Abstract
Rabies kills many people throughout the developing world every year. The murine monoclonal antibody (mAb) 62-71-3 was recently identified for its potential application in rabies postexposure prophylaxis (PEP). The purpose here was to establish a plant-based production system for a chimeric mouse-human version of mAb 62-71-3, to characterize the recombinant antibody and investigate at a molecular level its interaction with rabies virus glycoprotein. Chimeric 62-71-3 was successfully expressed in Nicotiana benthamiana. Glycosylation was analyzed by mass spectroscopy; functionality was confirmed by antigen ELISA, as well as rabies and pseudotype virus neutralization. Epitope characterization was performed using pseudotype virus expressing mutagenized rabies glycoproteins. Purified mAb demonstrated potent viral neutralization at 500 IU/mg. A critical role for antigenic site I of the glycoprotein, as well as for two specific amino acid residues (K226 and G229) within site I, was identified with regard to mAb 62-71-3 neutralization. Pseudotype viruses expressing glycoprotein from lyssaviruses known not to be neutralized by this antibody were the controls. The results provide the molecular rationale for developing 62-71-3 mAb for rabies PEP; they also establish the basis for developing an inexpensive plant-based antibody product to benefit low-income families in developing countries.—Both, L., van Dolleweerd, C., Wright, E., Banyard, A. C., Bulmer-Thomas, B., Selden, D., Altmann, F., Fooks, A. R., Ma, J. K.-C. Production, characterization, and antigen specificity of recombinant 62-71-3, a candidate monoclonal antibody for rabies prophylaxis in humans.
Keywords: plant biotechnology, molecular pharming, PEP, tobacco
Rabies has the highest human case:fatality ratio of all infectious diseases, and it is widely accepted that there is no effective treatment after onset of symptoms (1–3). The causative agent is a negative-stranded RNA virus in the order Mononegavirales, family Rhabdoviridae, genus Lyssavirus (4). All mammals are susceptible and can transmit rabies virus (RV; ref. 5), and both canine and sylvatic (wildlife) circulation patterns are recognized (6). Most human exposures are associated with the bites of rabid animals, in particular, unvaccinated dogs, and transmission of RV in their saliva (7). Human rabies cases have also been attributed to probable aerosol exposures in laboratories or airborne exposures in caves with high densities of bats (8, 9). In addition, atypical transmission through butchering and processing of rabid animals, as well as human-to-human transmission by organ or tissue transplantation have been reported (10, 11).
Although viral spread to the central nervous system (CNS) and resulting encephalitis are almost invariably fatal, the disease is preventable through postexposure prophylaxis (PEP). Swift administration of PEP is virtually 100% effective in preventing the onset of symptoms and fatal clinical disease after exposure (12–17). Rabies PEP is based on 3 pillars: wound cleansing, administration of rabies vaccine, and infiltration of rabies immunoglobulins (RIGs) of either human or equine origin (HRIGs or ERIGs, respectively). However, insufficient access to RIGs restricts the administration of appropriate PEP across the developing world where the vast majority of the annual 55,000–70,000 rabies fatalities occur (18–22). To overcome the short supply and the safety issues with blood-derived RIG products, several human and murine monoclonal antibodies (mAbs) are being investigated (23–25). A recent report by the World Health Organization (WHO) Rabies Collaborating Centres described the identification of three novel combinations of mAbs to replace RIGs (6). Stringent criteria concerning the neutralizing activity, binding specificities to different epitopes, immunoglobulin isotype, and history of hybridomas were used to evaluate the suitability of several murine mAbs. Combinations of 2 mAbs, all including mAb 62-71-3, were assessed both in vitro and in vivo and were shown to have an equal or superior efficiency to HRIGs in the hamster PEP model (6).
The objective of the present study was to clone and express a chimeric (mouse-human) full-length IgG1 version of mAb 62-71-3, using plants as an inexpensive production alternative to existing mammalian systems, and to perform a detailed molecular characterization of the recombinant mAb. Initially, a phage-displayed single-chain variable fragment (scFv) of mAb 62-71-3 was expressed in Escherichia coli and tested to confirm that the sequences for heavy and light chains correctly encoded for an antibody with neutralizing potency toward the virus. A chimeric 62-71-3 full-length IgG was then cloned, expressed, and purified from Nicotiana benthamiana leaves. The plant-derived mAb was investigated using mass spectrometry for glycan analysis, RV glycoprotein enzyme-linked immunosorbent assay (ELISA), fluorescent antibody virus neutralization (FAVN) and pseudotype neutralization assay (PNA). Mutations in antigenic site I of the RV glycoprotein severely diminished neutralization by mAb 62-71-3, pointing to an important role of this epitope in the binding between the viral glycoprotein and the plant-derived antibody.
The work presented here confirms the molecular rationale of using mAb 62-71-3 as part of a mAb cocktail for rabies PEP. It also highlights the feasibility of using plants for the inexpensive production of mAbs for developing countries (26, 27). Plants constitute an economically feasible production platform that can easily be scaled up and that is amenable for transfer to the developing world (28). As plants are eukaryotic organisms, they possess a similar intracellular machinery to that of mammalian cells, so that complex proteins like antibodies are correctly folded and assembled (29, 30).
MATERIALS AND METHODS
Cloning and expression of the 62-71-3 phage-displayed scFv
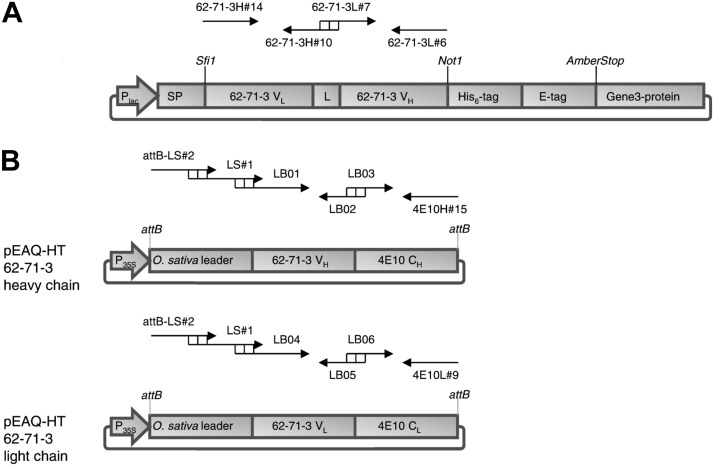
mAb 62-71-3 is a hybridoma-derived IgG2b antibody, originally generated by immunizing BALB/c mice with the rabies vaccine strain ERA (6, 31). The cDNA sequences for the variable regions of mAb 62-71-3 were received from Apotech (Lausanne, Switzerland). To confirm cloning of the correct variable region sequences, an scFv version of mAb 62-71-3 was initially expressed in E. coli. The variable regions of heavy and light chains were amplified by PCR and were connected with a flexible 15-aa linker, using the primers listed in Table 1 and the cloning strategy described in Supplemental Data. A schematic representation of the cloning strategy is shown in Fig. 1A.
Table 1.
PCR primers for cloning the phage-displayed 62-71-3 scFv
| Fragment | Primer |
|---|---|
| 62-71-3H#14 | ctatgcggcccagccggccatggctcaggtgcagctgaaggagtca |
| 62-71-3H#10 | accgctgccaccaccgccggagccaccgccacctgaggagactgtgagagtggt |
| 62-71-3L#7 | tccggcggtggtggcagcggtggcggcggttctgatgtccagatgacacagact |
| 62-71-3L#6 | tggtgctgcggccgcccgttttatttccagcttggtccc |
Figure 1.
Schematic representation of the constructs used in this study. A) Scheme of the phage-displayed 62-71-3 scFv for expression in E. coli. B) Scheme of the heavy and light chains of mAb 62-71-3 for expression in N. benthamiana. Primers and their relative location/orientation are indicated with black arrows.
Cloning and expression of chimeric mAb 62-71-3
The murine variable regions of mAb 62-71-3 were grafted on the constant regions of a human mAb (IgG1 κ), using the primers listed in Table 2 and the cloning strategy described in Supplemental Data. A schematic representation of the cloning strategy is shown in Fig. 1B. The heavy and light chain sequences of the chimeric 62-71-3 were then shuttled from the entry vector pDONR into the Gateway destination vector pEAQ-HT-Dest3 for expression in plants (32) or with the Gateway destination vectors pcDNA-Dest40 and pEF-Dest51 (Invitrogen, Carlsbad, CA, USA) for expression in mammalian cells. Agrobacterium tumefaciens cultures (strain LBA4404) transformed with either the heavy-chain or light-chain vectors were each adjusted to optical density at 600 nm (OD600) = 1 by diluting the cells with infiltration buffer (10 mM MES and 10 mM MgCl2, pH 5.6) and combined. The cells were incubated in the dark (2 h, room temperature) before infiltration of N. benthamiana plants with a 1-ml syringe without needle (2–3 leaves/plant). Soluble leaf extracts were prepared by grinding leaf tissue in a mortar and centrifugation.
Table 2.
PCR primers for cloning the 62-71-3 IgG
| Fragment | Primer |
|---|---|
| attB1-OryzaLS#2 | ggggacaagtttgtacaaaaaagcaggctcaaccatggggaagcaaatggccgccctgtgtggctttctc |
| OryzaLS#1 | agcaaatggccgccctgtgtggctttctcctcgtggcgttgctctggctcacgcccgacgtc |
| LB01 | cgttgctctggctcacgcccgacgtcgcgcatggtcaggtgcagctgaaggagtca |
| LB02 | accgatgggcccttggtggaggctgaggagactgtgagagtggt |
| LB03 | accactctcacagtctcctcagcctccaccaagggcccatcggt |
| 4E10H#15 | ggggaccactttgtacaagaaagctgggtctttacccggagacagggagaggct |
| LB04 | cgttgctctggctcacgcccgacgtcgcgcatggtgatgtccagatgacacagact |
| LB05 | acagatggtgcagccacagtccgttttatttccagcttggt |
| LB06 | accaagctggaaataaaacggactgtggctgcaccatctgt |
| 4E10L#9 | ggggaccactttgtacaagaaagctgggtcggtacctaacactctcccctgttgaagctcttt |
The HEK293-derived mAb 62-71-3 was generated by mixing 1 μg of the heavy- and light-chain plasmids with Fugene 6 (Roche, Basel, Switzerland) according to manufacturer's instructions and transfection of 70% confluent 293T-17 cells in a 6-well plate containing 2 ml medium/well (DMEM plus 15% FBS and 1% Pen/Strep). The plates were incubated at 37°C (5% CO2), and medium was changed after 24 h. After 2–3 d, the supernatants were collected, passed through a 0.45-μm filter, and stored at 4°C.
SDS-PAGE and Western blot analysis
The soluble fraction of a plant extract was passed through Miracloth (EMD Millipore, Billerica, MA, USA) and a 0.45-μm filter, and the mAb was purified by protein G (Sigma, Gillingham, UK) affinity chromatography. Both the purified mAb and the crude plant extract were analyzed by SDS-PAGE and semidry Western blot analysis. SDS-PAGE was performed using the Invitrogen Minigel system and Invitrogen NuPAGE buffers. Electrophoresis was carried out in 4–12% gradient gels (Invitrogen), which were stained with Coomassie brilliant blue or subjected to Western blotting. Transfer to the membrane (Amersham Hybond-ECL; Amersham Biosciences, Little Chalfont, UK) was carried out in a semidry system (Invitrogen). The membrane was blocked with milk (3%), incubated with horseradish peroxidase (HRP)-coupled antibodies at a concentration of 1:10,000, and developed (Amersham ECL Plus Western blotting detection kit, Amersham Hyperfilm ECL).
RV glycoprotein ELISA
A commercial ELISA kit (Bio-Rad Platelia Kit; Bio-Rad, Hemel Hempstead, UK) was used to investigate recombinant antibody binding to its target antigen. The ELISA is based on RV glycoprotein coated on the plate and detection of antibody using HRP-coupled protein A. Clarified plant extract supernatants (at 3, 5, and 7 d postinfiltration) were applied to the wells, and the assay was run according to the manufacturer's instructions.
The RV glycoprotein ELISA was also used for competition experiments, as described recently (33), with minor modifications. Briefly, plates were incubated with saturating amounts of purified plant-derived mAbs 62-71-3 and E559 for 1 h at room temperature. For the initial production of the plant-derived mAb E559, the E559 hybridoma was obtained from Dr. Thomas Müller [Friedrich Loeffler Institut(FLI), Wusterhausen, Germany; ref. 6). The hybridoma heavy- and light-chain sequences for this murine IgG1 antibody were obtained by RT-PCR, engineered into chimeric mouse-human sequences, and cloned into the plant expression vector pL32. The chimeric heavy- and light-chain sequences of mAb E559 were then coexpressed in N. benthamiana by agroinfiltration and purified as described above for mAb 62-71-3.
For the competition experiment, the hybridoma-derived mAb E559 was biotinylated with the No-Weigh Sulfo-NHS-LC-Biotin (Pierce, Rockford, IL, USA), according to manufacturer's instructions, and 50 μl of biotinylated mAb E559 (2.5 μg/ml) was then added to each well, incubated for 5 min at room temperature, and rinsed 5 times with 100 μl of Tris-buffered saline with 0.1% Tween (TBST). Subsequently, wells were incubated for 1 h at room temperature with 50 μl of a 1:5,000 dilution of streptavidin-HRP (Sigma). Wells were rinsed, and HRP activity was detected by the addition of 3,3′,5,5′-tetramethylbenzidine dihydrochloride substrate (Sigma). Color development was allowed to proceed for 10 min at room temperature before the addition of 2 M H2SO4 to terminate the reaction. The OD was measured at 450 nm on an ELISA plate reader.
Generation of lentiviral pseudotype viruses
DNA plasmids encoding the HIV gag-pol, luciferase reporter gene, and lyssavirus glycoproteins were used as previously reported (33). Briefly, pseudotype viruses were generated by mixing 1 μg of the HIV gag-pol plasmid, 1 μg of the glycoprotein plasmid, and 1.5 μg of the reporter gene plasmid with Fugene 6 (Roche) and transfection of 70% confluent 293T-17 cells in a 6-well plate containing 2 ml medium/well (DMEM plus 15% FBS and 1% Pen/Strep). The plates were incubated at 37°C (5% CO2), and medium was changed after 24 h. After 2 d, the supernatants containing pseudotype virus were collected, passed through a 0.45-μm filter, and frozen at −80°C.
PNA
The concentration of the purified plant-derived mAb was measured with the bicinchoninic acid (BCA) protein assay kit (Pierce). The phage-displayed 62-71-3 scFv or plant-derived 62-71-3 IgG (5 μg/ml) was added to medium (DMEM plus 10% FCS and 1% Pen/Strep) and titrated in doubling dilutions across a 96-well plate, starting with a 1:10 or 1:20 dilution (final volume of 50 μl/well). Controls were cells only, virus only, and cells and virus, which were set up with appropriate amounts of medium. Pseudotype virus (50 μl) was added to each well (apart from cells-only control) at a dilution where the international reference serum (OIE+ serum) neutralizes 100% at 1:40 serum dilution [∼100 median tissue culture infective dose (TCID50)]. The plate was centrifuged (500 rpm, 5 s) and incubated at 37°C (5% CO2) for 1 h. Medium (100 μl) containing 2 × 104 BHK cells was added to each well (apart from virus-only control), making up a total volume of 200 μl/well. The plate was centrifuged (500 rpm, 5 s) and incubated at 37°C (5% CO2) for 48 h. Medium (115 μl) was removed from each well, and 75 μl BrightGlo (Promega, Madison, WI, USA) was added. After measuring absolute infection (light units) by the pseudotype virus with a luminometer, the relative neutralization was calculated, and titration curves were generated.
FAVN assay
The standard FAVN test was set up in a similar way to the PNA, starting with a 1:8 dilution of antibody. OIE+ and OIE− sera were included as controls for CVS. To assess neutralization of other lyssaviruses, a modified FAVN was performed as described previously (63). Briefly, for each sample, mAbs and the viruses were incubated at 37°C (5% CO2) for 1 h before adding 4 × 105 BHK cells to each well. The plates were then incubated at 37°C (5% CO2) for 48 h, fixed in 80% acetone, and air-dried. The staining was carried out by adding 50 μl of fluorescein isothiocyanate (FITC)-conjugated antibody (Centocor, Philadelphia, PA, USA), specific for the RV nucleoprotein, to each well. After a 30-min incubation at 37°C (5% CO2), each plate was washed 3 times with PBS. Excess PBS was removed by briefly inverting the microplates on absorbent paper, and the neutralizing titer was evaluated by fluorescent microscopy.
Glycan analysis of the plant-derived mAb 62-71-3
A glycoproteomic analysis was undertaken by in-gel digestion of S-carbamidomethylated sample and analysis by reverse-phase electrospray ionization mass spectrometry (RP-ESI-MS), as described previously (63). Tandem MS results were also subjected to Mascot MS/MS ion search (Matrix Science Ltd., London, UK; http://www.matrixscience.com).
Mutational analysis of the RV glycoprotein
Chimeric RV/Lagos bat virus (LBV) glycoproteins were generated by swapping antigenic sites I–IV of the RV strain CVS11 (accession no. EU352767) with those of LBV strain Nig56-RV1 (accession no. EF547431). Moreover, glycoproteins containing either a K226R, G229E, or N336S mutation within antigenic site I were generated by site-directed mutagenesis, using the cloning primers 5′-GCGCGCGGTACCGCCACCATGGTTCCTCAGGTTCTT-3′ and 5′-GCGCGCCTCGAGTTACAGTCTGATCTCACCTC-3′, annealing to opposite ends of the CVS11 glycoprotein. Mutagenesis primers were 5′-ATGCAGGCTCAGGTTATGTGGAG-3′ (forward) and 5′-CTCCACATAACCTGAGCCTGCAT-3′ (reverse) for the K226R mutation within antigenic site I, 5′-CAAGTTATGTGAAGTACTTGGACTTAG-3′ (forward) and 5′-CTAAGTCCAAGTACTTCACATAACTTG-3′ (reverse) for the G229E mutation within antigenic site I, and 5′-TCCGGACCTGGAGTGAGATCATCC-3′ (forward) and 5′-GGATGATCTCACTCCAGGTCCGGA-3′ (reverse) for the N336S mutation within antigenic site III. All mutated glycoproteins were shuttled into the plasmid pl18 via KpnI and XhoI restriction sites and expressed on the surface of lentiviral pseudotype viruses, as described above. Neutralization assays with these pseudotypes were undertaken by preincubating the phage-displayed 62-71-3 scFv or the plant-derived 62-71-3 IgG (1 μg/ml) with the pseudotypes before adding the BHK cells, as described above. Controls were cells only, virus only, cells and virus, and the plant-derived mAb E559, known to be directed against antigenic site II of the viral glycoprotein.
RESULTS
Characterization of phage-displayed 62-71-3 scFv
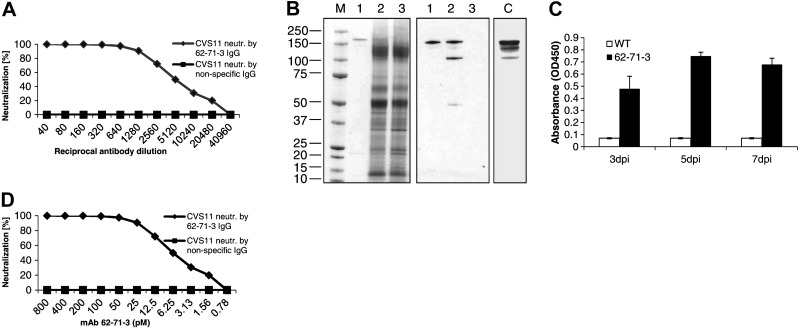
The cDNA sequences for the variable regions of the heavy and light chains of mAb 62-71-3 were cloned into a phagemid vector (Fig. 1A) to verify that the sequences correctly encoded for an antibody with neutralizing potency toward the virus. The phage-displayed 62-71-3 scFv was tested for its neutralization of a lentivirus (HIV) pseudotyped with the RV glycoprotein (strain CVS11) and demonstrated potent, dose-dependent neutralization of the pseudotype virus (Fig. 2A), confirming that the correct heavy and light chains were cloned. Controls included a nonspecific scFv (Fig. 2A), as well as cells only, virus only, and cells and virus (data not shown).
Figure 2.
Pseudotype virus neutralization by the 62-71-3 phage-displayed scFv and the plant-derived IgG. A) Titration curve showing the pseudotype virus neutralization by the phage-displayed 62-71-3 scFv and by a phage-displayed nonspecific scFv (control). B) SDS-PAGE (left panel) and Western blotting (middle and right panels) under nonreducing conditions. M denotes Coomassie gel with molecular mass marker in kDa. Lane 1, purified plant-derived 62-71-3 IgG; lane 2, plant extract from infiltrated plants; lane 3, plant extract from wild-type plants. C denotes control with HEK293-derived mAb 62-71-3. The Western blot was probed with an anti-human HC-specific antibody (Sigma). C) RV glycoprotein ELISA with plant samples harvested at various days post infiltration (dpi). D) Titration curve showing the pseudotype virus neutralization by the plant-derived 62-71-3 IgG and by a nonspecific IgG (control). Titrations were repeated 2 times; each panel shows one representative run.
Production and antigen-binding properties of plant-derived chimeric mAb 62-71-3
After the initial neutralization experiments with the 62-71-3 scFv, a chimeric antibody was constructed (Fig. 1B) and expressed in N. benthamiana. Infiltrated N. benthamiana leaves showed robust expression of 62-71-3 IgG (estimated as ∼3–4% of total soluble protein by quantitative ELISA and in excess of 100 mg/kg fresh tissue). The antibody was purified from the soluble plant extract fraction by affinity chromatography with protein G, which binds the human Fc region of the chimeric antibody heavy chain. The crude plant extract and the purified antibody were analyzed by nonreducing SDS-PAGE and Western blot analysis (Fig. 2B). Probing Western blots with an anti-HC antibody revealed the presence of mAb 62-71-3 in plant extracts and in the purified samples. The corresponding band was also detected in HEK293 supernatants after cotransfection with the 62-71-3 heavy- and light-chain vectors (Fig. 2B).
To confirm the specific antigen recognition of the plant-derived mAb, a functional ELISA was performed on plant extracts harvested at different time points after agroinfiltration. Samples derived from infiltrated leaves demonstrated specific binding to the RV glycoprotein, while samples derived from wild-type leaves showed no relevant antigen binding (Fig. 2C). This experiment confirmed that mAb 62-71-3 was correctly assembled in planta and that substituting the original murine antibody Fc region with its human counterpart did not abrogate its antigen-binding activity.
Neutralization of live viruses and pseudotype viruses
The plant-derived 62-71-3 IgG was tested for its neutralization of a rabies pseudotype virus (strain CVS11) and demonstrated potent neutralization (Fig. 2D). The concentration of the purified plant-derived mAb was measured with the bicinchoninic acid (BCA) protein assay kit (Pierce). A nonspecific IgG had no neutralizing activity. The plant-derived 62-71-3 also demonstrated excellent specific neutralization activity against BBLV, KELEV, and an E559 mAb escape mutant (6), weak neutralization against Pasteur virus, but no neutralization of Duvenhage virus or LBV (data not shown).
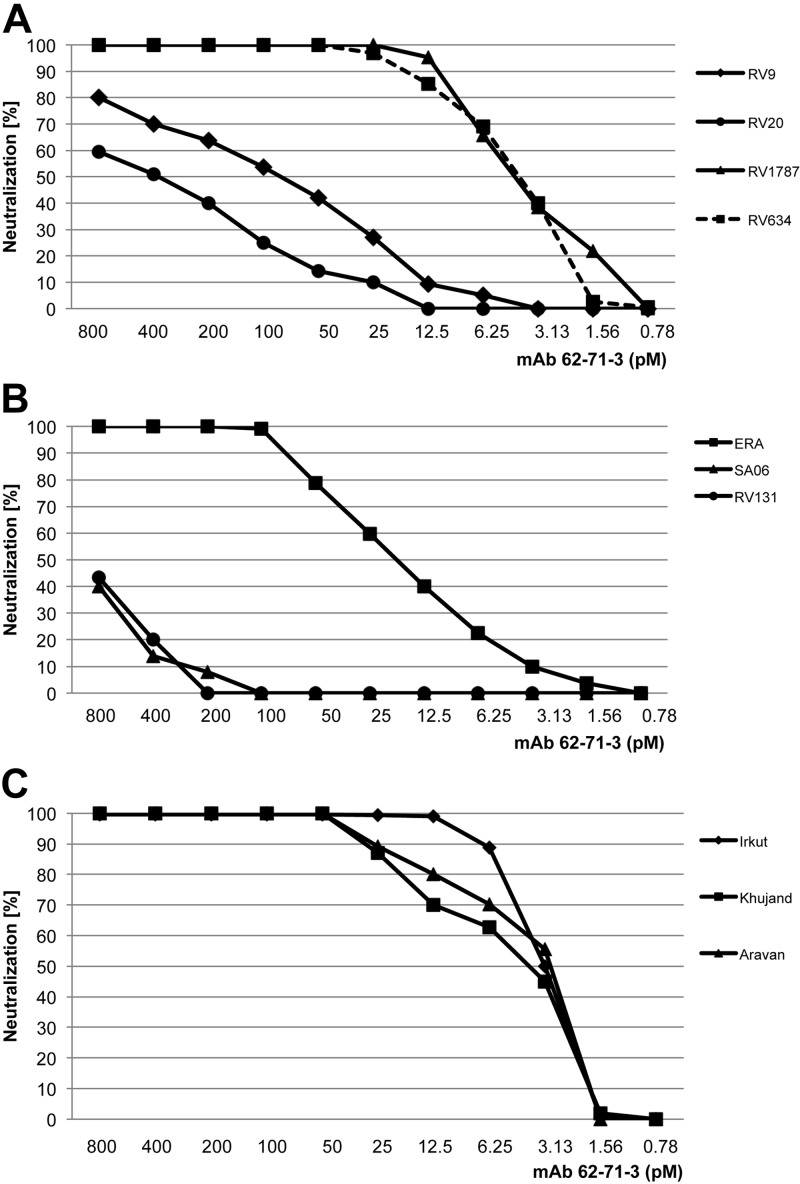
The breadth of neutralization was further investigated using lentiviruses pseudotyped with a panel of lyssavirus glycoproteins from different phylogroup I viruses and some related Eurasian lyssaviruses. In addition to the CVS11 strain, the plant-derived IgG demonstrated potent neutralization of several different lyssaviruses, including ERA (RV), RV1787, and RV634 (Fig. 3A, B). Diminished neutralization was observed for the two Duvenhage virus strains SA06 and RV131 (Fig. 3B) and the two European bat lyssavirus type 1 strains RV9 and RV20 (Fig. 3A). The three Eurasian lyssaviruses Irkut, Khujand, and Aravan were all strongly neutralized (Fig. 3C). We also tested RV neutralization by the plant-derived purified mAb with the FAVN assay. The plant-derived antibody demonstrated a neutralizing titer of ∼500 IU/mg for strain CVS11, while no neutralization was observed for LBV, an RV-related phylogroup II virus (negative control).
Figure 3.
Neutralization of different lyssavirus pseudotypes by the plant-derived 62-71-3 IgG. A) Neutralization of RV (strain ERA) and DUVV (strains SA06, RV131). B) Neutralization of EBLV1 (strains RV9, RV20), EBLV2 (strain RV1787), and ABLV (strain RV634). C) Neutralization of the Eurasian lyssaviruses. Titrations were repeated 2 times; each panel shows one representative run.
Glycoproteomic analysis
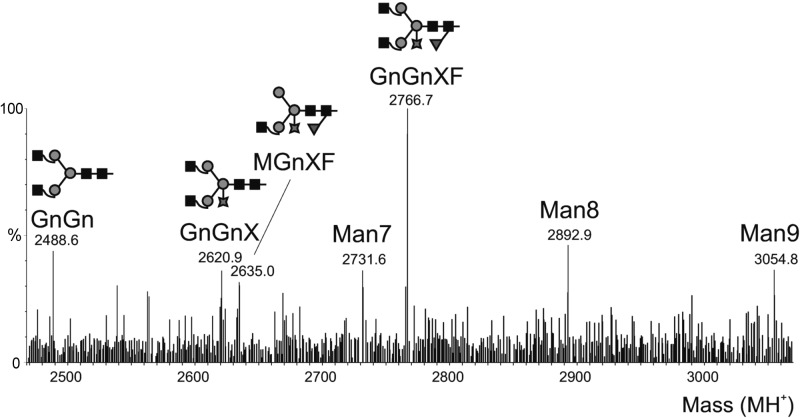
Sequence analysis of heavy and light chains of mAb 62-71-3 predicted the presence of a single potential N-linked glycosylation site in the antibody Fc region. The plant-derived antibody was subjected to glycoproteomic analysis by RP-ESI-MS. Glycopeptides comprising the Fc glycosylation site EEQYNSTYR (N-linked glycosyation site is underscored) were identified at the early retention time characteristic for this peptide (44, 63). The glycan analysis revealed that mAb 62-71-3 displayed glycan compositions typical of plant glycoproteins, with predominantly complex type glycans containing xylose and fucose, which are presumed to be the β1,2-linked xylose residues attached to the β-linked mannose and the α1,3-fucose residue linked to the Asn-linked N-acetyl-glucosamine (Fig. 4). Tandem MS results were subjected to Mascot MS/MS ion search, which confirmed the sample to contain essentially mAb 62-71-3.
Figure 4.
Glycoproteomic analysis of the plant-derived 62-71-3 IgG by in-gel digestion of S-carbamidomethylated sample and RP-ESI-MS. Deconvoluted spectrum of the glycopeptide elution region of the Fc glycopeptide. Masses correspond to oligomannosidic (Man7 etc.) and complex-type structures. Squares, circles, stars, and triangles represent N-acetylglucosamine, mannose, xylose, and fucose, respectively.
Competition ELISA
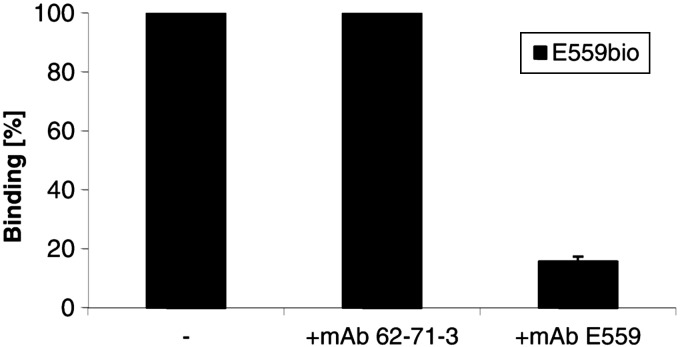
To verify that the binding of mAb 62-71-3 does not depend on antigenic site II, the immunodominant epitope in mice, a competition experiment with the site II-specific mAb E559 was performed, using a protocol adopted from Marissen et al. (34). RV glycoprotein ELISA plate wells were incubated with saturating amounts of purified plantibodies before adding biotinylated hybridoma-derived mAb E559. As expected, the binding of biotinylated E559 was blocked when plates were preincubated with plant-derived mAb E559 (Fig. 5). In contrast, binding was not blocked when the 62-71-3 plant-derived antibody or no antibody (buffer only, negative control) were added, indicating that mAb 62-71-3 does not compete with the site II-specific mAb E559.
Figure 5.
Competition ELISA. Saturating amounts of unlabeled plant-derived mAbs (62-71-3 or E559) or no mAb (−) were allowed to bind to RV glycoprotein before addition of biotinylated hybridoma-derived competitor E559. Binding is expressed as the percentage of the binding of the biotinylated antibody alone.
Mutational analysis of the RV glycoprotein
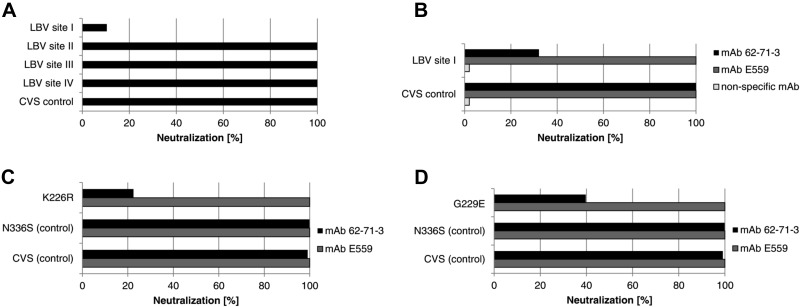
To investigate the antibody-antigen interaction in more detail, a set of RV glycoprotein mutants was analyzed regarding their neutralization by the 62-71-3 scFv. These mutants were based on CVS11 pseudotype viruses, each containing a separate replacement of one of the 4 major antigenic sites by the corresponding region from a phylogroup II virus (LBV.Nig56-RV1). The neutralization experiments with these mutated pseudotypes showed that neutralization by the 62-71-3 scFv was severely diminished when antigenic site I was altered (Fig. 6A). In contrast, mutations in other antigenic sites did not diminish neutralization by the 62-71-3 scFv. A similar experiment was undertaken with the plant-expressed 62-71-3 IgG (Fig. 6B). To demonstrate that the effects of mutating antigenic site I were specific for neutralization by mAb 62-71-3, we also included the plant-derived mAb E559, which targets antigenic site II of the viral glycoprotein (6). This chimeric version of mAb E559 was also cloned and expressed in planta (unpublished results) and purified from plant leaves by protein G affinity chromatography, for a direct comparison with the plant-derived purified 62-71-3 IgG. While mAbs E559 and 62-71-3 both showed potent and complete neutralization of a rabies pseudotype virus containing the CVS11 wild-type glycoprotein (CVS control), only mAb 62-71-3 showed diminished neutralization of the pseudotype virus containing the CVS11 glycoprotein mutated in site I, and a 100% neutralizing titer could not be defined for this mutant.
Figure 6.
Mutational analysis of the RV glycoprotein using the E. coli-derived 62-71-3 scFv and the plant-derived 62-71-3 IgG. A) Phage-displayed 62-71-3 scFv was tested against a set of pseudotype viruses containing either the CVS11 wild-type glycoprotein (CVS control) or mutated CVS11 glycoproteins containing the antigenic sites of LBV (LBV site I/II/III/IV) within the CVS backbone. B) Plant-derived mAbs 62-71-3 and E559 and a nonspecific mAb were tested regarding their neutralization of pseudotype viruses containing either the CVS11 wild-type glycoprotein or a mutated CVS11 glycoprotein containing LBV antigenic site I (aa 226–231). C) Role of antigenic site I residue K226 was investigated by testing the plant-derived mAbs 62-71-3 and E559 regarding their neutralization of pseudotypes containing glycoproteins with a K226R mutation. Controls included pseudotypes containing the CVS11 wild-type glycoprotein (CVS control) or containing the CVS glycoprotein containing a nonspecific mutation within antigenic site III (N336S control). D) Neutralization of a site I mutant containing a G229E mutation by mAbs 62-71-3 and E559.
Amino acid residues present in antigenic site I were then compared for the lyssavirus pseudotypes assessed. Antigenic site I comprises 6 amino acid residues at positions 226–231 of the mature viral glycoprotein (without signal peptide). Alignments of the antigenic site I sequences from the different glycoproteins used in Fig. 3 and 4 revealed conservation of the 3 core amino acids L227, C228, and G229 (Table 3). However, whereas lyssavirus genotypes containing the residue K226 within antigenic site I were completely neutralized by mAb 62-71-3, lyssaviruses containing R226 showed diminished neutralization, and no 100% neutralizing titer could be defined for this point mutant (Fig. 6C). To confirm the critical role of residue K226, we investigated the 2 plant-derived mAbs 62-71-3 and E559 regarding their neutralization of pseudotypes containing glycoproteins with a K226R (lysine to arginine) mutation (Fig. 6D). Controls included pseudotypes containing the CVS11 wild-type glycoprotein or containing the CVS11 glycoprotein harboring a nonspecific mutation in antigenic site III residue N336, which has previously been found to be critical for neutralization by mAbs targeting site III (24, 57). This residue was mutated to the serine found in LBV (N336S) and had no effect on neutralization by mAb 62-71-3 (Fig. 6C). In contrast, the K226R mutation diminished neutralization and confirmed the critical role of this residue for neutralization by mAb 62-71-3 (Fig. 6C). We also investigated the glycine residue (G229) that is part of the conserved region of antigenic site I. This residue has previously been found mutated in viral escape mutants resistant to a mAb-targeting antigenic site I (34), so we prepared CVS11 pseudotype virus harboring a G229E point mutation that corresponds to the escape mutant previously described (34). Similar to the K226R mutation, diminished neutralization by mAb 62-71-3 was observed with the G229E mutation, but not for mAb E559 (Fig. 6D).
Table 3.
Alignment of antigenic site I residues of the tested lyssaviruses
| Strain | Neutralization | Site I |
|---|---|---|
| CVS (RV) | + | KLCGVL |
| ERA (RV) | + | KLCGVL |
| RV131 (DUVV) | − | RLCGIS |
| SA06 (DUVV) | − | RLCGIS |
| RV9 (EBLV1) | − | RLCGVP |
| RV20 (EBLV1) | − | RLCGVP |
| RV1787 (EBLV2) | + | KLCGIS |
| RV634 (ABLV) | + | KLCGIS |
| Irkut | + | KLCGMA |
| Aravan | + | KLCGVM |
| Khujand | + | KLCGVS |
| *** |
See Fig. 3. Asterisks below alignments indicate conserved residues. Neutralization: + indicates complete (100%) neutralization; − indicates that 100% neutralization could not be determined.
DISCUSSION
Rabies occurs mainly among low-income families in Africa and Asia, and an inexpensive antibody product would be highly desirable for implementing appropriate PEP. Because of high costs and short supply, replacement of plasma-derived HRIGs and ERIGs remains a priority (35, 36), and a useful alternative is offered through the development of mAbs directed against the RV glycoprotein (37, 38). To overcome the short supply of RIGs, the WHO Rabies Collaborating Centres previously set out to investigate several mAbs for inclusion into an antibody cocktail (6, 35). In a WHO consultation in 2002, the mAbs 62-71-3, E559, 1112-1, D8, M777, and M727 were initially proposed (35). mAb 1112-1 was eventually excluded because of intellectual property reasons (6), and mAb D8 was excluded due to insufficient strain coverage (unpublished results). The remaining mAbs were shown to target the same epitope on the viral glycoprotein, namely antigenic site II, with the exception of mAb 62-71-3 (6). Three novel antibody cocktails were designed, all containing mAb 62-71-3 as an essential component (6).
In the present study, we report the cloning of a chimeric IgG version of mAb 62-71-3 and expression in a plant system. Initially, a phage-displayed scFv version of mAb 62-71-3 was produced and tested for its neutralizing potency against a rabies pseudotype virus. This step confirmed that the variable region sequences correctly encoded for an antibody with neutralizing potency toward the virus. A full-length chimeric antibody was then cloned and expressed in N. benthamiana. The expression levels of mAb 62-71-3 were estimated by quantitative ELISA and yielded ∼3–4% of total soluble protein. This is ∼100 mg/kg fresh tissue weight. Although these expression levels were satisfactory for the current study and do not necessarily require any additional improvements for large-scale production, further enhancement of expression would be desirable and might be achieved by codon-optimization and by using different expression vectors (two strategies that we are currently further investigating for mAb 62-71-3).
The original murine 62-71-3 IgG was converted here into a chimeric mAb with human antibody constant region sequences in order to improve half-life and reduce immunogenicity in humans. The resulting chimeric 62-71-3 IgG was shown to bind to its antigen in a functional ELISA and to neutralize a rabies pseudotype virus, confirming correct assembly and functionality of the antibody. We also confirmed the neutralizing activity of the plant-derived mAb in the FAVN assay and observed strong RV neutralization (500 IU/mg), similar to the previously reported potency of its murine hybridoma-derived counterpart (6).
To further corroborate the strong neutralizing potency by the plant-derived purified mAb 62-71-3, a panel of different pseudotype viruses was tested. Several lyssavirus pseudotypes were strongly neutralized, with the exception of two Duvenhage strains (SA06 and RV131) and two European bat lyssavirus type 1 strains (RV9 and RV20). These results are also consistent with previous observations of mAb 62-71-3 produced in hybridoma cells (6), which suggested that the hybridoma-derived mAb broadly neutralizes different genotypes but is ineffective at neutralizing the genotypes Duvenhage virus and European bat lyssavirus type 1.
The purified recombinant mAb was then subjected to mass spectroscopy and was shown to be glycosylated with typical plant complex glycan structures. Although mammalian and plant cells share similar cellular machinery for antibody assembly, their post-translational modifications, including the processing of glycans, are not completely identical. While the processing in the ER is conserved among all eukaryotic species and is restricted to oligomannose type N-glycans, the processing in the Golgi varies between species, resulting in differences between plant-derived proteins and their mammalian counterparts (39). Plant glycans usually do not contain sialic acid residues and may contain β(1,2)-xylose residues attached to the α-linked mannose of the glycan core and α (1,3)-fucose residues linked to the proximal N-acetyl-glucosamine (40, 41). While it cannot be excluded that these plant-specific glycans may potentially be antigenic or allergenic in humans (42), the glycosylation of plant-derived antibodies does not seem to interfere with successful passive antibody therapy (43, 44).
Competition experiments with biotinylated mAb E559 demonstrated that mAb 62-71-3 does not depend on antigenic site II, the immunodominant epitope in mice. To shed more light on the antibody-antigen interaction, a mutational analysis of the RV glycoprotein was undertaken. Lafon et al. (45) initially proposed an antigenic structure for the RV glycoprotein, defining several major epitopes. Antigenic site I has been defined as an epitope complex between residues 226 and 231 (46, 34), originally identified using a single mAb, which binds to an epitope, including residue 231 within antigenic site I (45, 46). Antigenic site II comprises a discontinuous epitope, including residues 34–42 and residues 198–200 (47). This antigenic site is targeted by most murine antibodies and mutations within these residues can result in heavily reduced pathogenicity of the virus (47). Antigenic site III comprises residues 330–338 and harbors two charged residues, K330 and R333, which are linked to viral pathogenicity and neuroinvasion (48, 49). Antigenic site IV contains overlapping linear epitopes with key residues 251 and 264 (50, 51). In addition, a few minor epitopes have been identified, e.g., between residues 342 and 343 (minor site a) and between residues 14 and 17 (45, 52).
Most studies on the antibody epitopes within the RV glycoprotein have been undertaken by sequencing viral escape mutants or by competition assays (53, 54). A few other studies have relied on investigating the interaction between antibodies and peptides derived from the viral glycoprotein, e.g., peptides derived by cleaving the glycoprotein with cyanogen bromide (55), short peptides expressed in yeast (56), as well as synthetic peptides for PEP-SCAN experiments (34). In the present study, we made use of pseudotype viruses containing chimeric RV/LBV glycoproteins generated by swapping the antigenic sites of RV strain CVS11 (phylogroup I lyssavirus) with those of LBV strain Nig56-RV1 (phylogroup II virus). It is well established that neutralizing antibodies (including mAb 62-71-3) targeting phylogroup I viruses are not effective at neutralizing phylogroup II viruses (57, 58). The neutralization assays indicated that virus neutralization by the 62-71-3 scFv and the plant-derived 62-71-3 IgG specifically involve antigenic site I.
To examine more closely how differences in antigenic site I residues account for the differences in neutralization observed with different lyssavirus pseudotypes, their glycoprotein sequences were compared. The alignment revealed diverse antigenic site I residues of lyssaviruses belonging to phylogroup I and related Eurasian lyssaviruses (59–61). Although lyssaviruses containing the antigenic site I residue K226 (lysine) were completely neutralized by mAb 62-71-3, lyssaviruses containing the residue R226 (arginine) demonstrated diminished neutralization. To obtain further insight into the role of antigenic site I residues K226 or R226, we tested the 2 plant-derived mAbs 62-71-3 and E559 regarding their neutralization of pseudotypes containing glycoproteins with a K226R mutation. These experiments corroborated the critical role of K226 for neutralization by mAb 62-71-3. Moreover, pseudotypes containing a G229E mutation, which has previously been found in viral escape mutants resistant to a mAb targeting antigenic site I (34), showed diminished neutralization.
Previous studies indicated that mAb 62-71-3 was able to neutralize viral escape mutants of mAb E559 (directed against site II; ref. 6), similar to another mAb (mAb D1) known to target antigenic site III. This observation led the researchers (6) to suggest that mAb 62-71-3 targets an epitope different from antigenic site II, possibly antigenic site III, as it is well established that the vast majority of mAbs bind to sites II or III (48). Unfortunately, no viral escape mutants could be generated for mAb 62-71-3 in vitro (6), so that its epitope has been unknown so far. While this previous study (6) already suggested that the epitope of mAb 62-71-3 does not overlap with the epitope (site II) of mAb E559, our present study provides the first systematic epitope analysis. Relatively few antibodies specific for antigenic site I have been described to date (47, 59), and the neutralization assays undertaken in our current study demonstrate that antigenic site I, including residue K226 is critical for neutralization mAb 62-71-3. The demonstration of the binding specificity of mAb 62-71-3 to the RV glycoprotein at a molecular level provides an important rationale for including mAb 62-71-3 in RV-neutralizing antibody cocktails. The results provided here also indicate that mAb 62-71-3 would be a particularly good complement to antibodies targeting site II, such as mAb E559, and that mAbs specific to site III could now also be considered for inclusion in an antirabies cocktail product.
The neutralization experiments with pseudotypes containing mutated glycoproteins cannot differentiate between direct and indirect effects, i.e., it is currently not clear whether the diminished neutralization observed with pseudotypes harboring site I mutations is due to the absence of the targeted epitope or due to indirect/allosteric effects (i.e., certain amino acid residues in site I might mask neighboring epitopes that might be the real target of mAb 62-71-3). Nevertheless, our results explain why mAb 62-71-3 is relatively ineffective at neutralizing certain genotypes, in particular, those belonging to phylogroup II. Notably, many of these particular genotypes have very little clinical relevance, so the lack of neutralization of these genotypes does not pose a significant problem in the further development of the plant-derived mAb 62-71-3.
Future work should now focus on the evaluation of additional mAbs regarding their potential to complement mAb 62-71-3. The development of a combination of mAbs targeting distinct, nonoverlapping epitopes and covering a broad panel of RV isolates would be highly desirable. This study has focused on the molecular characterization of mAb 62-71-3, but two other features of this study targeted facilitating the clinical development of this antibody. First, we have focused on the partial humanization of mAb 62-71-3. Although using a murine mAb in PEP is not contraindicated, the use of human sequences is generally preferred. Second, we have explored a powerful plant expression system to generate the recombinant mAb. For almost 2 decades, the potential of plants for production of mAbs has been proposed (62). Recent advances in regulatory approval of plant biotechnologies and manufacturing scaleup now take this potential much closer to reality. There is now a real prospect of producing mAbs for rabies PEP in plants to exploit low costs, massive scalability, and potential technology transfer to resource-poor regions where rabies affects low-income families.
Supplementary Material
Acknowledgments
The authors thank Marie Paule Kieny (World Health Organization, Geneva, Switzerland) and Charles E. Rupprecht (U.S. Centers for Disease Control and Prevention, Atlanta, GA, USA) for providing the 62-71-3 variable domain sequences and Polymun Scientific Immunbiologische Forschung (Klosterneuberg, Austria) for providing the 4E10 constant domain sequences. The authors also thank Thomas Müller [Friedrich Loeffler Institut(FLI), Wusterhausen, Germany] for the E559 hybridoma, Robin Weiss (University College London, London, UK) for help with the PNA, and Derek Healy [Animal Health and Veterinary Laboratories Agency (AHVLA), Weybridge, UK] for help with the FAVN. The authors are grateful to the EU-funded Pharma-Planta, Future-Pharma, and COST (FAO94) projects and the Dr. Hadwen Trust for Humane Research, as well as the Hotung Foundation.
This work was supported by St. George's, University of London (London, UK), and AHVLA.
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- ELISA
- enzyme-linked immunosorbent assay
- ERIG
- equine rabies immunoglobulin
- FAVN
- fluorescent antibody virus neutralization
- HRIG
- human rabies immunoglobulin
- HRP
- horseradish peroxidase
- LBV
- Lagos bat virus
- mAb
- monoclonal antibody
- OD
- optical density
- PEP
- postexposure prophylaxis
- PNA
- pseudotype neutralization assay
- RIG
- rabies immunoglobulin
- RP-ESI-MS
- reverse-phase electrospray ionization mass spectrometry
- RV
- rabies virus
- scFv
- single-chain variable fragment;
REFERENCES
- 1. Knobel D. L., Cleaveland S., Coleman P. G., Fevre E. M., Meltzer M. I., Miranda M. E., Shaw A., Zinsstag J., Meslin F. X. (2005) Re-evaluating the burden of rabies in Africa and Asia. Bull. World Health Organ. 83, 360–368 [PMC free article] [PubMed] [Google Scholar]
- 2. Both L., Banyard A. C., van Dolleweerd C., Horton D. L., Ma J. K., Fooks A. R. (2012) Passive immunity in the prevention of rabies. Lancet Infect. Dis. 12, 397–407 [DOI] [PubMed] [Google Scholar]
- 3. Mallewa M., Fooks A. R., Banda D., Chikungwa P., Mankhambo L., Molyneux E., Molyneux M. E., Solomon T. (2007) Rabies encephalitis in malaria-endemic area, Malawi Africa Emerg. Infect. Dis. 13, 136–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rupprecht C. E., Hanlon C. A., Hemachudha T. (2002) Rabies re-examined. Lancet Infect. Dis. 2, 327–343 [DOI] [PubMed] [Google Scholar]
- 5. Hemachudha T., Laothamatas J., Rupprecht C. E. (2002) Human rabies: a disease of complex neuropathogenetic mechanisms and diagnostic challenges. Lancet Neurol. 1, 101–109 [DOI] [PubMed] [Google Scholar]
- 6. Müller T., Dietzschold B., Ertl H., Fooks A. R., Freuling C., Fehlner-Gardiner C., Kliemt J., Meslin F. X., Franka R., Rupprecht C. E., Tordo N., Wanderler A. L., Kieny M. P. (2009) Development of a mouse monoclonal antibody cocktail for post-exposure rabies prophylaxis in humans. PLoS Negl. Trop. Dis. 3, e542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rupprecht C. E., Gibbons R. V. (2004) Clinical practice. Prophylaxis against rabies. N. Engl. J. Med. 351, 2626–2635 [DOI] [PubMed] [Google Scholar]
- 8. Winkler W. G. (1968) Airborne rabies virus isolation. Bull. Wildl. Dis. Assoc. 4, 37–40 [DOI] [PubMed] [Google Scholar]
- 9. Constantine D. G. (1967) Rabies Transmission by Air in Bat Caves, U.S. Public Health Service, Washington, DC [Google Scholar]
- 10. Fishbein D. B., Robinson L. E. (1993) Rabies. N. Engl. J. Med. 329, 1632–1638 [DOI] [PubMed] [Google Scholar]
- 11. Wertheim H. F. L., Nguyen T. Q., Nguyen K. A. T., de Jong M. D., Taylor W. R. J., Le T. V., Nguyen H. H., Nguyen H. T., Farrar J., Horby P., Nguyen H. D. (2009) Furious rabies after an atypical exposure. PLoS Med. 6, e1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. World Health Organization (September 2012) Rabies Fact Sheet No. 99. World Health Organization, Geneva, Switzerland; http://www.who.int/mediacentre/factsheets/fs099/en/ [Google Scholar]
- 13. Warrell M. (2010) Rabies and African bat lyssavirus encephalitis and its prevention. Int. J. Antimicrob. Agents 36, 47–52 [DOI] [PubMed] [Google Scholar]
- 14. Satpathy D. M., Sahu T., Behera T. R. (2005) Equine rabies immunoglobulin: a study on its clinical safety. J. Indian Med. Assoc. 103, 241–242 [PubMed] [Google Scholar]
- 15. Shantavasinkul P., Tantawichien T., Wacharapluesadee S., Jeamanukoolkit A., Udomchaisakul P., Chattranukulchai P., Wongsaroj P., Khawplod P., Wilde H., Hemachudha T. (2010) Failure of rabies postexposure prophylaxis in patients presenting with unusual manifestations. Clin. Infect. Dis. 50, 77–79 [DOI] [PubMed] [Google Scholar]
- 16. Manning S. E., Rupprecht C. E., Fishbein D., Hanlon C. A., Lumlertdacha B., Guerra M., Meltzer M. I., Dhankhar P., Vaidya S. A., Jenkins S. R., Sun B., Hull H. F. (2008) Human rabies prevention—United States, 2008: recommendations of the Advisory Committee on Immunization Practices. MMWR Recomm. Rep. 57, 1–28 [PubMed] [Google Scholar]
- 17. Rupprecht C. E., Briggs D., Brown C. M., Franka R., Katz S. L., Kerr H. D., Lett S., Levis R., Meltzer M. I., Schaffner W., Cieslak. P. R. (2009) Evidence for a 4-dose vaccine schedule for human rabies post-exposure prophylaxis in previously non-vaccinated individuals. Vaccine 27, 7141–7148 [DOI] [PubMed] [Google Scholar]
- 18. Warrell M. J. (2003) The challenge to provide affordable rabies post-exposure treatment. Vaccine 21, 706–709 [DOI] [PubMed] [Google Scholar]
- 19. Sudarshan M. K., Mahendra B. J., Madhusudana S. N., Ashwoath-Narayana D. H., Rahman A., Rao N. S., Meslin F. X., Lobo D., Ravikumar K., Gangaboraiah B. (2006) An epidemiological study of animal bites in India: results of a WHO-sponsored national multicentric rabies survey. J. Commun. Dis. 38, 32–39 [PubMed] [Google Scholar]
- 20. Hampson K., Dobson A., Kaare M., Dushoff J., Magoto M., Sindoya E., Cleaveland S. (2008) Rabies exposures, post-exposure prophylaxis and deaths in a region of endemic canine rabies. PLoS Negl. Trop. Dis. 2, e339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dodet B. (2009) The fight against rabies in Africa: from recognition to action. Vaccine 27, 5027–5032 [DOI] [PubMed] [Google Scholar]
- 22. Fooks A. R. (2005) Rabies remains a ‘neglected disease’. Euro. Surveill. 10, 1–2 [PubMed] [Google Scholar]
- 23. Goudsmit J., Marissen W. E., Weldon W. C., Niezgoda M., Hanlon C. A., Rice A. B., Kruif J., Dietzschold B., Bakker A. B., Rupprecht C. E. (2006) Comparison of an anti-rabies human monoclonal antibody combination with human polyclonal anti-rabies immune globulin. J. Infect. Dis. 193, 796–801 [DOI] [PubMed] [Google Scholar]
- 24. Sloan S. E., Hanlon C., Weldon W., Niezgoda M., Blanton J., Self J., Rowley K. J., Mandell R. B., Babcock G. J., Thomas W. D., Jr., Rupprecht C. E., Ambrosino D. M. (2007) Identification and characterization of a human monoclonal antibody that potently neutralizes a broad panel of rabies virus isolates. Vaccine 25, 2800–2810 [DOI] [PubMed] [Google Scholar]
- 25. Muhamuda K., Madhusudana S. N., Ravi V. (2007) Use of neutralizing murine monoclonal antibodies to rabies glycoprotein in passive immunotherapy against rabies. Hum. Vaccin. 3, 192–195 [DOI] [PubMed] [Google Scholar]
- 26. Tacket C. O., Mason H. S., Losonsky G., Estes M. K., Levine M. M., Arntzen C. J. (2000) Human immune responses to a novel Norwalk virus vaccine delivered in transgenic potatoes. J. Infect. Dis. 1, 302–305 [DOI] [PubMed] [Google Scholar]
- 27. McCormick A. A., Reddy S., Reinl S. J., Cameron T. I., Czerwinkski D. K., Vojdani F., Hanley K. M., Garger S. J., White E. L., Novak J., Barrett J., Holtz R. B., Tusé D., Levy R. (2008) Plant-produced idiotype vaccines for the treatment of non-Hodgkin's lymphoma: safety and immunogenicity in a phase I clinical study. Proc. Natl. Acad. Sci. U. S. A. 29, 10131–10136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ma J.K., Drake P. M., Chargelegue D., Obregon P., Prada A. (2005) Antibody processing and engineering in plants, and new strategies for vaccine production. Vaccine 23, 1814–1818 [DOI] [PubMed] [Google Scholar]
- 29. Ko K., Koprowski H. (2005) Plant biopharming of monoclonal antibodies. Virus Res. 111, 93–100 [DOI] [PubMed] [Google Scholar]
- 30. Ko K., Tekoah Y., Rudd P. M, Harvey D. J., Dwek R. A., Spitsin S., Hanlon C. A., Rupprecht C., Dietzschold B., Golovkin M., Koprowski H. (2003) Function and glycosylation of plant-derived antiviral monoclonal antibody. Proc. Natl. Acad. Sci. U. S. A. 100, 8013–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hanlon C. A., Kuzmin I. V., Blanton J. D., Weldon W. C., Manangan J. S., Rupprecht C. E. (2005) Efficacy of rabies biologics against new lyssaviruses from Eurasia. Virus Res. 111, 44–54 [DOI] [PubMed] [Google Scholar]
- 32. Sainsbury F., Thuenemann E. C., Lomonossoff G. P. (2009) pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7, 682–693 [DOI] [PubMed] [Google Scholar]
- 33. Wright E., Temperton N. J., Marston D. A., McElhinney L. M., Fooks A. R., Weiss R. A. (2008) Investigating antibody neutralization of lyssaviruses using lentiviral pseudotypes: a cross-species comparison. J. Gen. Virol. 9, 2204–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marissen W. E., Kramer R. A., Rice A., Weldon W. C., Niezgoda M., Faber M., Slootstra J. W., Meloen R. H., Clijsters-van der Horst M., Visser T. J., Jongeneelen M., Thijsse S., Throsby M., de Kruif J., Rupprecht C. E., Dietzschold B., Goudsmit J., Bakker A. B. (2005) Novel rabies virus-neutralizing epitope recognized by human monoclonal antibody: fine mapping and escape mutant analysis. J. Virol. 79, 4672–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. World Health Organization (2002) WHO Consultation on a Rabies Monoclonal Antibody Cocktail for Rabies Post Exposure Treatment. WHO, Geneva, 23–24 May 2002, World Health Organization, Geneva, Switzerland; http://www.who.int/rabies/vaccines/en/mabs_final_report.pdf [Google Scholar]
- 36. Bakker A. B. H., Python C., Kissling C. J., Pandya P., Marissen W. E., Brink M. F., Lagerwerf F., Worst S., van Corven E., Kostense S., Hartmann K., Weverling G. J., Uytdehaag F., Herzog C., Briggs D. J., Rupprecht C. E., Grimaldi R., Goudsmit J. (2008) First administration to humans of a monoclonal antibody cocktail against rabies virus: safety, tolerability, and neutralizing activity. Vaccine 26, 5922–5927 [DOI] [PubMed] [Google Scholar]
- 37. Champion J. M., Kean R. B., Rupprecht C. E., Notkins A. L., Koprowski H., Dietzschold B., Hooper D. C. (2000) The development of monoclonal human rabies virus-neutralizing antibodies as a substitute for pooled human immune globulin in the prophylactic treatment of rabies virus exposure. J. Immunol. Methods 235, 81–90 [DOI] [PubMed] [Google Scholar]
- 38. Prosniak M., Faber M., Hanlon C. A., Rupprecht C. E., Hooper D. C., Dietzschold B. (2003) Development of a cocktail of recombinant-expressed human rabies virus-neutralizing monoclonal antibodies for postexposure prophylaxis of rabies. J. Infect. Dis. 188, 53–56 [DOI] [PubMed] [Google Scholar]
- 39. Tekoah Y., Ko K., Koprowski H., Harvey D. J., Wormald M. R., Dwek R. A., Rudd P. M. (2004) Controlled glycosylation of therapeutic antibodies in plants. Arch. Biochem. Biophys. 2, 266–278 [DOI] [PubMed] [Google Scholar]
- 40. Bardor M., Faveeuw C., Fitchette A. C., Gilbert D., Galas L., Trottein F., Faye L., Lerouge P. (2003) Immunoreactivity in mammals of two typical plant glyco-epitopes, core alpha(1,3)-fucose and core xylose. Glycobiology 6, 427–434 [DOI] [PubMed] [Google Scholar]
- 41. Gomord V., Fitchette A. C., Menu-Bouaouiche L., Saint-Jore-Dupas C., Plasson C., Michaud D., Faye L. (2010) Plant-specific glycosylation patterns in the context of therapeutic protein production. Plant Biotechnol. J. 5, 564–587 [DOI] [PubMed] [Google Scholar]
- 42. Van Ree R., Cabanes-Macheteau M., Akkerdaas J., Milazzo J. P., Loutelier-Bourhis C., Rayon C., Villalba M., Koppelman S., Aalberse R., Rodriguez R., Faye L., Lerouge P. (2000) β(1,2)-xylose and alpha(1,3)-fucose residues have a strong contribution in IgE binding to plant glycoallergens. J. Biol. Chem. 15, 11451–11458 [DOI] [PubMed] [Google Scholar]
- 43. Ma J. K., Hikmat B. Y., Wycoff K., Vine N. D., Chargelegue D., Yu L., Hein M. B., Lehner T. (1998) Characterization of a recombinant plant monoclonal secretory antibody and preventive immunotherapy in humans. Nat. Med. 5, 601–606 [DOI] [PubMed] [Google Scholar]
- 44. Zeitlin L., Olmsted S. S., Moench T. R., Co M. S., Martinell B. J., Paradkar V. M., Russell D. R., Queen C., Cone R. A., Whaley K. J. (1998) A humanized monoclonal antibody produced in transgenic plants for immunoprotection of the vagina against genital herpes. Nat. Biotechnol. 13, 1361–1364 [DOI] [PubMed] [Google Scholar]
- 45. Lafon M., Wiktor T. J., Macfarlan R. I. (1983) Antigenic sites on the CVS rabies virus glycoprotein: analysis with monoclonal antibodies. J. Gen. Virol. 4, 843–851 [DOI] [PubMed] [Google Scholar]
- 46. Benmansour A., Leblois H., Coulon P., Tuffereau C., Gaudin Y., Flamand A., Lafay F. (1991) Antigenicity of rabies virus glycoprotein. J. Virol. 8, 4198–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Prehaud C., Coulon P., LaFay F., Thiers C., Flamand A. (1988) Antigenic site II of the rabies virus glycoprotein: structure and role in viral virulence. J. Virol. 1, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Seif I., Coulon P., Rollin P. E., Flamand A. (1985) Rabies virulence: effect on pathogenicity and sequence characterization of rabies virus mutations affecting antigenic site III of the glycoprotein. J. Virol. 3, 926–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dietzschold B., Wunner W. H., Wiktor T. J., Lopes A. D., Lafon M., Smith C. L., Koprowski H. (1983) Characterization of an antigenic determinant of the glycoprotein that correlates with pathogenicity of rabies virus. Proc. Natl. Acad. Sci. U. S. A. 1, 70–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dietzschold B., Gore M., Marchadier D., Niu H. S., Bunschoten H. M., Otvos L., Jr., Wunner W. H., Ertl H. C., Osterhaus A. D., Koprowski H. (1990) Structural and immunological characterization of a linear virus-neutralizing epitope of the rabies virus glycoprotein and its possible use in a synthetic vaccine. J. Virol. 8, 3804–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Luo T. R., Minamoto N., Ito H., Goto H., Hiraga S., Ito N., Sugiyama M., Kinjo T. (1997) A virus-neutralizing epitope on the glycoprotein of rabies virus that contains Trp251 is a linear epitope. Virus Res. 1, 35–41 [DOI] [PubMed] [Google Scholar]
- 52. Mansfield K. L., Johnson N., Fooks A. R. (2004) Identification of a conserved linear epitope at the N terminus of the rabies virus glycoprotein. J. Gen. Virol. 11, 3279–3283 [DOI] [PubMed] [Google Scholar]
- 53. Bakker A. B. H., Marissen W. E., Kramer R. A., Rice A. B., Weldon W. C., Niezgoda M., Hanlon C. A., Thijsse S., Backus H. H., de Kruif J., Dietzschold B., Rupprecht C. E., Goudsmit J. (2005) Novel human monoclonal antibody combination effectively neutralizing natural rabies virus variants and individual in vitro escape mutants. J. Virol. 79, 9062–9068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hultberg A., Temperton N. J., Rosseels V., Koenders M., Gonzalez-Pajuelo M., Schepens B., Ibañez L. I., Vanlandschoot P., Schillemans J., Saunders M., Weiss R. A., Saelens X., Melero J. A., Verrips C. T., Van Gucht S., de Haard H. J. (2011) Llama-derived single domain antibodies to build multivalent, superpotent and broadened neutralizing anti-viral molecules. PLoS One 6, e17665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dietzschold B., Wiktor T. J., Macfarlan R., Varrichio A. (1982) Antigenic structure of rabies virus glycoprotein: ordering and immunological characterization of the large CNBr cleavage fragments. J. Virol. 2, 595–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lafay F., Benmansour A., Chebli K., Flamand A. (1996) Immunodominant epitopes defined by a yeast-expressed library of random fragments of the rabies virus glycoprotein map outside major antigenic sites. J. Gen. Virol. 2, 339–346 [DOI] [PubMed] [Google Scholar]
- 57. Badrane H., Bahloul C., Perrin P., Tordo N. (2001) Evidence of two Lyssavirus phylogroups with distinct pathogenicity and immunogenicity. J. Virol. 75, 3268–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Horton D. L., McElhinney L. M., Marston D. A., Wood J. L., Russell C. A., Lewis N., Kuzmin I. V., Fouchier R. A., Osterhaus A. D., Fooks A. R., Smith D. J. (2010) Quantifying antigenic relationships among the lyssaviruses. J. Virol. 84, 11841–11848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Arai Y. T., Kuzmin I. V., Kameoka Y., Botvinkin A. D. (2003) New lyssavirus genotype from the Lesser Mouse-eared Bat (Myotisblythi), Kyrghyzstan. Emerg. Infect. Dis. 9, 333–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kuzmin I. V., Orciari L. A., Arai Y. T., Smith J. S., Hanlon C. A., Kameoka Y., Rupprecht C. E. (2003) Bat lyssaviruses (Aravan and Khujand) from Central Asia: phylogenetic relationships according to N, P, and G gene sequences. Virus Res. 97, 65–79 [DOI] [PubMed] [Google Scholar]
- 61. Botvinkin A. D., Poleschuk E. M., Kuzmin I. V., Borisova T. I., Gazaryan S. V., Yager P., Rupprecht C. E. (2003) Novel lyssaviruses isolated from bats in Russia. Emerg. Infect. Dis. 9, 1623–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hiatt A., Cafferkey R., Bowdish K. (1989) Production of antibodies in transgenic plants. Nature 342, 76–78 [DOI] [PubMed] [Google Scholar]
- 63. Stadlmann J., Pabst M., Kolarich D., Kunert R., Altmann F. (2008) Analysis of immunoglobulin glycosylation by LC-ESI-MS of glycopeptides and oligosaccharides. Proteomics 8, 2858–2871 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.