels, were then subjected to analysis.
Multiple regression analysis of RMR on fat-free mass (FFM), fat mass (FM), and age was performed using the REG or GLM procedures in SAS (version 9.1; SAS Institute, Cary, NC). In modeling determinants for RMR, data were log transformed where appropriate to obtain approximate normal distributions.
Results
Greater contribution of FM to RMR in women
We first investigated sex differences in whole-body metabolism by analyzing the relative contributions of FM (used as a proxy for adipose tissue) and FFM to RMR in women (n = 405) and men (n = 257) from the SOS Sib Pair cohort (Supplemental Table 1) using a multiple regression analysis. For both sexes, FFM and FM were positively correlated with RMR, whereas age showed a negative correlation, as expected (Table 1). When regressing RMR on fat mass, the slope of the regression line (b), which indicate the impact level of FM on RMR (kilocalories per day−1 per kilogram FM) was greater in women compared with men (0.3803 and 0.2550, respectively; Table 1). The statistical significance of this sex difference was verified in a model that included a sex-by-FM interaction term (P = .0019). The sex difference persisted after including estradiol and testosterone levels, waist to hip ratio, and HOMA-IR in the model (P value for sex by FM interaction = .0002). Thus, women have a higher metabolic rate per kilogram adipose tissue than men independent of sex steroids, body fat distribution, and insulin sensitivity.
Table 1.
Contribution of FFM, FM, and Age to RMR in Women and Men
| Factor | Women (n = 405) |
Men (n = 257) |
||||
|---|---|---|---|---|---|---|
| Standardized ba | Partial R2 | P Value | Standardized ba | Partial R2 | P Value | |
| FFM | 0.5113 | 0.4910 | <.0001 | 0.5858 | 0.4249 | <.0001 |
| FM | 0.3803 | 0.0966 | <.0001 | 0.2550 | 0.0467 | <.0001 |
| Age, y | −0.0786 | 0.0038 | .0534 | −0.0984 | 0.0061 | .0860 |
The table shows the results of a multiple regression analysis including FFM, FM, and age in the SOS Sib Pair cohort.
Kilocalories per day−1 per kilogram FFM or kilocalories per day−1 per kilogram FM or kilocalories per day−1 per year (age).
Sex-based partitioning of sc adipose tissue expression profiles
To search for possible explanations for the observed sex differences in adipose tissue energy expenditure, we analyzed microarray expression profiles from sc adipose tissue from 262 women and 113 men from the SOS Sib Pair offspring cohort (Supplemental Table 1) by singular value decomposition analysis. There was a clear separation of the adipose tissue gene expression in men and women on the basis of a significant differential expression of 5746 transcripts (Qvalue < 10−2) (Fig. 1A). This separation remained after exclusion of the 265 transcripts encoded by the sex chromosomes (Fig. 1B).
Figure 1.
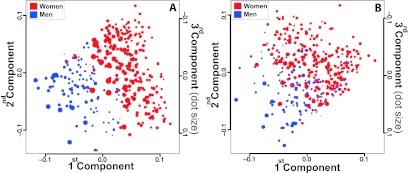
Separation of adipose tissue gene expression profiles of men (blue) and women (red) using singular value decomposition analysis. The results are illustrated in a pseudo-three-dimensional plot of the first three principal components of the singular value decomposition analysis. Each principal component accounts for as much as possible of the variability in gene expression between men and women. The first component is represented by the x-axis, the second component is represented by the y-axis, and the third component is represented by dot sizes. Each dot in the figure represents a subject from the SOS Sib Pair offspring cohort. A, Separation was based on 5746 transcripts with significant differential expression between the sexes. B, Same analysis as in A after the exclusion of 265 transcripts encoded by the sex chromosomes.
Higher expression of genes involved in mitochondrial function in adipose tissue in women
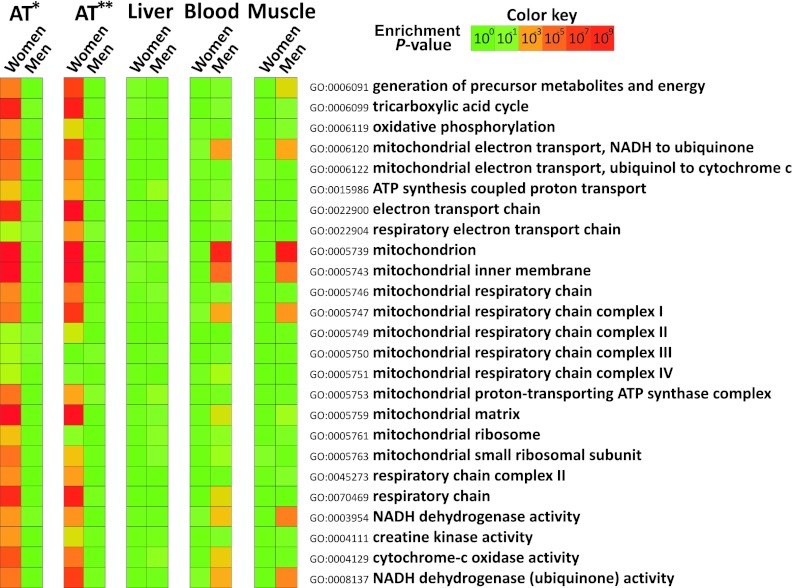
To identify key biological processes (defined by groups of genes with specific functions) associated with the sex difference in adipose tissue gene expression, we performed an integrated analysis of the microarray data from the SOS Sib Pair offspring cohort with GO information (see Supplemental Data Set 1). We found that expression of genes associated with mitochondrial function was significantly higher in women than in men (Fig. 2), as defined by general GO terms (e.g., mitochondrion) and more specific GO terms (e.g., oxidative phosphorylation and respiratory electron transport chain). We refer to the overrepresentation of these GO terms as a mitochondrial gene signature.
Figure 2.
Heat map of GO terms constituting the mitochondrial gene signature in different tissues from women and men. The color of each box in the heat map indicates the enrichment P value for the overrepresentation of the specific GO term. AT*, The adipose tissue expression profiles from the SOS Sib Pair study offspring cohort (Gene Expression Omnibus database no. GSE27916); AT**, adipose tissue expression profiles in the Icelandic cohort (GSE7965) and blood expression profiles (GSE7965) are from the study by Emilsson et al (16). Expression profiles from skeletal muscle (GSE9676) and liver (GSE9588) were from Welle et al (17) and Schadt et al (18), respectively.
We repeated the integrated analysis on a publically available adipose tissue expression data set from a large Icelandic cohort (391 women and 283 men) and again observed a pronounced mitochondrial gene signature in women (Fig. 2). By contrast, we observed enrichment in some mitochondrial GO terms in men (Fig. 2) when we performed integrated analyses on publically available global expression profiles from blood (458 women and 554 men) and skeletal muscle (15 women and 15 men). No sex difference in the expression of the mitochondrial gene signature was observed for liver (193 women and 234 men) (Fig. 2).
To further analyze the sex difference in the expression of the mitochondrial gene signature in adipose tissue, the cumulative distribution of the fold changes between men and women of the individual genes included in the various GO terms was analyzed. Although the fold changes between men and women were modest for genes defined by mitochondrial GO terms, the cumulative fold change distributions were significantly different from the sex distribution of the nonmitochondrial genes (Fig. 3A; P value derived from KS test < 9.92 × 10−17). Similar results were obtained after adjustment for FM, waist circumference, and HOMA-IR (Supplemental Fig. 1). Among the genes in the mitochondrial gene signature, a group of peroxisome proliferator-activated receptor-γ coactivator 1α (PGC 1α)-responsive genes involved in oxidative phosphorylation, referred to as Oxphos-CR genes (24), were tested in the same way. The Oxphos-CR genes displayed a larger magnitude of fold changes compared with the GO terms in the mitochondrial gene signature (Fig. 3A). The overexpression of Oxphos-CR genes in adipose tissue of women is also illustrated in Fig. 3, B and C. Measurement of the expression of eight Oxphos-CR genes by real-time PCR in a subset of the SOS Sib Pair cohort (Supplemental Table 1) confirmed the significantly higher expression in women compared with men (Fig. 3D).
Figure 3.
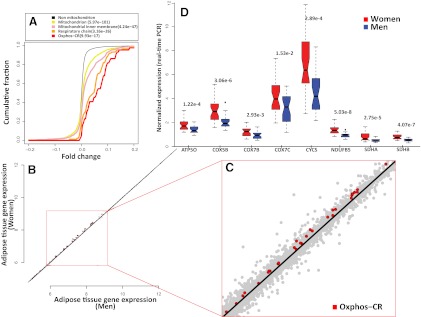
Adipose tissue overexpression of the mitochondrial gene signature and Oxphos-CR genes in women. A, Empirical cumulative distribution of fold changes for selected GO terms in the mitochondrial gene signature and Oxphos-CR genes. Positive fold changes indicate higher expression in women. Scatter plot (B) and magnification of scatter plot (C) for the adipose tissue expression of Oxphos-CR genes in women and men. Red dots represent Oxphos-CR genes, whereas gray dots represent all other genes on the microarray. Gene expression levels and fold changes are displayed using a log2 scale. D, Adipose tissue expression of 8 Oxphos-CR genes measured by real-time PCR. The analysis was performed in a subset of the SOS Sib Pair offspring cohort consisting of 30 women (red) and 28 men (blue). The Oxphos-CR genes are indicated by the gene symbol and data are displayed as box and whisker plots.
To investigate the possible effect of weight loss on the adipose tissue expression of the mitochondrial signature, we analyzed microarray profiles at before-diet intervention (week 0) and after 16 weeks in 24 obese subjects given a VLCD (Supplemental Table 1). Cumulative fold change analysis of the genes belonging to the broad GO term mitochondrion showed that the expression was higher in women than in men before weight loss (P value derived from KS test = 1.54 × 10−52). In the whole cohort, the expression of the mitochondrial gene signature was significantly reduced after weight loss (Fig. 4A). In addition, analysis of the cumulative distribution of the fold changes of genes defined by the GO term mitochondrion showed that the expression in response to weight loss was greater in women than in men (P = .0019) (Fig. 4B). At week 16, men had lost more weight than women, whereas there was no sex difference in the change in BMI (Fig. 4C).
Figure 4.
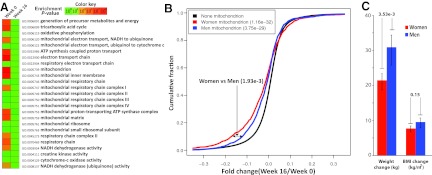
Expression of the mitochondrial gene signature during diet-induced weight loss. A, Heat map of GO terms constituting the mitochondrial gene signature before and after 16 weeks of diet-induced weight loss. B, Empirical cumulative distribution of fold changes for the GO term mitochondrion analyzed separately for women (n = 6) and men (n = 18). Negative fold changes indicate lower expression after weight loss. C, Changes in body weight and BMI in men and women after 16 weeks of diet.
Higher expression of UCP1 in adipose tissue in women
White adipocytes have few mitochondria, whereas brown adipocytes have high mitochondrial density, rendering them capable of converting stored energy to heat (25). We next performed integrated GO analysis of expression profiles of perithyroid adipose tissue containing islets with BAT and paired sc adipose tissue taken from 9 patients undergoing surgery in the thyroid area. As expected, we observed an enrichment of the mitochondrial gene signature in the BAT-containing samples (Fig. 5A). We therefore hypothesized that the sex differences in metabolic rate and mitochondrial gene signature in adipose tissue reflect a higher number of brown adipocytes in sc adipose tissue in women. In agreement with our hypothesis, the microarray expression profiles from the SOS Sib Pair offspring cohort showed that expression of the brown adipocyte marker gene UCP1 (26) was higher in sc adipose tissue from women compared with men (P = .013, Q = 0.069). To verify these results, we performed real-time PCR of UCP1 expression in a subset of the SOS Sib Pair study and in sc adipose tissue biopsies from the Mölndal Metabolic study (Supplemental Table 1). The expression of UCP1 was 5-fold and 6-fold higher (P = 7.43 × 10−7 and P = .00256, respectively) in the women compared with the men in the 2 studies (Fig. 5, B and C).
Figure 5.
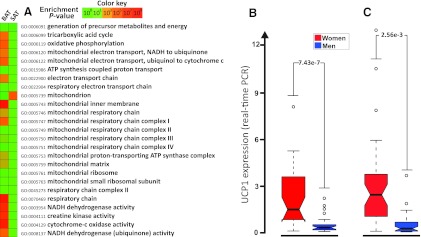
Expression of the mitochondrial gene signature and UCP1 in human adipose tissue. A, Heat map of GO terms constituting the mitochondrial gene signature in BAT-containing perithyroid adipose tissue (BAT) and paired sc adipose tissue (SAT) samples (n = 9). The color of each box in the heat map indicates the enrichment P value for the overrepresentation of the specific GO term. UCP1 expression in subcutaneous adipose tissue in a subset of the SOS Sib Pair study (30 women, 28 men) (B) and from women (n = 39) and men (n = 44) in the Mölndal Metabolic Study (39 women, 44 men) (C), analyzed by real-time PCR. UCP1 expression is displayed as box and whisker plot.
Discussion
The well-known sex differences in obesity and obesity-associated diseases may be related to functional differences within the adipose tissue. In this study we show that the metabolic rate per kilogram adipose tissue is higher in women than in men. To explore the molecular mechanisms behind this sex difference, we analyzed global adipose tissue gene expression and searched for differences in the expression of functional groups of genes. Our study, which is the first to report sex differences in global adipose tissue gene expression patterns in large cohorts, showed that women have increased expression of genes involved in mitochondrial function compared with men. Furthermore, the expression of such genes is down-regulated by weight loss in both men and women, but the reduction is greater in women. In addition, the sex difference in adipose tissue gene expression included an increased expression of the established BAT marker UCP1 in women.
RMR and basal metabolic rate (BMR) are estimates of the amount of energy needed to maintain vital processes in a resting and fasting state and vary between individuals, depending on both the size and the metabolic rate of all tissues in the body. It is well established that FFM, comprising brain, internal organs, and skeletal muscle, is the major determinant of BMR (27). The brain and internal organs such as liver, kidney, and heart have very high metabolic rates with a joint account of about 70%–80% of BMR but constitute only 5% of the body weight. Skeletal muscle, which has approximately 20-fold lower metabolic rate compared with the internal organs but much higher total weight, accounts for about 15% of BMR (27). The metabolic rate in adipose tissue is much lower compared with internal organs and muscle, and the importance of adipose tissue for the variation in whole-body energy expenditure has therefore not been extensively studied. However, Weyer et al (28) have shown that, after FFM, FM is the phenotypic characteristic contributing most to the variability of energy expenditure. In the general population, adipose tissue accounts for only about 6% of the variability of BMR (29). Nevertheless, the total as well as the relative amount of adipose tissue varies more than that of any other organ, and its impact on BMR is therefore higher in morbidly obese subjects.
Both BMR and body composition differ substantially between men and women. For example, men have larger muscle mass than women, whereas women therefore have higher proportion of their FFM as high metabolic-rate organs such as liver, kidney, and brain (30). The contribution of FM to BMR is greater in women than in men (31), and it has been assumed that this reflects the higher body fat content in women compared with men. However, our analysis shows that adipose tissue has higher metabolic rate per kilogram in women than in men, suggesting that the higher impact of FM on BMR in women partly is caused by differences within the adipose tissue that influence its energy turnover.
The practical significance of the sex difference in adipose tissue contribution to energy metabolism remains to be determined, but there are some situations in which it may have an impact. For example, it is well known that BMR is reduced during weight loss (32). Higher energy turnover in female adipose tissue suggests that loss of a certain amount of adipose tissue mass will result in a greater reduction in energy expenditure in women than in men, a mechanism that would counteract further weight loss. In theory, the sex difference in energy turnover in adipose tissue could therefore result in sex differences in the outcomes of weight loss programs because more metabolically active tissue is lost in women than in men. However, the regulation of body weight occurs through complex feedback systems that are likely to mask the clinical importance of such sex differences.
Analysis of global adipose tissue gene expression showed that female adipose tissue displayed a mitochondrial gene signature characterized by increased expression of numerous genes involved in mitochondrial function. The biological relevance of the mitochondrial gene signature is supported by the down-regulation in response to weight loss, a situation when RMR is reduced (32). The most distinct difference in expression was observed for Oxphos-CR genes, a group of PGC 1α-responsive genes. PGC 1α is a transcriptional coactivator implicated in mitochondriogenesis (33). Fat oxidation and the generation of energy in the form of ATP take place in mitochondria, and this organelle therefore plays a central role in energy metabolism. Each mitochondrion has multiple copies of a circular mitochondrial DNA genome that encodes 13 proteins that are essential for the respiratory function. However, most proteins necessary for the mitochondrial functions are derived from nuclear genes (34), and the increased mitochondrial signature in women was due to the increased expression of genes from nuclear DNA.
Metabolically active cells, such as those of internal organs, contain thousands of mitochondria, whereas cells with a low metabolic rate, such as white adipocytes, contain only a few. One interpretation of the increased expression of the mitochondrial signature in women is the adipose tissue in women contains more mitochondria or more active mitochondria. Adipose tissue is a complex tissue that consists of adipocytes and various stromavascular cells. Although the white adipocyte is the main cell type, sc adipose tissue from adult humans has been reported to contain some brown adipocytes (35). In contrast to white adipocytes, brown adipocytes are mitochondria-rich cells with multiple small lipid droplets. The brown adipocytes have a unique ability to generate heat instead of ATP through uncoupling of oxidative phosphorylation (25). This process is dependent on the expression of UCP1, a protein that allows protons to leak through the inner membrane of the mitochondria. Our data show that the adipose tissue expression of UCP1 is markedly higher in women compared with men. This suggests that adipose tissue in women has a higher capacity to burn calories by converting the energy to heat, something that may in part explain the higher contribution of adipose tissue to BMR in women than in men. UCP1 expression is considered a specific marker for BAT, and we and others have recently demonstrated that in histological sections of human adipose tissue containing both white and brown adipocytes, UCP1 is specifically expressed in the cells with brown adipocyte morphology (14, 26, 36). The increased adipose tissue expression of UCP1 suggests that women have more brown adipocytes in their subcutaneous adipose tissue.
Studies in rodents have clearly demonstrated that BAT is an important regulator of body temperature and body weight (25), but its role in humans is not as clear. The idea that BAT disappears early in life and therefore lacks importance in adults has recently been challenged by several studies. Specific BAT depots have been detected in the neck region and in central parts of the body of human adults (37). Furthermore, BAT activity in these depots is higher in lean than in obese subjects (38, 39), and BAT activity correlates with resting metabolic rate (39), indicating that BAT plays a role in adult human metabolism. Interestingly, glucose uptake in BAT depots is higher in women compared with men (38). The existence of a sex difference is also supported by rodent studies showing that female rats have a larger percentage of BAT depots per body mass (40). Our study suggests that the increased amount of BAT in women is not restricted to specific BAT depots but also includes increased number of brown adipocytes in their sc adipose tissue.
In this study we show that each kilogram of adipose tissue contributes more to resting metabolic rate in women than in men, and sex differences in adipose tissue gene expression suggest that this may in part be due to increased number of brown adipocytes. Given the higher adiposity in women, we conclude that the relative contribution of adipose tissue RMR to total RMR is greater in women than in men. Although FFM is the major determinant of energy expenditure, the impact of small sex differences in the contribution of FM to BMR may be substantial for conditions and diseases that develop slowly over several decades. Our findings may therefore have implications for the well-established sex differences in obesity and its health consequences.
Supplementary Material
Acknowledgments
We thank Rosie Perkins for editing the manuscript and Jenny M. Hoffman and Bengt E. Nilsson for valuable help in the project.
This study was supported by Grants K2010-55X-11285-13 and K2008-65X-20753-01-4 from the Swedish Research Council; Grant GR079534 from the Wellcome Trust; the Swedish Foundation for Strategic Research; the Chalmers Foundation; the Knut and Alice Wallenberg Foundation; Sahlgrenska Center for Cardiovascular and Metabolic Research; Bioinformatics Infrastructure for Life Sciences (BILS); and the Swedish federal government under the Läkarutbildningsavtalet/Avtal om läkarutbildning och forskning (LUA/ALF) agreement.
The computational analyses were performed on resources provided by the Swedish National Infrastructure for Computing (SNIC) at C3SE.
Disclosure Summary: The authors declare no conflict of interest.
Footnotes
- BAT
- Brown adipose tissue
- BMI
- body mass index
- BMR
- basal metabolic rate
- FFM
- fat-free mass
- FM
- fat mass
- GO
- Gene Ontology
- HOMA-IR
- homeostasis model assessment-insulin resistance
- KS
- Kolmogorov-Smirnov statistics
- PGC 1α
- peroxisome proliferator-activated receptor-γ coactivator 1α
- RMR
- resting metabolic rate
- UCP1
- uncoupling protein 1
- VLCD
- very low-calorie diet.
References
- 1. Smith SR, Lovejoy JC, Greenway F, et al. Contributions of total body fat, abdominal subcutaneous adipose tissue compartments, and visceral adipose tissue to the metabolic complications of obesity. Metabolism. 2001;50(4):425–435 [DOI] [PubMed] [Google Scholar]
- 2. Lemieux S, Prud'homme D, Bouchard C, Tremblay A, Despres JP. Sex differences in the relation of visceral adipose tissue accumulation to total body fatness. Am J Clin Nutr. 1993;58(4):463–467 [DOI] [PubMed] [Google Scholar]
- 3. Bergman RN, Kim SP, Hsu IR, et al. Abdominal obesity: role in the pathophysiology of metabolic disease and cardiovascular risk. Am J Med. 2007;120(2 suppl 1):S3–S8; discussion S29–S32 [DOI] [PubMed] [Google Scholar]
- 4. Mathieu P, Poirier P, Pibarot P, Lemieux I, Despres JP. Visceral obesity: the link among inflammation, hypertension, and cardiovascular disease. Hypertension. 2009;53(4):577–584 [DOI] [PubMed] [Google Scholar]
- 5. Tarnopolsky MA. Sex differences in exercise metabolism and the role of 17-β estradiol. Med Sci Sports Exerc. 2008;40(4):648–654 [DOI] [PubMed] [Google Scholar]
- 6. Lofgren P, Hoffstedt J, Ryden M, et al. Major gender differences in the lipolytic capacity of abdominal subcutaneous fat cells in obesity observed before and after long-term weight reduction. J Clin Endocrinol Metab. 2002;87(2):764–771 [DOI] [PubMed] [Google Scholar]
- 7. Kolehmainen M, Vidal H, Ohisalo JJ, Pirinen E, Alhava E, Uusitupa MI. Hormone sensitive lipase expression and adipose tissue metabolism show gender difference in obese subjects after weight loss. Int J Obes Relat Metab Disord. 2002;26(1):6–16 [DOI] [PubMed] [Google Scholar]
- 8. Montague CT, Prins JB, Sanders L, Digby JE, O'Rahilly S. Depot- and sex-specific differences in human leptin mRNA expression: implications for the control of regional fat distribution. Diabetes. 1997;46(3):342–347 [DOI] [PubMed] [Google Scholar]
- 9. Kern PA, Di Gregorio GB, Lu T, Rassouli N, Ranganathan G. Adiponectin expression from human adipose tissue: relation to obesity, insulin resistance, and tumor necrosis factor-alpha expression. Diabetes. 2003;52(7):1779–1785 [DOI] [PubMed] [Google Scholar]
- 10. Gallagher D, Belmonte D, Deurenberg P, et al. Organ-tissue mass measurement allows modeling of REE and metabolically active tissue mass. Am J Physiol. 1998;275(2 Pt 1):E249–E258 [DOI] [PubMed] [Google Scholar]
- 11. Carlsson LM, Jacobson P, Walley A, et al. ALK7 expression is specific for adipose tissue, reduced in obesity and correlates to factors implicated in metabolic disease. Biochem Biophys Res Commun. 2009;382(2):309–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Larsson I, Lindroos AK, Peltonen M, Sjostrom L. Potassium per kilogram fat-free mass and total body potassium: predictions from sex, age, and anthropometry. Am J Physiol Endocrinol Metab. 2003;284(2):E416–E423 [DOI] [PubMed] [Google Scholar]
- 13. Gummesson A, Jernas M, Svensson PA, et al. Relations of adipose tissue CIDEA gene expression to basal metabolic rate, energy restriction, and obesity: population-based and dietary intervention studies. J Clin Endocrinol Metab. 2007;92(12):4759–4765 [DOI] [PubMed] [Google Scholar]
- 14. Svensson PA, Jernas M, Sjoholm K, et al. Gene expression in human brown adipose tissue. Int J Mol Med. 2011;27(2):227–232 [DOI] [PubMed] [Google Scholar]
- 15. Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264 [DOI] [PubMed] [Google Scholar]
- 16. Emilsson V, Thorleifsson G, Zhang B, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452(7186):423–428 [DOI] [PubMed] [Google Scholar]
- 17. Welle S, Tawil R, Thornton CA. Sex-related differences in gene expression in human skeletal muscle. PLoS One. 2008;3(1):e1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schadt EE, Molony C, Chudin E, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6(5):e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patil KR, Nielsen J. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc Natl Acad Sci USA. 2005;102(8):2685–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oliveira AP, Patil KR, Nielsen J. Architecture of transcriptional regulatory circuits is knitted over the topology of bio-molecular interaction networks. BMC Syst Biol. 2008;2:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3 [DOI] [PubMed] [Google Scholar]
- 22. Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodological. 1995;57(1):289–300 [Google Scholar]
- 23. Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci USA. 2000;97(18):10101–10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273 [DOI] [PubMed] [Google Scholar]
- 25. Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84(1):277–359 [DOI] [PubMed] [Google Scholar]
- 26. Virtanen KA, Lidell ME, Orava J, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360(15):1518–1525 [DOI] [PubMed] [Google Scholar]
- 27. Muller MJ, Bosy-Westphal A, Kutzner D, Heller M. Metabolically active components of fat-free mass and resting energy expenditure in humans: recent lessons from imaging technologies. Obes Rev. 2002;3(2):113–122 [DOI] [PubMed] [Google Scholar]
- 28. Weyer C, Snitker S, Rising R, Bogardus C, Ravussin E. Determinants of energy expenditure and fuel utilization in man: effects of body composition, age, sex, ethnicity and glucose tolerance in 916 subjects. Int J Obes Relat Metab Disord. 1999;23(7):715–722 [DOI] [PubMed] [Google Scholar]
- 29. Johnstone AM, Murison SD, Duncan JS, Rance KA, Speakman JR. Factors influencing variation in basal metabolic rate include fat-free mass, fat mass, age, and circulating thyroxine but not sex, circulating leptin, or triiodothyronine. Am J Clin Nutr. 2005;82(5):941–948 [DOI] [PubMed] [Google Scholar]
- 30. Geer EB, Shen W. Gender differences in insulin resistance, body composition, and energy balance. Gend Med. 2009;6(Suppl 1):60–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Buchholz AC, Rafii M, Pencharz PB. Is resting metabolic rate different between men and women? Br J Nutr. 2001;86(6):641–646 [DOI] [PubMed] [Google Scholar]
- 32. Van Gaal LF, Vansant GA, De Leeuw IH. Factors determining energy expenditure during very-low-calorie diets. Am J Clin Nutr. 1992;56(1 suppl):224S–229S [DOI] [PubMed] [Google Scholar]
- 33. Wu Z, Puigserver P, Andersson U, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–124 [DOI] [PubMed] [Google Scholar]
- 34. Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 5:297–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heaton JM. The distribution of brown adipose tissue in the human. J Anat. 1972;112(Pt 1):35–39 [PMC free article] [PubMed] [Google Scholar]
- 36. Zingaretti MC, Crosta F, Vitali A, et al. The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. FASEB J. 2009;23(9):3113–3120 [DOI] [PubMed] [Google Scholar]
- 37. Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007;293(2):E444–E452 [DOI] [PubMed] [Google Scholar]
- 38. Cypess AM, Lehman S, Williams G, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360(15):1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360(15):1500–1508 [DOI] [PubMed] [Google Scholar]
- 40. Rodriguez-Cuenca S, Pujol E, Justo R, et al. Sex-dependent thermogenesis, differences in mitochondrial morphology and function, and adrenergic response in brown adipose tissue. J Biol Chem. 2002;277(45):42958–42963 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.