Abstract
Shewanella oneidensis exhibits a remarkable versatility in anaerobic respiration, which largely relies on its diverse respiratory pathways. Some of these are expressed in response to the existence of their corresponding electron acceptors (EAs) under aerobic conditions. However, little is known about respiration and the impact of non-oxygen EAs on the physiology of the microorganism when oxygen is present. Here we undertook a study to elucidate the basis for nitrate and nitrite inhibition of growth under aerobic conditions. We discovered that nitrate in the form of NaNO3 exerts its inhibitory effects as a precursor to nitrite at low concentrations and as an osmotic-stress provider (Na+) at high concentrations. In contrast, nitrite is extremely toxic, with 25 mM abolishing growth completely. We subsequently found that oxygen represses utilization of all EAs but nitrate. To order to utilize EAs with less positive redox potential, such as nitrite and fumarate, S. oneidensis must enter the stationary phase, when oxygen respiration becomes unfavorable. In addition, we demonstrated that during aerobic respiration the cytochrome bd oxidase confers S. oneidensis resistance to nitrite, which likely functions via nitric oxide (NO).
Introduction
Shewanella oneidensis is a facultative Gram-negative anaerobe with remarkable anaerobic respiration abilities that allow the use of a diverse array of terminal electron acceptors (EA), including fumarate, nitrate, nitrite, thiosulfate, trimethylamine N-oxide (TMAO), dimethylsulfoxide (DMSO), Fe(III), Mn(III) and (IV), Cr(VI), and U(VI) [1]. In order to reduce these diverse EAs, the S. oneidensis genome encodes a large number of respiratory pathways, some of which have been elucidated over the last two decades [1]–[8].
In the case of nitrate and nitrite respiration, S. oneidensis has many unique properties. The genome of S. oneidensis possesses operons napDAGHB and nrfA for periplasmic nitrate reductase (NAP) and nitrite reductase (NRF), respectively [3]. Surprisingly, NapC and NrfH or the complex of NrfBCD, which in many bacteria are essential and dedicated electron transport components of NAP and NRF respectively, were missing [9], [10]. To complete these pathways, nitrate and nitrite terminal reductases recruit CymA, a cytoplasmic membrane electron transport protein, to play the role of both NapC and NrfH for electron transport from the menaquinone pool. One consequence of sharing CymA is that reduction of nitrite to ammonium does not commence until nitrate is thoroughly exhausted, resulting in a characteristic two-step reduction of nitrate [3].
Nitrate (as a precursor to nitrite) and nitrite, on one hand, have been used for centuries as preservative in meat products to inhibit the growth of bacterial pathogens and these antibacterial effects are attributed to NO formation [11]–[13]. S. oneidensis is capable of producing NO in the presence of either nitrate or nitrite under microaerobic/anaerobic conditions although the enzymatic machinery for NO synthesis is unknown [14]. NO interferes with biological processes by either interacting with protein cofactors, such as Fe-S clusters, heme, and lipoamide, or by promoting the formation of reactive nitrogen species (RNS) [11], [15], [16]. On the other hand, both nitrate and nitrite can support the growth of S. oneidensis as sole EA under anaerobic conditions although biomass production is rather limited [3]. Two mM nitrite facilitates growth most effectively, resulting in an OD600 up to 0.06, which cannot be reliably measured [2], [3]. Nitrate per se supports a greater biomass but it is rapidly converted to nitrite, which hampers further growth, also resulting in an extremely low maximum cell density (less than 0.1 of OD600) [2], [3]. As a consequence, mutants lacking any of the genes involved in nitrate/nitrite respiration or its regulation have further limited growth and cannot be readily studied.
Following the elucidation of anaerobic nitrate and nitrite respiration in S. oneidensis [3], we have undertaken a study of the regulatory proteins mediating these processes. In order to obtain adequate biomass for the characterization of mutants impaired in regulation of nitrate/nitrite respiration, we tested various aerobic cultivation conditions, including shake flasks (uncontrolled batch), controlled batch bioreactors, and chemostats [17]. Although reduction of nitrate to nitrite occurred in controlled cultures, reduction of nitrite to ammonium did not, and thus unsuitable for studies of nitrite respiration in S. oneidensis. Surprisingly, in batch cultures both reduction processes preceded without compromising biomass production. This simple cultivation method is useful for studies of the nitrate/nitrite respiration regulatory systems and of the mechanism underlying the differences in nitrite respiration. In this study, we investigated the impact of nitrate and nitrite on the physiology of S. oneidensis under aerobic conditions in order to understand their inhibitory mechanisms as well as the basis for oxygen repression on respiration of EAs with less positive redox potential, such as nitrite.
Results
Inhibitory Effect of Nitrate on Aerobic Growth of S. oneidensis
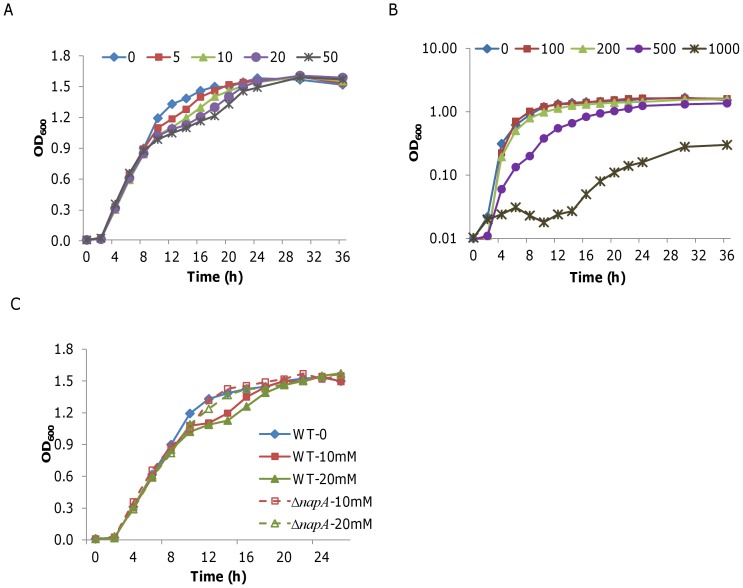
Under anaerobic conditions, S. oneidensis is able to survive in the presence of up to 100 mM nitrate but grows to a maximum cell density (less than 0.1 of OD600) only with less than 4 mM [2], [3]. To evaluate the impact of nitrate on aerobic cultures of S. oneidensis, we monitored the bacterial growth in LB-NaClless (containing one tenth of NaCl, ∼17 mM Na+ to reduce the impact of Na+) media supplemented with NaNO3 at various levels. When NaNO3 was added at low concentrations (5–50 mM), growth of S. oneidensis started to deviate slightly from the standard growth curve at ∼0.8 of OD600 (Fig. 1A). Growth was significantly reduced when NaNO3 exceeded 200 mM and was barely detectable in the presence of 1 M (Fig. 1B). Notably, the maximum cell densities were indistinguishable at all test concentrations below 1 M. These data suggest that NaNO3 affects growth of S. oneidensis under aerobic conditions by two distinct mechanisms, which are dose-dependent.
Figure 1. Impact of various nitrate/chloride compounds on growth of S. oneidensis under aerobic conditions.
A. Growth of S. oneidensis in the presence of NaNO3 (concentrations from 5 to 50 mM). B. Growth of S. oneidensis in the presence of NaNO3 (concentrations from 100 to 1000 mM). C. Growth of S. oneidensis wild type and the napA mutant in the presence of NaNO3 (10 and 20 mM). All experiments were performed at least three times. Error bars, representing the standard deviation of the mean, are all less than 3% and are omitted for clarity.
To separate these two mechanisms, the same experiment was carried out with NaCl (25–1000 mM). Growth of S. oneidensis was not affected by NaCl at concentrations less than 100 mM but resembled that resulted from NaNO3 at 200 mM or above (data not shown), implicating that NaNO3 at the high concentrations impedes growth through sodium which functions as a salt/osmotic stress agent. Second, the experiment was repeated with KNO3, which provides nitrate but not sodium. As expected, KNO3 at the concentrations of lower than 50 mM induced the aberrant growth similar to that with 50 mM NaNO3 (data not shown), thus confirming that the aberrant growth is specific to NaNO3. To determine whether nitrate per se accounts for the aberrant growth of S. oneidensis, we repeated the experiment with a strain (ΔnapA) lacking napA (encoding the large subunit of nitrate reductase), which is defective in nitrate reduction [3]. Results presented in Fig. 1C revealed that deletion of napA resulted in growth that was comparable to that of the wild type in the presence of nitrate, indicating that nitrate per se is not the cause for the aberrant growth of S. oneidensis.
Reduction of Nitrate in S. oneidensis during Aerobiosis
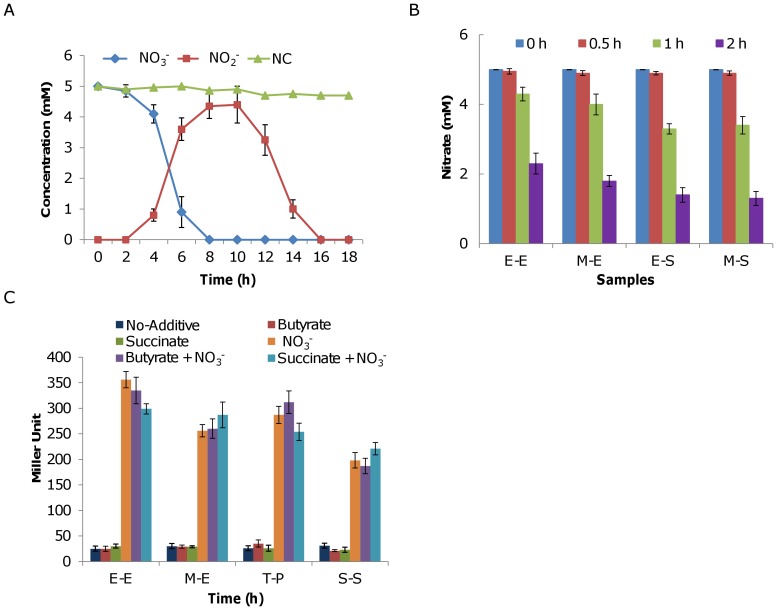
As nitrate is not the agent that directly impairs growth, it is conceivable that its reduced product, nitrite, most likely caused the observed phenotype given that the in vivo toxicity of nitrite is well-established. To gain a better understanding of nitrate and nitrite reduction mechanism in aerobic growing cultures, we examined concentrations of nitrate and nitrite in cultures supplemented with 5 mM NaNO3 (Fig. 2A). Nitrite reached detectable levels 4 hours after inoculation (∼0.2 of OD600), was maximum approximately 8 hours after inoculation (∼1.2 of OD600), remained at the same level for about 3 hours, and was gone within 16 hours, indicating that S. oneidensis cells are able to respire on nitrite when oxygen is present. However, reduction of nitrate to ammonium in the presence of oxygen is clearly distinct from the two-step process observed under anaerobic conditions where nitrite reduction begins immediately after nitrate is completely consumed [3], [18], suggesting that certain requirements must be met for aerobic growing cells to proceed with nitrite reduction.
Figure 2. Nitrate respiration under aerobic conditions in S. oneidensis.
A. Nitrate/nitrite assays of aerobic cultures. S. oneidensis grown in the presence of nitrate was collected at the indicated time points and the supernatants were assayed using IC. The ΔnapA strain was used as a negative control (NC). B. Nitrate respiration in cultures at different growth phases in the presence of 5 mM nitrate and assayed 30 min, 1 hour and 2 hours after addition of the chemical. Early exponential phase (E-E, ∼0.2 of OD600), mid-exponential phase (M-E, ∼0.4), entry into the stationary phase (E-S, ∼0.8), and 4 hour after the entry into the stationary phase (M-S, ∼1.6). C. Impacts of reduced and oxidized carbon sources on expression of the nap operon using a lacZ reporter system. Cultures of the wild type grown to the indicated phases were supplemented with a combination of 5 mM nitrate and 10 mM sodium succinate (oxidized carbon source) and/or 10 mM sodium butyrate (reduced carbon source), collected 1 h later, and assayed for β-galactosidase activity. Experiments were performed at least three times and error bars represent the standard deviation of the mean.
To test whether reduction of nitrate is dependent on the physiological status of cells, nitrate was added to cultures at various OD600 values: ∼0.2 (early exponential, E-E), ∼0.4 (mid-exponential, M-E), ∼0.8 (entry stationary, E-S), and ∼1.6 (∼4 h after the entry of stationary, M-S), and concentrations of nitrate and nitrite were then monitored by ion chromatography (IC). As shown in Fig. 2B, nitrate reduction occurred in all cultures, indicating that the process is independent of the growth phase of the cells.
In Paracoccus denitrificans, aerobic nitrate reduction by NAP is employed to dissipate excess redox energy during oxidative metabolism of reduced carbon substrates but not oxidized carbon substrates [19], [20]. On the contrary, aerobic nitrate reduction in Rhodobacter sphaeroides and Thiosphaera pantotropha is insignificant in regard of physiology and expression of nap is irrespective of oxygen, carbon source, and even nitrate [21], [22]. To test the role of aerobic nitrate reduction in S. oneidensis, experiments were repeated with the same medium supplemented with reduced carbon source butyrate or relatively oxidized carbon substrate succinate. Neither of these additives elicited a notable difference in growth or nitrate utilization (data not shown). Additionally, expression of nap in cells at different growth stages was examined using a Pnap-lacZ reporter system [17]. Significant changes in nap expression were not observed when any of these carbon sources was added solely whereas the expression was elevated considerably with supplemented nitrate (Fig. 2C). These results suggest that aerobic nitrate respiration of S. oneidensis may not have a role in dissipation of excess redox energy.
Inhibitory Effects of Nitrite on Growth of S. oneidensis Under Aerobic Conditions
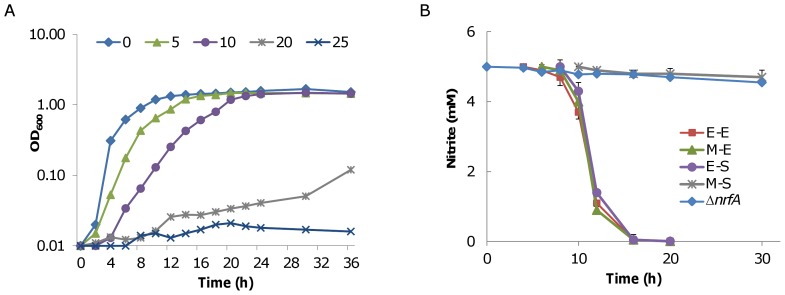
We then investigated the influence of nitrite on growth with NaNO2 as a nitrite provider under aerobic conditions. Not surprisingly, NaNO2 had a strong inhibitory effect on the growth of S. oneidensis. At 5 mM, there was an apparently extended lag time and a lowered growth rate whereas 25 mM completely prevented cells from growing (Fig. 3A). Since the concentrations of sodium used in the experiment is too low to elicit an osmotic effect, it is reasonable to conclude that the observed inhibitory effect results from nitrite. Meanwhile, it is worth noting that the maximum cell densities of these cultures were virtually the same if growth occurred (in the case of 20 mM, 72 h was needed to reach the maximum level).
Figure 3. Response to nitrite during aerobic growth of S. oneidensis.
A. Growth of S. oneidensis in the presence of NaNO2 (from 5 to 25 mM). Experiments were performed at least three times and error bars, representing the standard deviation of the mean, were all less than 3% and are omitted for clarity. B. Time-of-addition assay. 5 mM nitrite (final concentration) was added to aerobic growing cultures at E-E (∼0.2 of OD600), M-E (∼0.4), E-S (∼0.8), and M-S (∼0.8, 4 hour after E-S) as described in the text and the nitrite concentrations were determined at the indicated time points. Experiments were performed at least three times and error bars represent the standard deviation of the mean.
To validate the reduction of nitrite under aerobic conditions, we repeated the experiments with an nrfA in-frame deletion strain (ΔnrfA) [3]. As expected, this strain was unable to carry out nitrite reduction, indicating that the same NRF system responsible for respiration of nitrite under anaerobic conditions also catalyzes nitrite reduction under aerobic conditions (Fig. 3B). We then performed a time-of-addition assay to examine whether cells at different growth stages were responsive to nitrite in a similar pattern. Cultures at E-E (∼0.2, OD600), M-E (∼0.4), E-S (∼0.8), and M-S (∼1.6) were supplemented with 5 mM nitrite (final concentration), allowed to grow under identical conditions, and the concentrations of remaining nitrite were monitored (Fig. 3B). Interestingly, it was found that i) potential for nitrite respiration was limited in cultures prior to the stationary phase, and ii) respiration of nitrite did not occur until cells had entered the stationary phase, confirming that nitrite respiration correlates with the physiological status of the cells.
NrfA is not Sufficient for the Delay of Nitrite Reduction
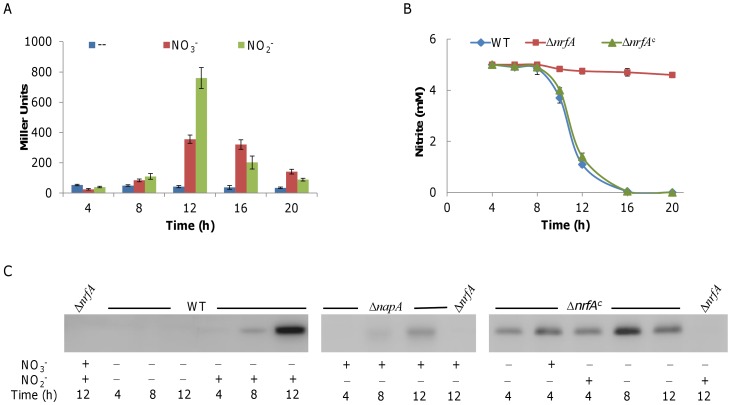
Discovery of an unexpected delay in nitrite reduction under aerobic conditions raised questions about the regulatory mechanisms controlling the process. To address this issue, we first examined the transcriptional level of nrfA at various growth stages using a PnrfA-lacZ reporter system [17]. Both nitrate (the ΔnapA strain was used to prevent nitrate reduction) and nitrite were able to induce expression of nrfA (Fig. 4A). In both cases the wild-type strain expressed nrfA slightly before mid-stationary phase but robustly afterwards. Independent qRT-PCR analysis validated this observation (Fig. S1A).
Figure 4. Expression of NrfA during aerobic growth of S. oneidensis.
A. A lacZ-based reporter analysis of the nrfA promoter. Time of expression of nrfA in the ΔnapA strain (used to block nitrate reduction) cultured aerobically with or without nitrate or nitrite. B. Nitrite respiration in aerobic growing cultures. Samples from cultures grown in the presence 5 mM nitrite were taken at the indicated time points and assayed for remaining nitrite. In both B and C, ΔnrfA c represents the nrfA mutant strain containing pHG102-nrfA, which allows constitutive expression of nrfA. C. Western blot analysis of NrfA in various samples. Cells grown for 4 (M-E), 8 (E-S), and 12 h (M-S) were collected and assayed. The ΔnrfA strain, grown in the presence of nitrate, nitrite, or both, was used as a negative control. To show induction of the nrfA gene by nitrate, the wild type (WT) was replaced by the ΔnapA strain in order to prevent interference from conversion to nitrite. Experiments were performed at least three times. In A and B, error bars represent the standard deviation of the mean.
The absence of NrfA prior to the stationary phase appears to be the cause for the delayed nitrite reduction. If so, robust expression of nrfA at the early growth stages should permit nitrite reduction. To test this, we transcriptionally fused nrfA to the arcA promoter which is constitutively active [18], [23] and introduced the construct into the ΔnrfA strain. Surprisingly, the ΔnrfA strain carrying the ParcA–controlled nrfA failed to consume nitrite before the stationary phase (Fig. 4B). As this ParcA–controlled nrfA complemented the ΔnrfA strain in its ability to respire on nitrite after the stationary phase, the expressed NrfA is therefore functional. To verify expression levels of NrfA in cells at various growing stages, we raised an antibody against the enzyme and used in cells at various growth stages. Consistent with the levels of the nrfA transcript, NrfA was present at an extremely low level in E-E cultures, increased significantly in cells entering the stationary phase, and maximized in M-S cultures (Fig. 4C). Apparently, the production of NrfA was inducible by either nitrate or nitrite, with the latter more efficient. More importantly, the Western blotting analysis confirmed that nrfA driven by ParcA was indeed expressed constitutively, independent of not only nitrate/nitrite but also of the growth phases. In total, these data indicate that NrfA per se is not accountable for the delayed reduction of nitrite.
Respiratory Status is Likely Responsible for the Delayed Nitrite Reduction in S. oneidensis
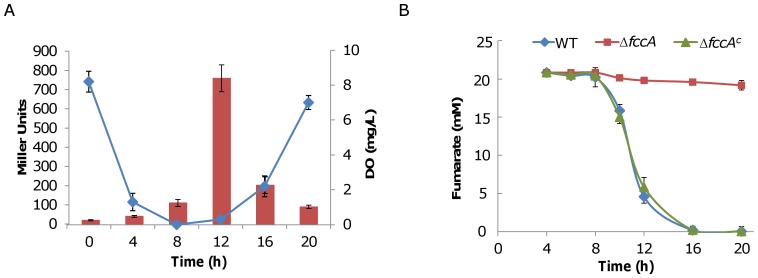
As the initiation of nitrite respiration is dependent on the growth phase, we speculated that the physiological status of S. oneidensis cells with respect to respiration may be critical to the phenotype. To test this, we measured the dissolved oxygen concentrations of cultures in the presence of nitrite. Consistent with previous findings [23], levels of dissolved oxygen in shaking cultures followed a ‘U’ curve, indicating that oxygen was saturated in the beginning, decreasing below detectable levels (exponential phase), and then increasing to saturation in the late-stationary phase (Fig. 5A). Saturation levels of oxygen in the beginning can be readily explained by the low number of cells in the culture whereas at the late stage it implies that oxygen consumption decreased substantially. Given that maximum nrfA expression coincides with the accumulation of oxygen in the culture, we hypothesize that S. oneidensis cells in the stationary phase have shifted to utilize EAs with a low reduction potential even when oxygen is available. To test this hypothesis, we cultured cells aerobically with 20 mM fumarate and assayed for fumarate reduction. As shown in Fig. 5B, with an fccA mutant as a negative control, cells started to respire on fumarate only after entry into the stationary phase [24]. An additional analysis of the fccA expression using qRT-PCR confirmed that fumarate failed to stimulate transcription of fccA until cells had entered the mid-stationary phase (Fig. S1B). In sum, it appears that S. oneidensis is unable to overcome oxygen repression in order to respire on EAs with the relatively low positive standard redox potential.
Figure 5. Characteristics of aerobic cultures of S. oneidensis .
A. Dissolved oxygen (line with diamond markers) in cultures used in Figure 4A. Expression of the nrfA gene (Bar) was included to show the relation of these parameters. B. Assay for remaining fumarate in aerobic growing cultures in the absence or presence of 20 mM. ΔfccAc represents the fccA mutant strain containing pHG101-fccA. Experiments were performed at least three times and error bars represent the standard deviation of the mean.
Cytochrome bd Oxidase Confers S. oneidensis Resistance to Nitrite and NO
The antibacterial effects of nitrite are generally attributed to NO, which is reportedly produced endogenously from nitrite in S. oneidensis as in many prokaryotes [11], [13]–[16], [25]. A common strategy adopted by many species to overcome NO toxicity is to express multiple oxidases, one of which is less sensitive to NO [11], [26]–[28]. The genome of S. oneidensis encodes two cytochrome c terminal oxidases: SO4606-4609 (caa 3-HCO) and SO2364-2361 (CcoN-O-Q-P, cbb3-HCO), and a quinol oxidase SO3286-3285 (CydA-B, bd-type) [6], [29]. As SO4608 has been annotated as coxG, we named SO4606, SO4607, and SO4609 coxB, coxA, and coxC, respectively based on sequence similarities to characterized analogues. In S. oneidensis, cbb3-HCO dominates respiration of oxygen whereas caa 3-HCO appears to be irrelevant [24], [29]. The bd-type oxidase, inferior to cbb3-HCO in oxygen respiration, is critical in the bacterial resistance to nitrite [30].
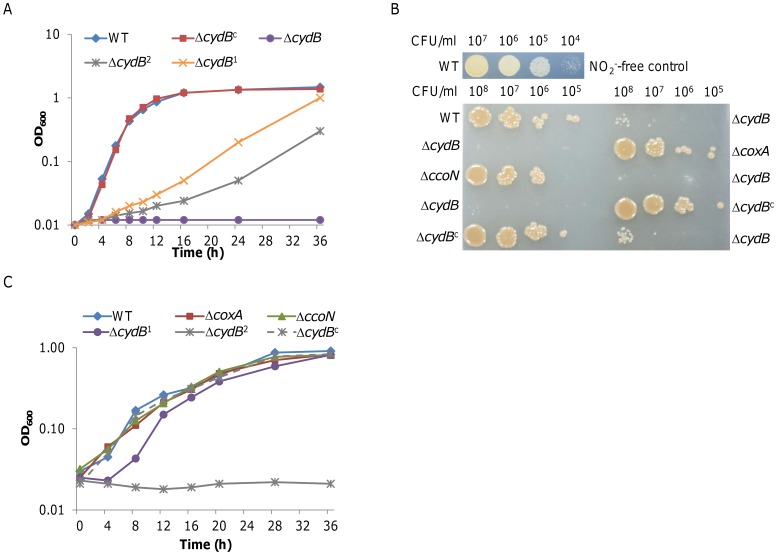
To assess the possible roles that these oxidases play in nitrite/NO resistance, in-frame deletions of coxA (encoding the essential subunit of caa 3-HCO), ccoN (encoding the essential subunit of cbb 3-HCO), and cydB (encoding the essential subunit of the bd-type oxidase) were characterized [29], [30]. The ΔcydB strain displayed significantly impaired growth when challenged by nitrite at 1 mM and failed to grow with 5 mM (Fig. 6A). The observed hypersensitivity of this strain can be restored to the wild type level by expressing cydB in trans. In contrast, strains lacking either coxA or ccoN behaved as the wild type in the presence of 5 mM nitrite (data not shown). These observations were supported by results from the nitrite susceptibility assay (Fig. 6B). The cydB mutant displayed a hypersensitivity to nitrite whereas other mutants were similar in resistance to the chemical in comparison with the wild type. Notably, nitrite showed a profound impact on the physiology of S. oneidensis, resulting in fuzzy-looking colonies.
Figure 6. Characteristics of S. oneidensis strains devoid of one of the oxidases.
A. Growth in the presence of 5 mM nitrite under aerobic conditions. Results with ΔcoxA and ΔccoN, which were identical to the wild type, are not shown. ΔcydB 1 and ΔcydB 2 represent that the ΔcydB strain grew in the presence of 1 and 2 mM nitrite, respectively. In all panels, ΔcydB c represents the cydB mutant strain containing pHG102-cydB. B. Sensitivity to nitrite of strains lacking one of the oxidases. Ten µl of each sample (from 105 to 108 CFU/ml) was spotted onto LB plates containing 5 mM nitrite. The photo was taken after 24 h incubation. The assay was repeated at least three times with similar results. WT cells spotted on the LB plates lacking nitrite were used as the reference for colony morphology. C. Growth of strains devoid of one of oxidases in air-tight tubes (microaerobic conditions as described in the text) in the presence of 750 µM NONOate. ΔcydB 1 and ΔcydB 2 represent that the ΔcydB strain grown in the presence of 750 and 1500 µM NONOate, respectively. In both A, and C, error bars, representing the standard deviation of the mean, are all less than 5% and are omitted for clarity.
We then tested the effects of NO on growth of these S. oneidensis strains. NO was introduced to 2 ml medium in 15-ml rubber-stopped Hungate tubes by adding DETA NONOate (t1/2, 20 h at 37°C and 56 h at 25°C), which releases NO slowly so there is a relatively steady NO concentration [14], [31]. Under these conditions, oxygen in the headspace enables cells to grow, allowing us to measure the inhibitory effects of NO. DETA NONOate at 1500 µM prohibited growth of the ΔcydB strain but not others (Fig. 6C). At 750 µM, DETA NONOate extended the lag time of the cydB mutant but showed no effect on growth of other mutants as well as the wild type. These data, collectively, indicate that i) nitrite, likely via NO, inhibits growth by interacting with terminal oxidases and ii) the cytochrome bd oxidase plays a crucial role in overcoming nitrite inhibition.
Discussion
S. oneidensis is well known for its respiratory versatility, capable of respiring on oxygen or a wide range of organic and inorganic substrates under anaerobic conditions. Interestingly, there is synthesis of some anaerobic respiratory systems in the presence of oxygen, although to much lesser extent than in the absence of oxygen [32]. This implies that multiple EAs may be utilized simultaneously. Indeed, our results demonstrate that both nitrate and nitrite can be respired in agitated aerobic batch cultures despite the delay in nitrite respiration. Under conditions where the inhibitory effect of nitrite is minimal, it is possible to obtain sufficient biomass for biochemical and genetic analyses and for analyzing the degree of defectiveness of various mutants. With batch cultures under aerobic conditions, we succeeded in characterizing systems that regulate nitrate/nitrite respiration [17].
In E. coli, regulation of respiratory enzyme synthesis is under hierarchical control, following the standard redox potential of the electron acceptor couples (e.g. O2> NO3 –>NO2 –>fumarate) [33]. As a consequence, EAs with the most positive standard redox potential are preferentially used, a dogma that could explains why nitrate respiration is able to escape oxygen repression in S. oneidensis. In terms of physiological significance, aerobic nitrate reduction in S. oneidensis, as in R. sphaeroides and T. pantotropha, appears to play no role in dissipation of excess redox energy as observed in P. pantotrophus [19]–[22]. In contrast, other respiratory oxidants such as nitrite, and a group of S and N oxides exemplified by TMAO and fumarate, which are less preferentially consumed [34], are able to induce expression of their corresponding terminal reductases only after cells have entered the mid-stationary phase. In cells constitutively expressing NrfA, S. oneidensis cells were still unable to respire on these EAs, most likely due to the fact that oxygen outcompetes other EAs for electrons in actively growing cells. Such oxygen repression was relieved in cells that were relatively inactive with oxygen. Consequently, in these cells oxygen is no longer preferred and other EAs with less positive redox potential are utilized to drive ATP generation.
In regard to the inhibitory effects of nitrate and nitrite on S. oneidensis, these are substantial differences. Nitrate per se apparently has little impact on cells and is converted to nitrite under aerobic conditions, which is very toxic. The antimicrobial activity of nitrite is likely attributed to NO as S. oneidensis is able to produce NO endogenously in the presence of either nitrate or nitrite and contains a complex NO signaling pathway [14], [31]. In prokaryotes, in addition to NO synthases which are involved in the oxidation of L-arginine to NO, copper- or heme-containing cytochrome cd 1 nitrite reductases and nitrate reductases (NarGHI) can also catalyze NO formation from nitrite [35]–[37]. Intriguingly, analogues to any of these enzymes have not been identified in the genome [2], [3], [14]. The enzymatic source of NO in S. oneidensis is not known and thus merits investigation.
NO elicits its pleiotropic effects on bacterial physiology by direct reaction with metallocofactors or through S-nitrosation of cysteine residues [38], [39]. To counteract NO stress, bacteria have evolved different strategies. A well understood mechanism involves a number of proteins that degrade NO such as flavogemoglobin, flavorubredoxin, and cytochrome c 552 nitrite reductase, the last of which has been implicated to be in place in S. oneidensis [11], [31], [40]–[42]. In addition to detoxification, bacteria may synthesize multiple proteins of the same function, one of which is less sensitive to NO. A good example is terminal oxidases [11]. Some bacteria such as Vibrio fischeri uses alternative oxidase (AOX), which is NO-inducible and plays a specific role in NO resistance [27]. For bacteria such as S. oneidensis and E. coli lacking specific oxidases in response to NO, one of the common oxidases may assume the role of overcoming nitrosative stress [11]. The cytochrome bd oxidase, which is bioenergetically inferior to cytochrome c oxidases and plays a dispensable role in aerobic respiration, elevates tolerance to stresses elicited by both nitrite and NO [43]. It has recently been demonstrated that cytochrome bd is unlike HCOs in that it quickly recovers activity upon NO depletion, thereby conferring a higher tolerance to NO [11], [28], [44].
It is worth noting that the S. oneidensis strains devoid of cytochrome bd appear more sensitive to nitrite than to NO. One explanation is that the concentration of NO from DETA NONOate (no more than 0.125 µM when 200 µM NONOate is supplied) is too low to provide an inhibitory effect equal to that of 5 mM nitrite [31]. In addition, NO may be rapidly consumed by cells as the NO concentrations in control media is between 0.5 and 3 µM [31].This can explain why the cytochrome bd null mutant has an extended lag phase but eventually grows up, presumably after NO is reduced to a tolerable level.
Methods
Bacterial Strains, Plasmids, PCR Primers, and Culture Conditions
A list of all bacterial strains and plasmids used in this study is given in Table 1. Primers used for generating PCR products are available upon request. Escherichia coli and S. oneidensis strains were grown in Luria-Bertani (LB, Difco, Detroit, MI) medium at 37°C and 30°C for genetic manipulation, respectively. Where needed, antibiotics were added at the following concentrations: ampicillin at 50 µg/ml, kanamycin at 50 µg/ml, and gentamycin at 15 µg/ml.
Table 1. Strains and plasmids used in this study.
| Strain or plasmid | Description | Reference or source |
| E. coli strain | ||
| DH5α | Host for regular cloning | Lab stock |
| WM3064 | Donor strain for conjugation; ΔdapA | W. Metcalf, UIUC |
| S. oneidensis strains | ||
| MR-1 | Wild-type | Lab stock |
| HG0848 | napA deletion mutant derived from MR-1; ΔnapA | [3] |
| HG0970 | fccA deletion mutant derived from MR-1; ΔfccA | [24] |
| HG2364 | ccoN deletion mutant derived from MR-1; ΔccoN | This study |
| HG3980 | nrfA deletion mutant derived from MR-1; ΔnrfA | [3] |
| HG3285 | cydB deletion mutant derived from MR-1; ΔcydB | This study |
| HG4607 | coxA deletion mutant derived from MR-1; ΔcoxA | This study |
| Plasmids | ||
| pDS3.0 | Apr, Gmr, derivative from suicide vector pCVD442 | Lab stock |
| pHG101 | Promoterless broad-host Kmr vector | [46] |
| pHG102 | pHG101 containing the S. oneidensis arcA promoter | [46] |
| pTP327 | lacZ reporter vector | [18] |
| pHG101-nrfA | nrfA is under the control of its own promoter within pHG101 | This study |
| pHG102-nrfA | nrfA is under the control of the arcA promoter within pHG102 | This study |
| pHG101-fccA | fccA is under the control of its own promoter within pHG101 | This study |
| pHG101-ccoN | ccoN is under the control of its own promoter within pHG101 | This study |
| pHG102-cydB | cydB is under the control of the arcA promoter within pHG102 | This study |
| pTP327-Pnap | pTP327 containing the S. oneidensis nap promoter | This study |
| pTP327-PnrfA | pTP327 containing the S. oneidensis nrfA promoter | This study |
Mutagenesis and Complementation of Mutant Strains
In-frame deletion strains were constructed using the Fusion PCR method as previously described [23]. In brief, two fragments flanking the targeted gene were amplified independently first and joined together by the second round of PCR. The resulting fusion fragment for each individual gene was introduced into plasmid pDS3.0. The resulting mutagenesis vector was transformed into E. coli WM3064, and then transferred into S. oneidensis by conjugation [45]. Integration of the mutagenesis construct into the chromosome was selected by gentamycin resistance and confirmed by PCR. Verified transconjugants were grown in LB broth in the absence of NaCl and plated on LB supplemented with 10% of sucrose. Gentamycin-sensitive and sucrose-resistant colonies were screened by PCR for deletion of the targeted gene. The deletion mutation was then verified by sequencing of the mutated region.
For complementation of genes next to their promoter, a fragment containing the targeted gene and its native promoter was generated by PCR and cloned into pHG101 [46]. For other genes, the targeted gene was amplified and inserted into the MCS of pHG102 under the control of the arcA promoter. Introduction of each verified complementation vector into the corresponding S. oneidensis mutant was done by mating with the appropriate E. coli WM3064 strain, and confirmed by plasmid extraction and restriction enzyme mapping.
Physiological Characterization of the Mutant Strains
Aerobic and anaerobic growth was assayed essentially the same as described elsewhere [3]. LB-NaClless (LB containing one tenth amount of NaCl) and M1 defined medium containing 0.02% (w/v) of vitamin-free Casamino Acids were used unless otherwise noted. For anaerobic growth, 20 mM lactate served as an electron donor with one of following : NaNO3 (4 mM), NaNO2 (2 mM), and fumarate (20 mM) as an electron acceptor. To test effects of different carbon sources on aerobic nitrate/nitrite reduction, sodium succinate (10 mM final concentration) and/or sodium butyrate (10 mM final concentration) were used. Growth of S. oneidensis strains was determined by monitoring an increase in OD600 in triplicate samples. For biochemical analyses, cells were grown in 30 ml of media supplemented with NaNO3/NaNO2 (5 mM), collected by centrifugation, frozen immediately in liquid-nitrogen, and stored at −80°C for qRT-PCR, β-Galactosidase activity assay, and Western blotting and supernatants were used directly for nitrate/nitrite assays.
β-Galactosidase Activity Assay
β-Galactosidase assays were performed using an assay kit (Beyotime, China) according to manufacturer’s instructions. Cells were washed with PBS (phosphate buffered saline) (137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4 1.76 mM KH2PO4, pH7.4), and treated with lysis buffer (0.25 M Tris/HCl, 0.5% Trion-X100, pH7.5). The resulting soluble protein was collected after centrifugation and used for enzyme assays. β-galactosidase activity was determined by monitoring color development at 420 nm using a Sunrise Microplate Reader (Tecan). The protein concentration of the cell lysates was determined using a Bradford assay with BSA as a standard (Bio-Rad). Activity was expressed in Miller units.
Quantitative RT-PCR (qRT-PCR) Analysis
Quantitative real-time reverse transcription-PCR (qRT-PCR) analysis was carried out with an ABI7300 96-well qRT-PCR system (Applied Biosystems) as described previously [47]. The expression of each gene was determined from three replicates in a single real-time qRT-PCR experiment. The Cycle threshold (CT) values for each gene of interest were averaged and normalized against the CT value of 16s rRNA, whose abundance was consistent from early exponential to stationary phase. The relative abundance (RA) of each gene compared to that of 16s rRNA was calculated using the equation RA = 2−ΔCT.
Immunoblotting Assay
Immunoblotting analysis was performed with rabbit polyclonal antibodies against NrfA essentially as previously described [17]. Cell pellets were washed once with PBS, and resuspended in PBS to an optical density of 0.5 at 600 nm (OD600) of PBS. The total protein concentration of the cell lysates was then determined by the bicinchoninic acid assay (Pierce Chemical). Samples were loaded onto SDS-10% polyacryl-amide gels and either stained with Coomassie brilliant blue or electrophoretically transferred to polyvinylidene difluoride (PVDF) according to the manufacturer’s instructions (Bio-Rad). The gels were blotted for 2 h at 60 V using a Criterion blotter (Bio-Rad). The blotting membrane was probed with anti-NrfA antibody followed by a 1∶5,000 dilution of goat anti-rabbit IgG-HRP (Horse radish peroxidase) (Roche Diagnostics) and was detected using a chemiluminescence Western blotting kit (Roche Diagnostics) in accordance with the manufacturer’s instructions. Images were visualized with the UVP Imaging System.
Chemical Assays
Culture supernatants were subjected to Ion Chromatography (IC) for determination of nitrate and nitrite concentrations essentially as previously described [3]. The assay was performed with IonPac® AS19 with Na2SO4 as the eluent at a concentration of 100 mM with a flow rate of 0.6 ml/min in ICS-3000 (Dionex, Sunnyvale, CA). Fumarate measurements were carried out using fumarate assay kit according to the manufacturer’s instructions (Biovision, CA, USA). A standard curve was prepared each time.
Plate Sensitivity Assay
Cells of S. oneidensis strains grown in LB at 30°C to an OD600 of ∼0.6 were adjusted to approximately 108 CFU/ml with fresh LB, followed by six 10-fold serial dilutions. Ten µl of each bacterial sample (from 105 to 108 CFU/ml) was spotted onto the LB plates containing 5 mM nitrite. All plates were incubated at 30°C and photographs were taken about 24 hours later when colonies of the wild type were fully developed. The assay was repeated at least for three times with similar results.
Supporting Information
Expression analysis using qRT-PCR. A. Transcription levels of nrfA in the samples used in Figure 4A. B. Transcription levels of fccA in cells grown with 20 mM fumarate to indicated time points. Experiments were performed at least three times and error bars represent the standard deviation of the mean.
(PDF)
Funding Statement
This research was supported by Major State Basic Research Development Program (973 Program: 2010CB833803), National Natural Science Foundation of China (31270097), and Natural Science Foundation of Zhejiang province (R3110096) and Major Program of Science and Technology Department of Zhejiang (2009C12061) to HG. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, et al. (2008) Towards environmental systems biology of Shewanella . Nat Rev Micro 6: 592–603. [DOI] [PubMed] [Google Scholar]
- 2. Cruz-Garcia C, Murray AE, Klappenbach JA, Stewart V, Tiedje JM (2007) Respiratory nitrate ammonification by Shewanella oneidensis MR-1. J Bacteriol 189: 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gao H, Yang ZK, Barua S, Reed SB, Romine MF, et al. (2009) Reduction of nitrate in Shewanella oneidensis depends on atypical NAP and NRF systems with NapB as a preferred electron transport protein from CymA to NapA. ISME J 3: 966–976. [DOI] [PubMed] [Google Scholar]
- 4. Gon S, Patte J-C, Dos Santos J-P, Méjean V (2002) Reconstitution of the trimethylamine oxide reductase regulatory elements of Shewanella oneidensis in Escherichia coli . J Bacteriol 184: 1262–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gralnick JA, Vali H, Lies DP, Newman DK (2006) Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proc Natl Acad Sci USA 103: 4669–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson WC, et al. (2002) Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis . Nat Biotechnol 20: 1118–1123. [DOI] [PubMed] [Google Scholar]
- 7. Maier TM, Myers JM, Myers CR (2003) Identification of the gene encoding the sole physiological fumarate reductase in Shewanella oneidensis MR-1. J Basic Microbiol 43: 312–327. [DOI] [PubMed] [Google Scholar]
- 8. Shirodkar S, Reed S, Romine M, Saffarini D (2011) The octahaem SirA catalyses dissimilatory sulfite reduction in Shewanella oneidensis MR-1. Environ Microbiol 13: 108–115. [DOI] [PubMed] [Google Scholar]
- 9. Jepson BJN, Marietou A, Mohan S, Cole JA, Butler CS, et al. (2006) Evolution of the soluble nitrate reductase: defining the monomeric periplasmic nitrate reductase subgroup. Biochem Soc Trans 34: 122–126. [DOI] [PubMed] [Google Scholar]
- 10. Simpson PJL, Richardson DJ, Codd R (2010) The periplasmic nitrate reductase in Shewanella: the resolution, distribution and functional implications of two NAP isoforms, NapEDABC and NapDAGHB. Microbiology 156: 302–312. [DOI] [PubMed] [Google Scholar]
- 11. Mason MG, Shepherd M, Nicholls P, Dobbin PS, Dodsworth KS, et al. (2009) Cytochrome bd confers nitric oxide resistance to Escherichia coli . Nat Chem Biol 5: 94–96. [DOI] [PubMed] [Google Scholar]
- 12. Sindelar JJ, Milkowski AL (2012) Human safety controversies surrounding nitrate and nitrite in the diet. Nitric Oxide 26: 259–266. [DOI] [PubMed] [Google Scholar]
- 13. Reddy D, Lancaster J, Cornforth D (1983) Nitrite inhibition of Clostridium botulinum: electron spin resonance detection of iron-nitric oxide complexes. Science 221: 769–770. [DOI] [PubMed] [Google Scholar]
- 14. Price MS, Chao LY, Marletta MA (2007) Shewanella oneidensis MR-1 H-NOX regulation of a histidine kinase by nitric oxide. Biochemistry 46: 13677–13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hyduke DR, Jarboe LR, Tran LM, Chou KJY, Liao JC (2007) Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli . Proc Natl Acad Sci USA 104: 8484–8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Richardson AR, Payne EC, Younger N, Karlinsey JE, Thomas VC, et al. (2011) Multiple targets of nitric oxide in the tricarboxylic acid cycle of Salmonella enterica Serovar Typhimurium. Cell Host Microbe 10: 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong Y, Wang W, Fu H, Zhou G, Shi M, Gao H (2012) A Crp-dependent two-component regulates nitrate and nitrite respiration in Shewanella oneidensis. PLoS ONE 10.1371/journal.pone.0051643. [DOI] [PMC free article] [PubMed]
- 18. Gao H, Wang X, Yang ZK, Chen J, Liang Y, et al. (2010) Physiological roles of ArcA, Crp, and EtrA and their interactive control on aerobic and anaerobic respiration in Shewanella oneidensis . PLoS ONE 5: e15295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Richardson DJ (2000) Bacterial respiration: a flexible process for a changing environment. Microbiology 146: 551–571. [DOI] [PubMed] [Google Scholar]
- 20. Sears HJ, Spiro S, Richardson DJ (1997) Effect of carbon substrate and aeration on nitrate reduction and expression of the periplasmic and membrane-bound nitrate reductases in carbon-limited continuous cultures of Paracoccus denitrificans Pd1222. Microbiology 143: 3767–3774. [DOI] [PubMed] [Google Scholar]
- 21. Gavira M, Roldán MD, Castillo F, Moreno-Vivián C (2002) Regulation of nap gene expression and periplasmic nitrate reductase activity in the phototrophic bacterium Rhodobacter sphaeroides DSM158. J Bacteriol 184: 1693–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moreno-Vivián C, Cabello P, Martínez-Luque M, Blasco R, Castillo F (1999) Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J Bacteriol 181: 6573–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao H, Wang X, Yang Z, Palzkill T, Zhou J (2008) Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses. BMC Genomics 9: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gao H, Barua S, Liang Y, Wu L, Dong Y, et al. (2010) Impacts of Shewanella oneidensis c-type cytochromes on aerobic and anaerobic respiration. Microbial Biotech 3: 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Crane BR, Sudhamsu J, Patel BA (2010) Bacterial nitric oxide synthases. Annu Rev Biochem 79: 445–470. [DOI] [PubMed] [Google Scholar]
- 26. Sarti P, Giuffrè A, Barone MC, Forte E, Mastronicola D, et al. (2003) Nitric oxide and cytochrome oxidase: reaction mechanisms from the enzyme to the cell. Free Radical Biol Med 34: 509–520. [DOI] [PubMed] [Google Scholar]
- 27. Dunn AK, Karr EA, Wang Y, Batton AR, Ruby EG, et al. (2010) The alternative oxidase (AOX) gene in Vibrio fischeri is controlled by NsrR and upregulated in response to nitric oxide. Mol Microbiol 77: 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Giuffrè A, Borisov VB, Mastronicola D, Sarti P, Forte E (2012) Cytochrome bd oxidase and nitric oxide: From reaction mechanisms to bacterial physiology. FEBS lett 586: 622–629. [DOI] [PubMed] [Google Scholar]
- 29.Zhou G, Yin J, Chen H, Hua Y, Sun L, Gao H (2013) Combined effect of loss of the caa 3 oxidase and Crp regulation drives Shewanella to thrive in redox-stratified environments. ISME J In Press. [DOI] [PMC free article] [PubMed]
- 30.Fu H, Chen H, Wang J, Zhou G, Zhang H, Zhang L, Gao H (2013) Crp-dependent cytochrome bd oxidase confers nitrite resistance to Shewanella oneidensis. Environ. Microbiol. DOI: 10.1111/1462–2920.12091. [DOI] [PubMed]
- 31. Plate L, Marletta MA (2012) Nitric oxide modulates bacterial biofilm formation through a multicomponent Cyclic-di-GMP signaling network. Mol Cell 46: 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baraquet C, Théraulaz L, Iobbi-Nivol C, Méjean V, Jourlin-Castelli C (2009) Unexpected chemoreceptors mediate energy taxis towards electron acceptors in Shewanella oneidensis . Mol Microbiol 73: 278–290. [DOI] [PubMed] [Google Scholar]
- 33. Goh E-B, Bledsoe PJ, Chen L-L, Gyaneshwar P, Stewart V, et al. (2005) Hierarchical control of anaerobic gene expression in Escherichia coli K-12: the nitrate-responsive NarX-NarL regulatory system represses synthesis of the fumarate-responsive DcuD-DcuR regulatory system. J Bacteriol 187: 4890–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Unden G, Bongaerts J (1997) Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta 1320: 217–234. [DOI] [PubMed] [Google Scholar]
- 35. Zumft WG (1997) Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61: 533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rinaldo S, Arcovito A, Brunori M, Cutruzzolà F (2007) Fast dissociation of nitric oxide from ferrous Pseudomonas aeruginosa cd 1 nitrite reductase: a novel outlook on the catalytic mechanism. J Biol Chem 282: 14761–14767. [DOI] [PubMed] [Google Scholar]
- 37. Gilberthorpe NJ, Poole RK (2008) Nitric oxide homeostasis in Salmonella typhimurium: roles of respiratory nitrate reductase and flavohemoglobin. J Biol Chem 283: 11146–11154. [DOI] [PubMed] [Google Scholar]
- 38.Bowman LAH, McLean S, Poole RK, Fukuto JM (2011) The diversity of microbial responses to nitric oxide and agents of nitrosative stress: Close cousins but not identical twins, p. 135–219. In K. P. Robert (ed.), Advances in Microbial Physiology, vol. Volume 59. Academic Press. [DOI] [PubMed]
- 39.Pullan ST, Monk CE, Lee L, Poole RK (2008) Microbial responses to nitric oxide and nitrosative stress: growth, “omic,” and physiological methods, p. 499–519. In K. P. Robert (ed.), Methods in Enzymology, vol. Volume 437. Academic Press. [DOI] [PubMed]
- 40.Angelo M, Hausladen A, Singel DJ, Stamler JS (2008) Interactions of NO with Hemoglobin: From Microbes to Man, p. 131–168. In K. P. Robert (ed.), Methods in Enzymology, vol. Volume 436. Academic Press. [DOI] [PubMed]
- 41. Justino MC, Vicente JB, Teixeira M, Saraiva LM (2005) New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J Biol Chem 280: 2636–2643. [DOI] [PubMed] [Google Scholar]
- 42. Poock SR, Leach ER, Moir JWB, Cole JA, Richardson DJ (2002) Respiratory detoxification of nitric oxide by the cytochromec nitrite reductase of Escherichia coli . J Biol Chem 277: 23664–23669. [DOI] [PubMed] [Google Scholar]
- 43. Borisov VB, Gennis RB, Hemp J, Verkhovsky MI (2011) The cytochrome bd respiratory oxygen reductases. Biochim Biophy Acta 1807: 1398–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Borisov VB, Forte E, Sarti P, Brunori M, Konstantinov AA, et al. (2007) Redox control of fast ligand dissociation from Escherichia coli cytochrome bd . Biochem Biophs Res Commun 355: 97–102. [DOI] [PubMed] [Google Scholar]
- 45. Saltikov CW, Newman DK (2003) Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci USA 100: 10983–10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu L, Wang J, Tang P, Chen H, Gao H (2011) Genetic and molecular characterization of flagellar assembly in Shewanella oneidensis . PLoS ONE 6: e21479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yuan J, Wei B, Shi M, Gao H (2011) Functional assessment of EnvZ/OmpR two-component system in Shewanella oneidensis . PLoS ONE 6: e23701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression analysis using qRT-PCR. A. Transcription levels of nrfA in the samples used in Figure 4A. B. Transcription levels of fccA in cells grown with 20 mM fumarate to indicated time points. Experiments were performed at least three times and error bars represent the standard deviation of the mean.
(PDF)