Abstract
Obscurins comprise a family of giant (∼870- to 600-kDa) and small (∼250- to 55-kDa) proteins that play important roles in myofibrillogenesis, cytoskeletal organization, and cell adhesion and are implicated in hypertrophic cardiomyopathy and tumorigenesis. Giant obscurins are composed of tandem structural and signaling motifs, including 2 serine/threonine kinase domains, SK1 and SK2, present at the COOH terminus of giant obscurin-B. Using biochemical and cellular approaches, we show for the first time that both SK1 and SK2 possess enzymatic activities and undergo autophosphorylation. SK2 can phosphorylate the cytoplasmic domain of N-cadherin, a major component of adherens junctions, and SK1 can interact with the extracellular domain of the β1-subunit of the Na+/K+-ATPase, which also resides in adherens junctions. Immunostaining of nonpermeabilized myofibers and cardiocytes revealed that some obscurin kinase isoforms localize extracellularly. Quantification of the exofacial expression of obscurin kinase proteins indicated that they occupy ∼16 and ∼5% of the sarcolemmal surface in myofibers and cardiocytes, respectively. Treatment of heart lysates with peptide-N-glycosidase F revealed that while giant obscurin-B localizes intracellularly, possessing dual kinase activity, a small obscurin kinase isoform that contains SK1 localizes extracellularly, where it undergoes N-glycosylation. Collectively, our studies demonstrate that the obscurin kinase domains are enzymatically active and may be involved in the regulation of cell adhesion.—Hu, L.-Y. R., Kontrogianni-Konstantopoulos, A. The kinase domains of obscurin interact with intercellular adhesion proteins.
Keywords: myosin light chain, N-cadherin, sodium-potassium ATPase
Obscurin is the third and most recently discovered member of the family of giant proteins expressed in striated muscles (1). It is a multidomain protein, composed of adhesion modules and signaling domains arranged mostly in tandem. The NH2 terminus of the prototypical form, referred to as obscurin-A (∼720 kDa), contains 55 immunoglobulin (Ig) and 3 fibronectin-III (Fn-III) domains, while its COOH terminus contains 4 additional Ig repeats and several signaling domains, including an isoleucine-glutamine motif and a conserved Src-homology 3 domain, adjacent to Rho-guanine nucleotide exchange factor and pleckstrin homology domains (1, 2). These are followed by 2 additional Ig domains and a nonmodular sequence of ∼400 aa that contains several copies of a consensus phosphorylation motif for ERK kinases (1).
The obscurin gene, OBSCN, also encodes 2 serine/threonine (Ser/Thr) kinase (SK) domains, referred to as SK1 and SK2, that belong to the myosin light chain kinase (MLCK) subfamily (3–4). Although these are found at the COOH terminus of a ∼870-kDa form of obscurin, namely, obscurin-B or giant MLCK, they are also expressed as smaller, alternatively spliced products containing one (∼55 kDa) or both (∼145 kDa) kinase domains (3, 5).
Members of the MLCK subfamily have been shown to play key roles in the maintenance of subcellular structures and the regulation of cell contractility (6). The cardiac-specific MLCK (cMLCK) has been implicated in sarcomeric organization and cardiac contraction. Consistent with this, ablation of cMLCK leads to diminished phosphorylation of myosin regulatory light chain and cardiac hypertrophy (7). Similarly, genetic ablation of the titin kinase domain, another member of the MLCK subfamily, results in impaired lateral assembly of myofilaments, muscle weakness, and embryonic lethality due to defects in heart development (8). Striated preferentially expressed gene (SPEG), previously suggested to derive from duplication of the OBSCN gene (4), also contains tandem SK domains; however, only the internal kinase domain appears to be active (9). Genetic disruption of the SPEG gene has been associated with the development of dilated cardiomyopathy (10). As the OBSCN and SPEG kinases share >40% identity and are both preferentially expressed in striated muscles (3, 9), they may be involved in similar cellular pathways.
While the exact roles of the OBSCN kinase domains have remained elusive for more than a decade, it has been shown that the transcript levels of SK2 increase dramatically in response to aortic stenosis (11). In the current study, we report that both OBSCN kinase domains possess enzymatic activities and can undergo autophosphorylation. The β1 subunit of Na+/K+ ATPase is a ligand of SK1, and N-cadherin is a substrate of SK2. While giant obscurin-B localizes intracellularly, a smaller OBSCN kinase isoform localizes extracellularly, where it may undergo glycosylation.
MATERIALS AND METHODS
Antibodies
ObsKin-1 and ObsKin-2 antibodies were generated in rabbits by injection of glutathione-S-transferase (GST)-tagged recombinant proteins, including domains Ig69/FN-III70 (aa 7480–7746, accession no. NP001164983) and FN-III70 (aa 8476–8570, accession no. A2AAJ9), respectively. Antisera were sequentially purified over GST and GST-Ig69/FN-III70 or GST and GST-FN-III70 columns conjugated to Sulfolink resin (Thermo Scientific, Waltham, MA, USA) or cyanogen bromide (Sigma-Aldrich, St. Louis, MO, USA), respectively.
ObsKin-1 (150 ng/ml), ObsKin-2 (300 ng/ml), and mouse polyclonal ObsKin-3 (1:2000, A02; Abnova, Taipei City, Taiwan) antibodies were used in immunoblotting experiments.
The following primary antibodies were used in immunofluorescence and the Duolink proximity ligation assay (PLA; Olink Bioscience, Uppsala, Sweden): mouse monoclonal antibodies to α-actinin (1:400, A7811; Sigma-Aldrich), myomesin (3 mg/ml, mMac, tissue culture supernatant; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA, USA), HAX-1 (3 mg/ml; BD Biosciences, Franklin Lakes, NJ, USA), Na+/K+ ATPase β1 (NKAβ1;1:50; Thermo Scientific), N-cadherin (1.2:100; BD Biosciences; recognizing the cytoplasmic domain of the protein), and N-cadherin (1:50; Santa Cruz Biotechnology, Santa Cruz, CA, USA; recognizing the extracellular domain of the protein), as well as rabbit polyclonal antibodies to HAX-1 (1:100; Santa Cruz Biotechnology), the first two Ig domains of titin (Titin-Z; 3 mg/ml; ref. 12), and obscurin kinases (ObsKin-1; 3 mg/ml).
Preparation of tissue homogenates and immunoblotting
Tissue lysates were prepared in either SDS/β-mercaptoethanol (β-ME) lysis buffer containing 10 mM Na2HPO4 (pH 7.4), 120 mM NaCl, 2 mM EDTA, 10 mM NaN3, 2% SDS, 1% β-ME (EMD, Darmstadt, Germany), and Complete mini-protease inhibitor cocktail tablets (Roche, Indianapolis, IN, USA) or in RIPA lysis buffer containing 10 mM Na2HPO4 (pH 7.4), 120 mM NaCl, 2 mM EDTA, 10 mM NaN3, 1% Nonidet P-40, 0.5% sodium deoxycholate, and Complete mini-protease inhibitor cocktail tablets (Roche), as described previously (13). Immunoreactive bands were detected with either the Tropix chemiluminescence kit (Life Technologies, Grand Island, NY, USA) or the ECL Plus Western blot detection kit (GE Healthcare, Little Chalfont, UK).
Preparation of primary cultures
Cultures of flexor digitorum brevis (FDB) myofibers
FDB muscles were dissected from adult FVB mice and incubated in MEM (Life Technologies) containing 10% FBS (Life Technologies) and 0.1% gentamicin sulfate (Teknova, Hollister, CA, USA) in the presence of 2 mg/ml collagenase type I (Sigma-Aldrich) at 37°C, 5% CO2 for 3 h. Following incubation, muscles were transferred to MEM containing 10% FBS and 0.1% gentamicin and gently triturated to obtain single myofibers.
Cultures of cardiocytes
Cultures of adult mouse cardiocytes were prepared as described previously (14).
Preparation of adult cardiac sections
Adult female FVB mice were sacrificed by perfusion with 4% paraformaldehyde (PFA) in PBS under anesthesia. Hearts were excised and embedded in 7.5% gelatin and 15% sucrose in PBS and gradually frozen using 2-methylbutane at −60°C. Cardiac sections (∼12–15 μm in thickness) were obtained with a Microm HM550 cryostat (Thermo Scientific).
Immunostaining, confocal microscopy, and quantification of extracellular staining
Frozen cardiac tissue sections were blocked with 1 mg/ml BSA and 1 mM NaN3 in PBS for 2 h at room temperature and incubated with the appropriate primary antibodies overnight at 4°C. Permeabilized cells were fixed with 2% PFA, treated with 0.1% Triton X-100, blocked in 1 mg/ml BSA for 30 min, and incubated with primary antibodies for 1 h at room temperature. Nonpermeabilized cells were blocked in 1 mg/ml BSA for 30 min, incubated with primary antibody for 1 h at room temperature, and then fixed in 2% PFA. Samples were counterstained with Alexa-488 goat anti-mouse or Alexa-568 goat anti-rabbit (1:200; Life Technologies), mounted with Vectashield (Vector Laboratories, Burlingame, CA, USA), and analyzed under a LSM510 confocal microscope with a ×63 objective (Carl Zeiss, Tarrytown, NY, USA).
ImageJ software (U.S. National Institutes of Health, Bethesda, MD, USA) was used to quantify the percentage of the outer sarcolemmal surface or the inner sarcoplasmic area of nonpermeabilized cells that was immunopositive for the obscurin kinases and N-cadherin. Approximately 20 regions from Z-stack images of each area were analyzed. Statistical significance was evaluated using Student's t test; error bars represent sem.
PLA and immunofluorescent microscopy
Frozen cardiac tissue sections were processed as above, with the exception that secondary antibodies were conjugated with PLA probes (Duolink in situ PLA kit; Olink Bioscience). Samples were analyzed with an Olympus IX51 fluorescent microscope (Olympus, Tokyo, Japan) under a ×20 objective.
Yeast 2-hybrid (Y2H) screening
SK1 and SK2 were amplified from cDNA prepared from mouse heart mRNA with the Superscript III First Strand Synthesis System for RT-PCR (Life Technologies). SK1 was also amplified from a human heart cDNA library (OriGene, Rockville, MD, USA). The primers used are listed in Table 1. SK1 and SK2 bait plasmids were constructed in the pGBKT7 vector. Y2H screening was performed in Y187 Saccharomyces cerevisiae pretransformed with a human heart cDNA Library (Matchmaker Gal4 Two-Hybrid System 3; Clontech, Mountain View, CA, USA). Absence of autoactivation by pGBKT7-SK1 and pGBKT7-SK2 was examined in AH109 cells before proceeding to library screening. We screened ∼1.22 and ∼1.08 × 106 colonies with the SK1 and SK2 bait constructs and obtained 37 and 42 positive colonies, respectively. These were selected on synthetic dextrose (SD)/−adenine (Ade)/−tryptophan (Trp)/−leucine (Leu)/−histidine (His) dropout plates supplemented with X-α-gal (Clontech) to select for α-galactosidase activity. To identify the SK1 and SK2 interacting partners (i.e., prey clones), positive colonies were grown overnight and digested with lyticase. Plasmid DNA was extracted using Chroma spin columns (Clontech), transformed in JM109 Escherichia coli cells, and fully sequenced. To identify the minimal interacting domains of bait and prey clones, deletion constructs were subcloned in pGBKT7 bait and pGADT7 prey vectors, respectively, verified by sequencing, and sequentially transformed in AH109 cells. Interaction strength was scored based on colony growth and color intensity.
Table 1.
Primers used in this work
| Primer | Sequence |
|---|---|
| Human | |
| SK1 full-length sense | 5′-ACGTGAATTCCCCTACAGCAGCCCC-3′ |
| SK1 full-length antisense | 5′-ACGTGTCGACTGCGCACCTGGGCCAG-3′ |
| Mouse | |
| SK1 full-length sense | 5′-ACGTGAATTCCCCTACAGCAGCCCCT-3′ |
| SK1 full-length antisense | 5′-ATTAGTCGACTCAGCGCACCTGGGCCA-3′ |
| SK1 catalytic sense | 5′-ACGTGAATTCCCCTACAGCAGCCCCT-3′ |
| SK1 catalytic antisense | 5′-ATTAGTCGACTCACAGCCATCCGCATTG-3′ |
| SK1 regulatory sense | 5′-GGGTGAATTCACAGAGGAGGGCCCC-3′ |
| SK1 regulatory antisense | 5′-ATTAGTCGACTCAGCGCACCTGGGCCA-3′ |
| SK2 full-length sense | 5′-ACGTGGATCCATCTAGATGCCGAAAAT-3′ |
| SK2 full-length antisense | 5′-ACGTCTGCAGATTCCCCTCGTAGGTG-3′ |
| SK2 catalytic sense | 5′-ACGTGGATCCATCTAGATGCCGAAAAT-3′ |
| SK2 catalytic antisense | 5′-ACGTCTGCAGTCAGAACCAGGGGT-3′ |
| SK2 regulatory sense | 5′-GGGTGGATCCATCTGAAATCCATGCCT-3′ |
| SK2 regulatory antisense | 5′-ACGTCTGCAGATTCCCCTCGTAGGTG-3′ |
| NKAβ1 clone A sense | 5′-ACGTGAATTCGAAAAGTACAAGGATTC-3′ |
| NKAβ1 clone A antisense | 5′-ACGTCTCGAGTCAGCTCTTAATTTCAA-3′ |
| NKAβ1 clone B sense | 5′-ACGTGAATTCGAAAAGTACAAGGATTC-3′ |
| NKAβ1 clone B antisense | 5′-ACGTCTCGAGTCACATCAGTGGGTA-3′ |
| NKAβ1 clone C sense | 5′-GCGCTGAATTCATGAAGTATAATCCA-3′ |
| NKAβ1 clone C antisense | 5′-ACGTCTCGAGTCAGCTCTTAATTTCAA-3′ |
| NKAβ1 clone D sense | 5′-ACGTGAATTCGGTCTCAATGATGAC-3′ |
| NKAβ1 clone D antisense | 5′-ACGTCTCGAGTCAGAGTTTGCCGTA-3′ |
| N-cad clone A sense | 5′-ACGTGAATTCGTTTTGGACAGAGAATCG-3′ |
| N-cad clone A antisense | 5′-ACGTCTCGAGTCAGTCGTCACCACC-3′ |
| N-cad clone B sense | 5′-ACGTGAATTCCAAGAGGCGGAGACCTGT-3′ |
| N-cad clone B antisense | 5′-ACGTCTCGAGTCAGTCGTCACCACC-3′ |
| N-cad clone C sense | 5′-ACGTGAATTCGCCATCATCGCTATCCTT-3′ |
| N-cad clone C antisense | 5′-ACGTCTCGAGTCAGTCGTCACCACC-3′ |
| N-cad clone D sense | 5′-ACGTGAATTCGTTTTGGACAGAGAATCG-3′ |
| N-cad clone D antisense | 5′-ACGTCTCGAGTCAAGGTAACACCTGAGG-3′ |
| N-cad clone E sense | 5′-ACGTGAATTCCAAGAGGCGGAGACCTGT-3′ |
| N-cad clone E antisense | 5′-ATTACTCGAGTCAGCCCGTGCCAAGCCC-3′ |
| N-cad clone F sense | 5′-ACGTGAATTCGCCATCATCGCTATCCTT-3′ |
| N-cad clone F antisense | 5′-ACGTCTCGAGTCACATCCATACCAC-3′ |
| N-cad clone G sense | 5′-ACGTGAATTCAAACGGCGGGATAAAG-3′ |
| N-cad clone G antisense | 5′-ACGTCTCGAGTCAGTCGTCACCACC-3′ |
Expression and purification of SK1 and SK2 from insect cells
To express SK1 and SK2 proteins in insect cells, mouse SK1 (aa 8595–8847, accession no. A2AAJ9) and SK2 (aa 7415–7668, accession no. A2AAJ9) were subcloned in pVL1393 vector (Invitrogen, Carlsbad, CA, USA) in frame with a 6xHis tag. Tni cells were infected with pVL1393-SK2 virus at multiplicity of infection (MOI) 3, while Sf9 cells were infected with pVL1393-SK1 virus at MOI 0.1. Transfected cell pellets were collected 3 d postinfection and lysed through sonification in lysis buffer containing 20 mM Tris-HCl (pH 8.0), 20 mM NaCl, 0.5 mM Tris (2-carboxyethyl) phosphine hydrochloride (TCEP-HCl; Thermo Scientific), 5% glycerol, 1% Nonidet P-40, and Complete mini-protease inhibitor cocktail tablets (Roche). Clarified lysates containing His-tagged SK2 were incubated with a Talon metal affinity resin (Clontech) in the presence of 6 M urea (Sigma-Aldrich) and 2 mM β-ME. Following extensive washes with 10 mM imidazole, the protein was eluted with 150 mM imidazole and 6 M urea, underwent buffer exchange in 50 mM Tris-HCl, 150 mM NaCl, and 1 mM TCEP-HCl in the presence of Halt protease inhibitors (Thermo Scientific), and was concentrated using Amicon Centricon columns (EMD Millipore, Billerica, MA, USA).
In parallel, clarified lysates containing His-tagged SK1 were incubated with a Talon metal affinity resin (Clontech) in the presence of 6 M urea (Sigma-Aldrich). The flow-through fraction, which contained significant amounts of the His-tagged SK1, was dialyzed in PBS overnight at 4°C and subjected to 20% ammonium sulfate precipitation. The soluble fraction was incubated with a Talon metal affinity resin in the presence of 6 M urea. Following extensive washes with gradually reduced urea solutions (5-0 M), His-tagged SK1 was eluted with 150 mM imidazole and 1 mM β-ME. Nontransfected cells underwent the same purification procedures described for each kinase, and served as negative controls. Recombinant SK1 and SK2 were analyzed by 4–12% SDS-PAGE, followed by SYPRO Ruby gel stain (Invitrogen), according to the manufacturer's instructions.
Tandem mass spectrometry
Purified fractions containing recombinant His-SK1 and His-SK2 were subjected to liquid chromatography (LC) and tandem mass spectrometry using a linear-trap quadrupole mass-spectrometer interfaced with a 2D nanoLC system (Thermo Fisher, Waltham, MA, USA; Mass Spectrometry Proteomics Facility, Johns Hopkins University, School of Medicine). Samples were reduced, alkylated with iodoacetamide and proteolyzed with trypsin. Peptides were fractionated by reverse-phase high-performance LC, and their sequences were identified using the Mascot software (http://www.matrixscience.com) to search the NCBI-nr 167 database, allowing for oxidation on methionine and carbamidomethylation on cysteine as variable modifications. Mascot search results (*.dat) were processed in Scaffold (http://www.proteomesoftware.com) to validate protein and peptide identifications.
In addition, we performed tandem mass spectrometry to confirm the identity of the ∼70/55-kDa immunoreactive band retained by the lectin column as an obscurin kinase isoform. To this end, following elution, the ∼70/55-kDa band was separated by SDS-PAGE, stained with Coomassie Brilliant Blue-R250, and excised with a sharp razor. The isolated protein band was reduced, alkylated, and in-gel digested with trypsin. The tryptic peptides were subjected to nanoLC (Dionex Ultimate 3000; Dionex Corp., Milford, MA, USA) and tandem mass spectrometry (AB SciexTOF/TOF 5800 System; Applied Biomics, Hayward, CA, USA). The resulting peptide mass and the associated fragmentation spectra were submitted to Mascot software to search the Swiss-Prot database, allowing for one missed cleavage, and oxidation on methionine and carbamidomethylation on cysteine as variable modifications.
In vitro kinase assays
We incubated 100–400 ng of purified SK1 or SK2 with 10 μg of the appropriate substrates and 1 μCi of [γ-32P] labeled ATP in kinase buffer containing 10 mM Na2HPO4, 10 mM MgCl2, 200 μM ATP, 50 mM β-glycerophosphate, 100 nM okadaic acid, 1 mM CaCl2, and 1 mM dithiothreitol, in the presence of Complete mini-protease inhibitor cocktail tablets, EDTA-free (Roche). After 1 h incubation at 30°C, reactions were stopped by addition of LDS sample buffer (Life Technologies), analyzed by 4–12% SDS-PAGE, and visualized by autoradiography.
In silico prediction of N- and O-glycosylation lectin-column chromatography and deglycosylation of cardiac lysates
Potential glycosylation sites of the obscurin kinase domains and the interkinase regions (accession no. ENSMUSP00000038264) were identified using the NetNGlyc1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/) and NetOGlyc3.0 (http://www.cbs.dtu.dk/services/NetOGlyc-3.0/) bioinformatics tools, respectively (Center for Biological Sequence Analysis of Technical University of Denmark, Lyngby, Denmark).
To confirm the presence of glycosylation sites, adult mouse heart homogenates were prepared in SB buffer, incubated with a lectin resin overnight at 4°C, washed, and eluted with buffers ME or SE (QProteome total glycoprotein kit; Qiagen; Venlo, Limberg, the Netherlands; buffers SB, ME, and SE, as well as detergents and protease inhibitors are provided by the kit). For PNGase F treatment, lysates and eluted fractions were precipitated with ice-cold acetone and denatured at 100°C for 10 min in 0.5% SDS and 40 mM dithiothreitol. Denatured samples were treated with 1000 U of PNGase F (New England Biolabs, Ipswich, MA, USA) in 50 mM sodium phosphate buffer (pH 7.5) in the presence of 10% Nonidet P-40 at 37°C for 2 h. Protein lysates were loaded to the lectin column, while eluted fractions were immediately analyzed by immunoblotting.
RESULTS AND DISCUSSION
Expression of obscurin kinase isoforms in striated muscles
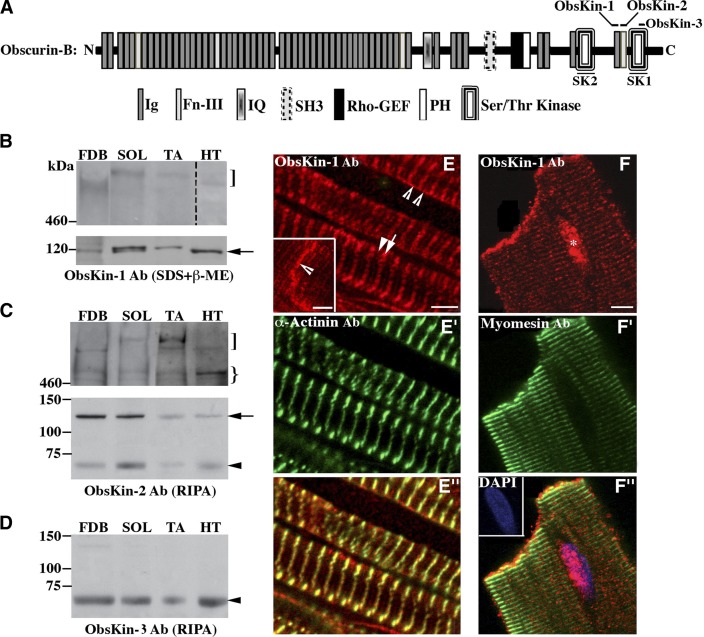
To study the localization of the obscurin kinase proteins, we generated 2 antibodies that recognize epitopes within Ig69 and Fn-III70 (ObsKin-1) or Fn-III70 (ObsKin-2), and obtained a commercial antibody (ObsKin-3) that targets epitopes within SK1 (Fig. 1A). All 3 antibodies are directed against sequences that are present in all putative (known and predicted) obscurin kinase isoforms (2, 3, 5). Immunoblot analysis using lysates from mouse skeletal and cardiac muscles demonstrated that obscurin-B (∼870 kDa) is present in all muscles examined (Fig. 1B, C, top panels, square brackets). Its expression, however, was significantly lower in cardiac than skeletal muscles. Obscurin-B migrated faster in lysates prepared from FDB and heart compared to soleus and tibialis anterior muscles. This apparent difference in the mobility of obscurin-B may be due to differential splicing of the OBSCN gene in the different muscles examined (2, 3), distinct post-translational modifications, or tissue-specific degradation of the protein. An additional immunoreactive band with a molecular mass of ∼600 kDa was detected with the ObsKin-2 antibody (Fig. 1C, top panel, curly bracket), which may result from partial degradation of obscurin-B or correspond to a novel obscurin kinase isoform. Consistent with this, the complete sequence of a novel obscurin kinase isoform of human origin was recently deposited in Ensembl database (accession no. ENSP00000355668) with a calculated molecular mass of ∼611 kDa that starts in Ig35 and contains both SK1 and SK2.
Figure 1.
Expression profile of obscurin kinase proteins. A) Schematic representation of giant obscurin-B denoting the epitopes used for antibody production. B–D) Immunoblots using protein homogenates prepared from skeletal [FDB, soleus (SOL), and tibialis anterior (TA)] and heart (HT) muscles. Note that the lane including heart lysates was exposed for a longer time period than those including skeletal muscle lysates. Obscurin-B (square bracket) is expressed in higher amounts in skeletal than in cardiac muscle. An immunoreactive band of ∼600 kDa (curly bracket) was consistently detected in skeletal and cardiac muscle lysates with the ObsKin-2 antibody and may represent a novel obscurin isoform or a degradation product of obscurin B. Moreover, 2 small obscurin isoforms of ∼120 kDa (arrow) and ∼70 kDa (arrowhead) were also present in all striated muscles examined. E–E″) Mouse cardiac tissue sections stained with ObsKin-1 (red; E) and α-actinin (green; E′) antibodies and merged image (E″). F–F″) Immunolabeling of adult rat cardiocytes with ObsKin-1 (red; F) and myomesin (green; F″) antibodies counterstained with DAPI (blue) and merged image (E″). Scale bars = 5 μm.
In addition to giant obscurin-B, we also detected smaller obscurin kinase proteins. Both ObsKin-1 and ObsKin-2 antibodies specifically and efficiently recognized 2 bands with molecular masses of ∼120 and ∼70 kDa in all muscles examined (Figs. 1B, C, arrow and arrowhead, and 4B, arrowhead); the ∼70 kDa band was also detected with the ObsKin-3 antibody (Fig. 1D, arrowhead). Earlier work has indicated the presence of 2 small obscurin kinase isoforms, containing solely SK1 (single kinase) or partial SK2 and SK1 (tandem kinase) (5). However, the exact translation initiation site of the single kinase and the domain composition of either isoform have yet to be experimentally confirmed. It is therefore likely that the ∼120- and ∼70-kDa bands that we consistently detect in skeletal and cardiac muscle homogenates may correspond to the tandem and single obscurin kinase isoforms, respectively. Molecular characterization of the transcripts that encode the 2 proteins will confirm this notion.
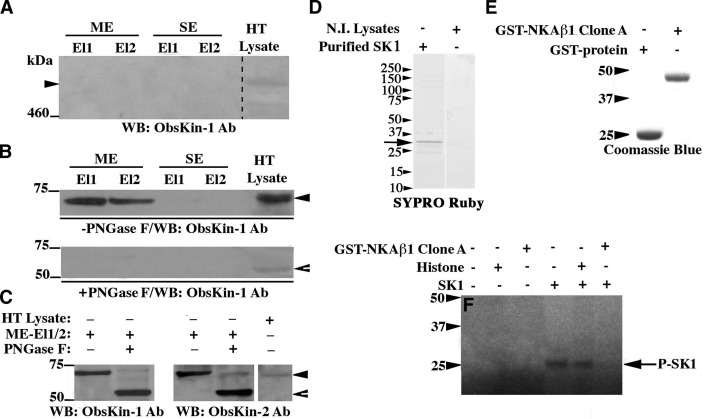
Figure 4.
A small obscurin kinase isoform undergoes glycosylation. A–C) Adult mouse heart lysates were incubated with lectin resins. Eluted fractions were immunoprobed for the presence of giant obscurin-B (A) or smaller obscurin kinase isoforms prior to (B, top panel) and after (C) treatment with PNGase F glycosidase. Protein lysates were also treated with PNGase F before they were applied to the lectin resin (B, bottom panel); ME and SE, elution buffers (see Materials and Methods), El1 and El2, eluted fractions 1 and 2. D) Purified His-tagged SK1 protein analyzed by SDS-PAGE and stained with SYPRO Ruby. E) Purified control GST-protein and recombinant NKAβ1 peptide fused to GST, separated by SDS-PAGE, and stained with Coomassie Brilliant Blue-R250. F) Autoradiogram of an in vitro kinase assay using His-tagged SK1 and different substrates; bands marked as P-SK1 represent autophosphorylated species of SK1. Notably, the autophosphorylation activity of SK1 is significantly diminished in the presence of recombinant NKAβ1, possibly due to direct binding of the latter to the catalytic part of the former, as our Y2H results indicated.
We then examined the subcellular distribution of the obscurin kinase proteins in sections of adult mouse cardiac muscle (Fig. 1E–E″) as well as cultures of adult rat cardiocytes (Fig. 1F–F″). Obscurin kinase proteins are present at the level of the sarcomeric M-band (Fig. 1E, arrowhead) and the Z disk (Fig. 1E, arrow), as shown by costaining with the Z-disk protein, α-actinin (Fig. 1E′), the sarcolemma (Fig. 1E, open arrowheads), and the intercalated disk (ICD; Fig. 1E, inset, open arrowhead). In addition, we repeatedly observed nuclear accumulation of obscurin kinases in adult cultured cardiocytes (Fig. 1F, asterisk).
While the presence of obscurin kinase proteins at the M band and Z disk is not surprising (13, 15), their nuclear concentration, sarcolemmal distribution, and presence at the ICD of cardiocytes are novel, although not unprecedented. Our group has previously reported nuclear localization of a small obscurin kinase isoform in mammary epithelial cells (16). Moreover, Carlsson et al. (17) described the presence of obscurins at the myotendenous junction. It is therefore apparent that the obscurin kinase proteins are present in multiple locations within muscle cells. Whether specific isoforms preferentially localize to select subcellular compartments is still unknown. However, given the absence of unique sequences in the obscurin kinase proteins that would allow the generation of isoform-specific antibodies, this may be a technically challenging question to answer.
Characterization of ligands and substrates of the obscurin kinase domains
To identify ligands and/or substrates of the obscurin kinase domains, we performed a Y2H screen using SK2 (Fig. 2A) or SK1 (Fig. 3A) as baits and an adult human cardiac cDNA library; a list of the obtained prey clones is shown in Table 2. All interactions were also examined using the mouse homologues of the bait and prey clones.
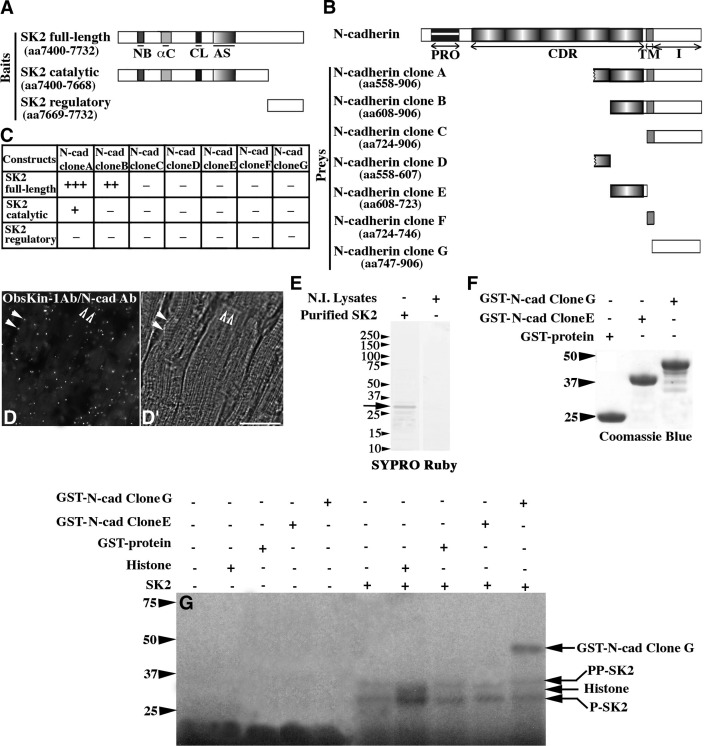
Figure 2.
N-cadherin is a substrate of SK2. A, B) Schematic diagrams illustrating the SK2 bait (A) and N-cadherin prey (B) constructs. αC, α-helix C; AS, activation segment; CDR, extracellular cadherin repeats; CL, catalytic loop; I, intracellular domain; NB, nucleotide binding domain; PRO, prodomain; TM, transmembrane region. C) Identification of minimal interacting domains using the Y2H system; relative strength of the identified interactions is indicated by + and − symbols. D, D′) Fluorescent (D) and bright field (D′) images obtained from the PLA assay. Scale bar = 30 μm. E) Purified His-tagged SK2 protein analyzed by SDS-PAGE and stained with SYPRO Ruby. F) Purified control GST-protein and recombinant N-cadherin peptides fused to GST separated by SDS-PAGE and stained with Coomassie Brilliant Blue-R250. G) Autoradiogram of an in vitro kinase assay using His-tagged SK2 and different substrates; bands denoted as P-SK2 and PP-SK2 represent SK2 species autophosphorylated to different extents.
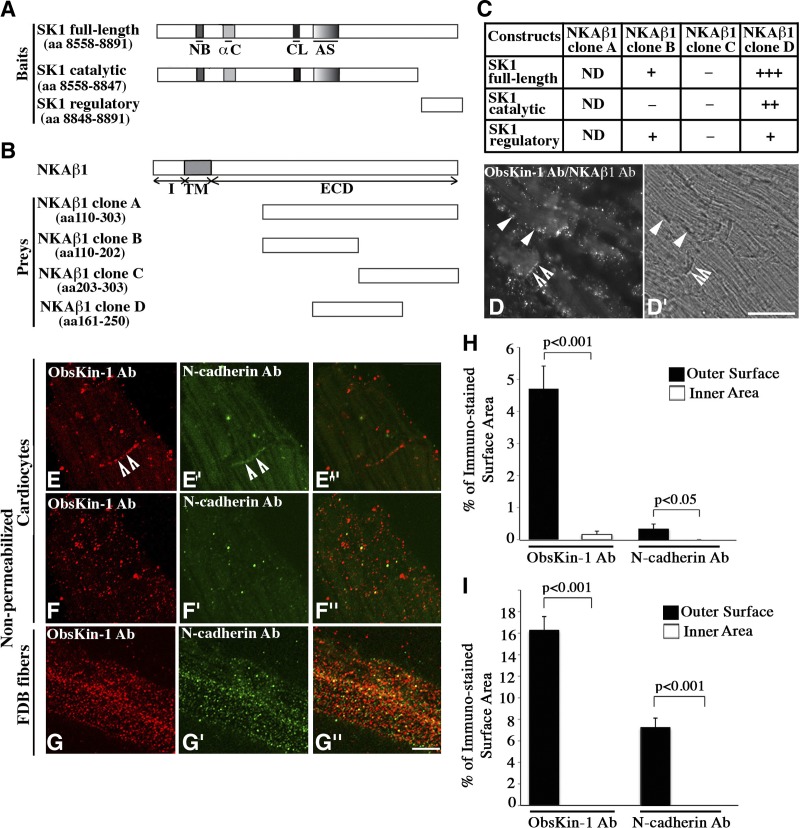
Figure 3.
Extracellular localization of obscurin kinase proteins in striated muscle cells. A, B) Schematic representations of SK1 bait (A) and NKAβ1 prey (B) constructs. αC, α-helix C; AS, activation segment; CL, catalytic loop; ECD, extracellular domain; I, intracellular domain; NB, nucleotide binding domain; TM, transmembrane domain. C) Identification of minimal interacting domains using the Y2H system; relative strength of the identified interactions is indicated by + and − symbols. ND, not determined due to autoactivation. D, D′) Fluorescent (D) and bright field (D′) images obtained from the PLA assay. Scale bar =30 μm. E–G″) Colabeling of nonpermeabilized cardiocytes (E–F″) and FDB fibers (G–G″) with ObsKin-1 (red; E, F, G) and N-cadherin (green; E′, F′, G′) antibodies that recognize epitopes in its extracellular domain and merged images (E″, F″, G″). Scale bar = 8 μm. H, I) Quantification of the percentage of the outer sarcolemmal surface and the inner sarcoplasmic area of nonpermeabilized cardiocytes (H) and FDB myofibers (I) immunostained with the ObsKin-1 and N-cadherin antibodies.
Table 2.
SK1 and SK2 preys identified by the Y2H screen
| Bait | Y2H prey clones |
|---|---|
| SK1 | Na+/K+ ATPase β1-subunit (NM_001677.3) |
| Galectin (NM_002305.3) | |
| Fibronectin-1 (NM_002026.2) | |
| γ-filamin (NM_001127487.1) | |
| Four and a half LIM domain-2 protein (NM_201557.3) | |
| SK2 | N-cadherin (NM_001792.2) |
| Galectin (NM_002305.3) | |
| Immunoglobulin superfamily with leucine repeats protein (NM_005545.3) | |
| Titin (NM_133378.3) |
We found N-cadherin as a potential interacting partner of SK2 (Fig. 2B). Deletion analysis demonstrated that sequences within both the extracellular and intracellular portions of N-cadherin are required to support binding to the catalytic portion of SK2 (Fig. 2C). Costaining of adult cardiac sections with ObsKin-1 and N-cadherin antibodies followed by confocal evaluation demonstrated their coincident distribution (Supplemental Fig. S1A–A″). Use of PLA in combination with immunofluorescent microscopy of cardiac sections colabeled with ObsKin-1 and N-cadherin antibodies confirmed that obscurin kinase proteins and N-cadherin localize in proximity (<40 nm) at the level of the sarcolemma and the ICD, suggesting the physiological relevance of their interaction (Fig. 2D–D′, closed and open arrowheads, respectively; positive and negative controls of the PLA assay are included in Supplemental Fig. S1C–E′).
Previous work has shown that N-cadherin undergoes extensive phosphorylation primarily on tyrosine residues (18). Phosphorylation of N-cadherin on Y860 located in its cytoplasmic domain by Src kinase abolishes binding to β-catenin at the plasma membrane. Freed β-catenin translocates to the nucleus and activates the transcription of target genes. Phosphorylation of serine or threonine residues of N-cadherin, however, is less understood. Hsu et al. (19) recently reported phosphorylation of S788, also located in the cytoplasmic domain of N-cadherin, in response to activation of the mTOR pathway. We therefore sought to examine whether N-cadherin is a substrate of SK2. To do so, we used the baculovirus system to produce the catalytic part of SK2 (2) coupled to a 6xHis tag (Fig. 2E, arrow), and the bacterial system to produce individual N-cadherin domains conjugated to GST (Fig. 2F). Following affinity purification of the recombinant proteins, we confirmed the purity of SK2 using tandem mass spectrometry. Even though trace amounts of contaminating proteins were detected, in addition to SK2, these mainly included ribosomal proteins and keratins. These results therefore demonstrated the absence of any known or putative kinases in the SK2 sample that was subsequently used in in vitro kinase assays (Fig. 2G). We found that SK2 possesses kinase activity, as it can phosphorylate a generic substrate of Ser/Thr kinases (i.e., histone). More important, SK2 undergoes autophosphorylation, which may be essential for its activation, and can efficiently and specifically phosphorylate the cytoplasmic domain of N-cadherin. This is the first evidence since the original identification of the OBSCN gene in 2001 demonstrating that SK2 is an active kinase that phosphorylates, and potentially regulates, a substrate involved in cell adhesion. It is therefore possible that obscurin-B, via SK2, may play key roles in modulating the strength of cell adhesion. Moreover, it is likely that obscurin-B, via SK2, may indirectly regulate the nucleocytoplasmic distribution of β-catenin, a potent player of the Wnt pathway, that directly binds to the cytoplasmic domain of N-cadherin depending on its phosphorylation status (reviewed in ref. 20). Identification of the exact amino acids in the cytoplasmic domain of N-cadherin phosphorylated by SK2, and the physiological importance of these modifications are the next goals of our studies.
Using the Y2H system, we also found that the β1 subunit of the Na+/K+ ATPase pump, NKAβ1, is a potential interacting partner of SK1 (Fig. 3B). Deletion analysis revealed that part of the extracellular domain of NKAβ1, including aa 161 to 250, is necessary and sufficient to support binding to the catalytic domain of SK1 (Fig. 3C). Confocal images of adult cardiac sections costained with ObsKin-1 and NKAβ1 antibodies that recognize its extracellular domain demonstrated their overlapping distribution (Supplemental Fig. S1B–B″). Use of the PLA assay in combination with immunofluorescent microscopy of cardiac sections colabeled with ObsKin-1 and NKAβ1 antibodies further verified that obscurin kinase proteins and NKAβ1 are located in proximity (<40 nm) at the level of the sarcolemma and the ICD (Fig. 3D–D′, closed and open arrowheads, respectively). It is worth mentioning that while labeling of cardiac tissue sections for obscurin kinases and NKAβ1 under confocal optics appears as continuous striations at the level of Z disks and M bands (Supplemental Fig. S1B–B″), the fluorescent signal obtained in the PLA assay appears dotted and is restricted to the sarcolemma and the ICD. This is due to the fact that the presence of fluorescent signal in the PLA assay does not necessarily depict the subcellular distribution of obscurin kinases and NKAβ1, but their proximity in select cellular compartments (i.e., the sarcolemma and the ICD).
The localization of NKAβ1 in adherens junctions and its role in mediating intercellular adhesion of epithelial cells are well established (21). Although its presence at the sarcolemma, the ICD and transverse tubules of cardiocytes has also been documented (22), its roles in cardiac structure and function has remained speculative. Recently part of the extracellular domain of NKAβ1, encompassing aa 198–207, was shown to mediate trans-dimerization of NKAβ1 and cell-cell adhesion (23). Interestingly, this segment is included within the minimal domain of NKAβ1 required to support binding to SK1 (Fig. 3C; residues 161–250). It is therefore possible that binding of SK1 to NKAβ1 close to its dimerization domain may modulate the strength of cell adhesion either through physical interaction or phosphorylation.
The ∼70-kDa obscurin kinase isoform localizes extracellularly and undergoes glycosylation
The interaction between SK1 and part of the extracellular domain of NKAβ1 suggested that some obscurin kinase proteins might localize extracellularly. To test this possibility, we stained nonpermeabilized primary cultures of adult mouse cardiocytes and FDB myofibers with ObsKin-1 and antibodies to either the extracellular domain of N-cadherin (Fig. 3E–G″) or myomesin, which labels sarcomeric M-bands (Supplemental Fig. S2A–B″). Z-stack sectioning using confocal optics revealed that the ObsKin-1 antibody stained specifically and efficiently ICDs and the outer sarcolemmal surface of cardiocytes and FDB fibers in a punctate pattern (Fig. 3E–G, open arrowheads indicate ICDs; and Supplemental Movies S1 and S2, respectively). Similarly, the N-cadherin antibody labeled mainly ICDs between cardiocytes and the outer sarcolemmal surface of FDB myofibers (Fig. 3E′-G′, open arrowheads indicate ICDs; and Supplemental Movies S1 and S2, respectively). Using ImageJ software, we quantified the percentage of the external surface area that was immunostained with the ObsKin-1 and N-cadherin antibodies. We found that obscurin kinase proteins occupy ∼5 and ∼16% of the outer sarcolemmal surface of cardiocytes and myofibers, respectively, while N-cadherin occupies ∼0.3 and ∼7% (Fig. 3H–I). Notably, neither the ObsKin-1 nor the N-cadherin antibody stained the sarcoplasm of nonpermeabilized cardiocytes and myofibers, as our quantification indicated (Fig. 3H–I). Similarly, the myomesin antibody failed to stain intracellular M bands in nonpermeabilized cardiocytes or FDB fibers (Supplemental Fig. S2A–B″), but not in permeabilized ones (Supplemental Fig. S2C–D″). Absence of primary antibodies completely abolished staining in either nonpermeabilized or permeabilized cells (Supplemental Fig. S2E–H″). Our findings therefore demonstrate that at least some obscurin kinase isoform containing SK1 may localize extracellularly where it can interact with the extracellular domain of NKAβ1.
Given that proteins localizing in the extracellular matrix often undergo glycosylation, we examined whether this is the case for any of the obscurin kinase isoforms. To do so, we incubated adult mouse cardiac homogenates with a lectin resin that preferentially binds N-glycans, and eluted fractions were analyzed by SDS-PAGE and immunoblotting (Fig. 4). We did not detect obscurin-B (Fig. 4A); however, we consistently detected a ∼70-kDa immunoreactive band in the eluted fractions (Fig. 4B, top panel, solid arrowhead). This is consistent with the identification of a ∼70-kDa band in skeletal and cardiac muscle homogenates, shown in Fig. 1C–D. Pretreatment of cardiac lysates with peptide-N-glycosidase F (PNGase F), which removes most types of N-glycans, abolished binding of the ∼70 kDa protein to the lectin resin (Fig. 4B, bottom panel). Interestingly, we observed a shift in the mobility of the ∼70-kDa protein in PNGase F-treated homogenates, migrating at ∼55 kDa (Fig. 4B, bottom panel, open arrowhead). To further assess this observation, we incubated cardiac lysates with the lectin resin as before, but treated the eluted fractions with PNGase F before fractionation by SDS-PAGE and immunoblotting. We observed a significant decrease in the apparent molecular mass of the ∼70-kDa protein following treatment with PNGase F to ∼55 kDa (Fig. 4C, closed and open arrowheads). Using mass spectrometry, we verified the identity of the ∼70/55-kDa protein as an obscurin kinase isoform by identifying sequences within the nonmodular region preceding SK1 (e.g., ASMAHISR and GRPEGPER). Previous work has indicated that the single obscurin kinase isoform containing SK1 has a calculated molecular mass of ∼55 kDa (5). It is therefore likely that the ∼70/55-kDa immunoreactive band that our experiments identified corresponds to the single obscurin kinase, which localizes in the extracellular matrix of cardiocytes and myofibers, where it may undergo glycosylation and can interact with the extracellular domain of NKAβ1.
The conventional secretion pathway of transmembrane and extracellular proteins involves recognition of a signal peptide present in the NH2 terminus of the newly synthesized protein that targets it to the endoplasmic reticulum-Golgi pathway. Examination of the obscurin kinase or the NKAβ1 sequences failed to identify such a signal in either of them. It is therefore likely that the small obscurin kinase isoform that localizes extracellularly is secreted through nonconventional pathways, similar to galectin-1 (24), which was also identified as a potential binding partner of SK1 in our Y2H screen (Table 2).
Our findings clearly indicate that NKAβ1 is an interacting partner of SK1. We therefore examined whether NKAβ1 is a substrate of SK1 by performing an in vitro kinase assay (Fig. 4F). As before, we used the baculovirus and bacterial systems to produce the catalytic part of SK1 (Fig. 4D, arrow) and the interacting domain of NKAβ1 (Fig. 4E), coupled to 6xHis and GST tags, respectively. The purity of SK1 was further verified with tandem mass spectrometry; similar to SK2, we detected trace amounts of contaminating ribosomal proteins and keratins, but no known or putative kinase. Unlike SK2, SK1 was unable to phosphorylate histone, suggesting that it has more stringent substrate specificity (Fig. 4F). Although SK1 did undergo autophosphorylation, indicating that it possesses kinase activity, it failed to phosphorylate recombinant GST-NKAβ1 (Fig. 4F). This result can be explained in many ways. It is likely that NKAβ1 is not a substrate of SK1, and that binding to SK1 regulates its accessibility to other kinases or interacting partners. It is also possible that the recombinant NKAβ1 peptide is misfolded in the bacterial system or that it requires additional modifications (i.e., glycosylation) prior to phosphorylation by SK1 (21). Alternatively, it is conceivable that SK1 needs to undergo further post-translational modification, potentially glycosylation, before it can phosphorylate NKAβ1. This is consistent with our findings demonstrating the presence of extensive N-glycosylation in the ∼70/55 kDa obscurin kinase isoform.
Our study shows for the first time that the obscurin kinase domains, SK1 and SK2, are active enzymes with distinct substrate specificities. In the context of giant obscurin-B, this finding indicates dual kinase activities within the same molecule. To date, the ribosomal s6 kinase (RSK) is the only other protein that contains 2 functional SK domains. In particular, the more NH2-terminal kinase shares sequence similarity with the PKA/PKG/PKC (AGC) subfamily of kinases, and the more COOH-terminal kinase belongs to the Ca2+/calmodulin-dependent protein kinase (CAMK) subfamily. While several substrates have been identified for the AGC RSK kinase, the only known function of the CAMK RSK kinase is to phosphorylate and activate the NH2-terminal AGC RSK kinase (25). Unlike the kinase domains of RSK, both obscurin SK1 and SK2 contain a putative calmodulin (CAM) binding regulatory motif (2). Since CAM is predicted to modulate the activities of both SK1 and SK2, it is unlikely that one may regulate the other through phosphorylation and sequential activation.
UNC-89, the OBSCN homologue of Ceanorhabditis elegans, also encodes a giant isoform, referred to as UNC-89 B, that contains tandem kinase domains, namely PK1 and PK2, that belong to the MLCK subfamily (26). Although the enzymatic activities of PK1 and PK2 are still speculative, homology modeling suggests that PK2 is catalytically active while PK1 is inactive. Both PK1 and PK2 bind small carboxyl-terminal domain (CTD) phosphatase-like-1 (27), while PK1 also binds LIM-9 protein, which is indirectly linked to the integrin adhesion complex through its interaction with UNC-97/PINCH (28, 29). The physiological significance of these interactions has yet to be established.
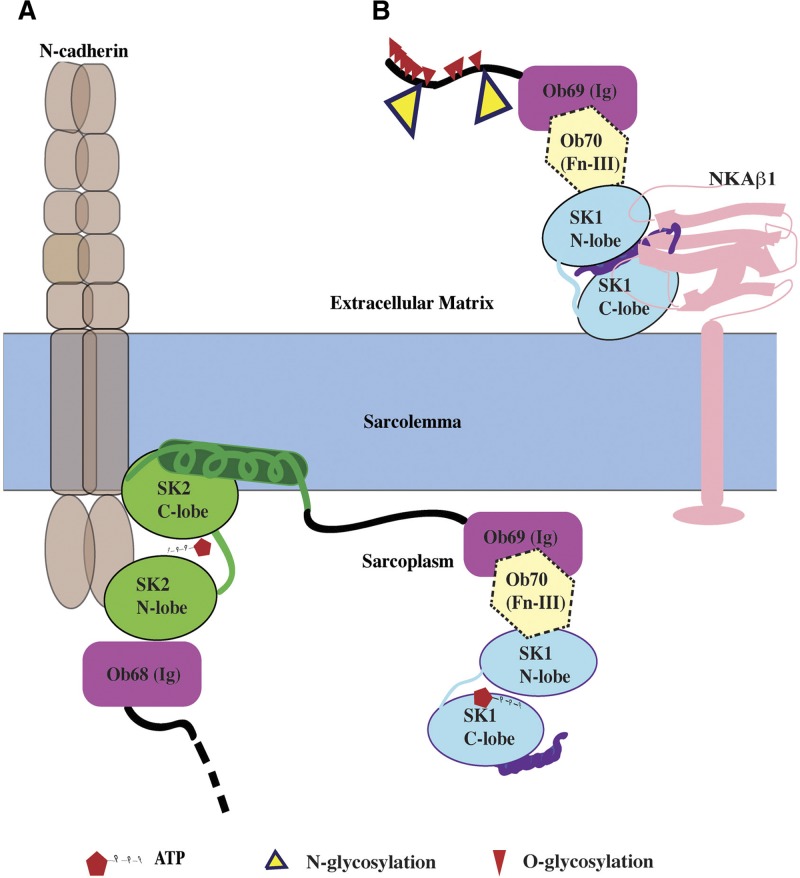
Obscurins have been previously implicated in the organization of sarcomeres and internal membranes (1, 30–32). Our study suggests a potential role of obscurin kinase proteins in cell adhesion. While obscurin-B appears to localize intracellularly where SK2 can phosphorylate the cytoplasmic domain of N-cadherin (Fig. 5A), a smaller obscurin kinase isoform containing SK1 is localized extracellularly, where it can interact with the extracellular domain of NKAβ1 (Fig. 5B). The physiological importance of these interactions in regulating cardiac development and contractility will be the next goals of our studies.
Figure 5.
Topology of obscurin kinase proteins. A) SK2 domain of obscurin-B (or of other intracellular obscurin kinase isoforms) may phosphorylate the cytoplasmic domain of N-cadherin at ICD. B) A small obscurin kinase isoform (∼70/55 kDa) containing SK1 localizes extracellularly, where it can undergo glycosylation and interact with the β1 subunit of NKA. N- and O-glycosylation sites were predicted using NetNGlyc1.0 and NetOGlyc3.1 software, respectively.
Supplementary Material
Acknowledgments
The authors thank Drs. J. W. Lederer, B. Prosser, and M. Bamgboye (University of Maryland) for providing the primary cultures of adult mouse cardiocytes.
This work was supported by U.S. National Institutes of Health training grants 5T32GM08181-23 and 2T32AR7592-16 (L.-Y.R.H.) and the American Heart Association (A.K.K.).
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- β-ME
- β-mercaptoethanol
- CAMK
- Ca2+/calmodulin-dependent protein kinase
- cMLCK
- cardiac-specific MLCK
- FDB
- flexor digitorum brevis
- Fn-III
- fibronectin-III
- GST
- glutathione-S-transferase
- His
- histidine
- ICD
- intercalated disk
- Ig
- immunoglobulin
- LC
- liquid chromatography
- MLCK
- myosin light chain kinase
- NKAβ1
- Na+/K+ ATPase β1
- PFA
- paraformaldehyde
- PLA
- proximity ligation assay
- PNGase F
- peptide-N-glycosidase F
- RSK
- ribosomal s6 kinase
- Ser
- serine
- SK
- serine/threonine kinase
- SPEG
- striated preferentially expressed gene
- Thr
- threonine
- Y2H
- yeast 2-hybrid
REFERENCES
- 1. Kontrogianni-Konstantopoulos A., Ackermann M. A., Bowman A. L., Yap S. V., Bloch R. J. (2009) Muscle giants: molecular scaffolds in sarcomerogenesis. Physiol. Rev. 89, 1217–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fukuzawa A., Idowu S., Gautel M. (2005) Complete human gene structure of obscurin: implications for isoform generation by differential splicing. J. Muscle Res. Cell Motil. 26, 427–434 [DOI] [PubMed] [Google Scholar]
- 3. Russell M. W., Raeker M. O., Korytkowski K. A., Sonneman K. J. (2002) Identification, tissue expression and chromosomal localization of human obscurin-MLCK, a member of the titin and Dbl families of myosin light chain kinases. Gene 282, 237–246 [DOI] [PubMed] [Google Scholar]
- 4. Sutter S. B., Raeker M. O., Borisov A. B., Russell M. W. (2004) Orthologous relationship of obscurin and Unc-89: phylogeny of a novel family of tandem myosin light chain kinases. Dev. Genes Evol. 214, 352–359 [DOI] [PubMed] [Google Scholar]
- 5. Borisov A. B., Raeker M. O., Russell M. W. (2008) Developmental expression and differential cellular localization of obscurin and obscurin-associated kinase in cardiac muscle cells. J. Cell. Biochem. 103, 1621–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kamm K. E., Stull J. T. (2011) Signaling to myosin regulatory light chain in sarcomeres. J. Biol. Chem. 286, 9941–9947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ding P., Huang J., Battiprolu P. K., Hill J. A., Kamm K. E., Stull J. T. (2010) Cardiac myosin light chain kinase is necessary for myosin regulatory light chain phosphorylation and cardiac performance in vivo. J. Biol. Chem. 285, 40819–40829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weinert S., Bergmann N., Luo X., Erdmann B., Gotthardt M. (2006) M line-deficient titin causes cardiac lethality through impaired maturation of the sarcomere. J. Cell Biol. 173, 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hsieh C. M., Fukumoto S., Layne M D., Maemura K., Charles H., Patel A., Perrella M. A., Lee M. E. (2000) Striated muscle preferentially expressed genes α and β are two serine/threonine protein kinase derived from the same gene as the aortic preferentially expressed gene-1. J. Biol. Chem. 275, 36966–36973 [DOI] [PubMed] [Google Scholar]
- 10. Liu X., Ramjiganesh T., Chen Y. H., Chung S. W., Hall S. R., Schissel S. L., Padera R. F., Liao R., Ackerman K. G., Kajstura J., Leri A., Anversa P., Yet S. F., Layne M. D., Perella M. A. (2009) Disruption of striated preferentially expressed gene locus leads to dilated cardiomyopathy in mice. Circulation 119, 261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Borisov A. B., Raeker M. O., Kontrogianni-Konstantopoulos A., Yang K., Kurnit D.M., Bloch R. J., Russell M. W. (2003) Rapid response of cardiac obscurin gene cluster to aortic stenosis: differential activation of Rho-GEF and MLCK and involvement in hypertrophic growth. Biochem. Biophys. Res. Commun. 310, 910–918 [DOI] [PubMed] [Google Scholar]
- 12. Ackermann M. A., Hu L.Y., Bowman A. L., Bloch R. J., Kontrogianni-Konstantopoulos A. (2009) Obscurin interacts with a novel isoform of MyBP-C slow at the periphery of the sarcomeric M-band and regulates thick filament assembly. Mol. Biol. Cell 20, 2963–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kontrogianni-Konstantopoulos A., Jones E. M., van Rossum D. B., Bloch R. J. (2003) Obscurin is a ligand for small ankyrin 1 in skeletal muscle. Mol. Biol. Cell 14, 1138–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prosser B. L., Ward C. W., Lederer W. J. (2011) X-ROS signaling: rapid mechano-chemo transduction in heart. Science 333, 1440–1445 [DOI] [PubMed] [Google Scholar]
- 15. Bowman A. L., Kontrogianni-Konstantopoulos A., Hirsch S. S., Geisler S. B., Gonzalez-Serratos H., Russell M. W., Bloch R. J. (2007) Different obscurin isoforms localize to distinct sites at sarcomeres. FEBS Lett. 581, 1549–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perry N. A., Shriver M., Mameza M. G., Grabias B., Balzer E., Kontrogianni-Konstantopoulos A. (2012) Loss of giant obscurins promotes breast epithelial cell survival through apoptotic resistance. FASEB J. 26, 2764–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carlsson L., Yu J. G., Thornell L. E. (2008) New aspects of obscurin in human striated muscles. Histochem. Cell Biol. 130, 91–103 [DOI] [PubMed] [Google Scholar]
- 18. Qi J., Wang J., Romanyak O., Siu C. H. (2006) Involvement of Src family kinases in N-cadherin phosphorylation and beta-catenin dissociation during transendothelial migration of melanoma cells. Mol. Biol. Cell 17, 1261–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hsu P. P., Kang S. A., Rameseder J., Zhang Y., Ottina K. A., Lim D., Peterson T. R., Choi Y., Gray N. S., Yaffe M. B., Marto J. A., Sabatini D. M. (2011) The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heuberger J., Birchmeier W. (2010) Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb. Perspect. Biol. 2, a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vagin O., Dada L. A., Tokhtaeva E., Sachs G. (2012) The Na-K-ATPase α1β1 heterodimer as a cell adhesion molecule in epithelia. Am. J. Physiol. Cell Physiol. 302, C1271–C1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McDonough A. A., Zhang Y., Shin V., Frank J. S. (1996) Subcellular distribution of sodium pump isoform subunits in mammalian cardiac myocytes. Am. J. Physiol. 270, C1221–C1227 [DOI] [PubMed] [Google Scholar]
- 23. Tokhtaeva E., Sachs G., Sun H., Dada L. A., Sznajder J. I., Vagin O. (2012) Identification of the amino acid region involved in the intercellular interaction between the β1 subunits of Na+/K+-ATPase. J. Cell Sci. 125, 1605–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nickel W., Rabouille C. (2009) Mechanisms of regulated unconventional protein secretion. Nat. Rev. Mol. Cell Biol. 10, 148–155 [DOI] [PubMed] [Google Scholar]
- 25. Romeo Y., Zhang X., Roux P. P. (2012) Regulation and function of the RSK family of protein kinases. Biochem. J. 441, 553–569 [DOI] [PubMed] [Google Scholar]
- 26. Small T. M., Gernert K. M., Flaherty D. B., Mercer K. B., Borodovsky M., Benian G. M. (2004) Three new isoforms of Caenorhabditis elegans UNC-89 containing MLCK-like protein kinase domains. J. Mol. Biol. 342, 91–108 [DOI] [PubMed] [Google Scholar]
- 27. Qadota H., McGaha L. A., Mercer K. B., Stark T. J., Ferrara T. M., Benian G. M. (2008) A novel protein phosphatase is a binding partner for the protein kinase domains of UNC-89 (obscurin) in Caenorhabditis elegans. Mol. Biol. Cell 19, 2424–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xiong G., Qadota H., Mercer K. B., McGaha L. A., Oberhauser A. F., Benian G. M. (2009) A LIM-9 (FHL) /SCPL-1 (SCP) complex interacts with the C-terminal protein kinase regions of UNC-89 (obscurin) in Caenorhabditis elegans muscle. J. Mol. Biol. 386, 976–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qadota H., Mercer K. B., Miller R. K., Kaibuchi K., Benian G. M. (2007) Two LIM domain proteins and UNC-96 link UNC-97/PINCH to myosin thick filaments in Caenorhabditis elegans muscle. Mol. Biol. Cell 18, 4317–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Spooner P.M., Bonner J., Maricq A.V., Benian G. M., Norman K. R. (2012) Large isoforms of UNC-89 (obscurin) are required for muscle cell architecture and optimal calcium release in Caenorhabditis elegans. PLoS ONE 7, e40182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kontrogianni-Konstantopoulos A., Catino D. H., Strong J. C., Sutter S., Borisov A. B., Pumplin D. W., Russell M. W., Bloch R. J. (2006) Obscurin modulates the assembly and organization of sarcomeres and the sarcoplasmic reticulum. FASEB J. 20, 2102–2111 [DOI] [PubMed] [Google Scholar]
- 32. Lange S., Ouyang K., Meyer G., Cui L., Cheng H., Lieber R. L., Chen J. (2009) Obscurin determines the architecture of the longitudinal sarcoplasmic reticulum. J. Cell Sci. 122, 2640–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.