Abstract
Introduction
Hypodontia, hypohidrosis, sparse hair and characteristic faces are the main characters of X-linked hypohidrotic ectodermal dysplasia (XLHED) which is caused by genetic ectodysplasin A (EDA) deficiency. Heterozygous female carriers tend to have mild to moderate XLHED phenotype, even though 30% of them present no obvious symptom.
Methods
A large Chinese XLHED family was reported and the entire coding region and exon–intron boundaries of EDA gene were sequenced. To elucidate the mechanism for carriers’ tempered phenotype, we analyzed the methylation level on four sites of the promoter of EDA by the pyrosequencing system.
Results
A known frameshift mutation (c.573–574 insT) was found in this pedigree. Combined with the pedigrees we reported before, 120 samples comprised of 23 carrier females from 11 families and 97 healthy females were analyzed for the methylation state of EDA promoter. Within 95% confidence interval (CI), 18 (78.26%) carriers were hypermethylated at these 4 sites.
Conclusion
Chinese XLHED carriers often have a hypermethylated EDA promoter.
Introduction
Mutations in EDA gene can lead to X-linked hypohidrotic ectodermal dysplasia (XLHED), the most common form of ectodermal dysplasias (EDs). The incidence is less than one in per 100’000 [1]. Mutant EDA affects cell signaling transduction or cell migration during the epithelial-mesenchymal inductive process. The structures of ectodermal origin are affected. Patients with XLHED have prominent clinical features: sparse hair, eyelashes and eyebrows, small, misshapen or missing teeth, diminished sweating with a history of high fevers in hot weather, decreased salivary secretions, and a characteristic special facial appearance. Facial features include prominent forehead, narrow and short maxillary regions, small palatal depth, small cranial length, and depressed nasal root and bridge of the nose. However most heterozygous carriers only show minor to moderate degrees of these abnormalities [2].
DNA methylation refers to the biological process that a methyl group added to cytosine that stands directly before a guanine molecule by DNA methyltransferases after DNA duplication. In living cells, methylation has been reported as one of the most common covalent modifications of DNA. It has both epigenetic and mutagenic effects on specific gene expression, cell differentiation, chromatin inactivation, embryo growth, and cancer.
EDA contains a large CpG island in its promoter. CpG islands located in gene promoters represent a major target for DNA hypermethylation, which impairs transcription upon regional or specific methylation events. It has been confirmed that promoter CpG island hypermethylation contributed to gene silencing by inhibiting the binding of certain transcription factors to their recognition sequence, attracting methylated DNA-binding proteins, and/or through chromatin remodeling [3]–[11]. However, whether aberrant methylation is related to XLHED carriers’ phenotype has been controversial.
In the present study, we report a causative EDA mutation (c. 573–574 insT) in a Chinese XLHED family. We investigated the methylation of EDA promoter of this family’ carrier as well as other 22 carriers we reported before [12], [13] to study correlations between the phenotype of carriers and the methylation state of the promoter. EDA gene in eighteen (78.26%) of the carriers were hypermethylated, and this result demonstrated that there was a correlation between being a XLHED carrier and the hypermethylation status of the EDA promoter.
Materials and Methods
Ethical Approval
This study was approved by the Institutional Review Board (IRB) of Hospital and School of Stomatology, Wuhan University. Written informed consents were obtained from all participants or their guardians.
Nomenclature
Gene mutation nomenclature used in this study follows the recommendations of den Dunnen and Antonarakis Gene symbols used in this article follow the protocol created by the HUGO gene nomenclature committee [15].
DNA Sample Collection
All probands were outpatient cases of School and Hospital of Stomatology, Wuhan University. Two professional dentists examined the patients respectively according to the classical diagnosis criteria. Comprehensive physical examinations and panoramic radiograph films were taken thoroughly. Pilocarpine iontophoresis sweat test was used to evaluate the function of sweat gland. – to ++ was scored for normal to absolutely no sweat. After a definite diagnosis was obtained, patients’ family members were interviewed and examined. 5–10 ml blood samples were collected with EDTA 2Na+ and heparin as anticoagulant. DNA was isolated from leukocytes using the standard sodium dodecyl sulphate–proteinase K–phenol/chloroform method After quality accessing, DNA was frozen at −20°C.
Polymerase Chain Reaction (PCR) and Mutation Screening
The entire EDA coding region and exon–intron boundaries of patients and their relatives were amplified using the same primers used in the previous report Amplified fragments were purified with a PCR purification kit (Omega, USA) according to the manufacturer’s protocol. DNA sequences were obtained from both strands with an ABI PRISM 3730 genetic analyzer. Sequence analysis was performed by the CHROMAS program and BLAST program of the National Center for Biotechnology Information (NCBI). After identifying nucleotide variants in the EDA gene, 200 unrelated controls were examined respectively.
Quantification of EDA Promoter’s Methylation State by Pyrosequencing
Bisulfite modification of the genome was processed with the CpGenome DNA Modification Kit (Intergen Company, Purchase, NY). The potential methylation sites analyzed in this study were “CGgctgaggcagaCGcagCGgctccCG” located in EDA promoter. Specific amplification and sequencing primers were designed by PSQ Assay Design 1.0. One of the PCR primers was labeled with biotin. Blank sample was used as negative control. 97 clinically healthy subjects who had similar ages as carriers were recruited as normal controls.
Pyrosequencing analysis was conducted with PYRO MARK ID (BIOTAGE). The figures and data were analyzed by PSQ96MA 2.1 software. Peak value in the sequencing picture means allele frequency of the DNA sample. The percentage of residual C (Cm) showed how much of each site has been methylated. To insure the reliability of the results, all of the samples underwent pyrosequencing twice.
Statistical Analysis
Taking advantage of parameter estimation, 95% confidence interval (CI) was employed to do statistical analysis. Within CI, carriers were divided into 3 groups, hypermethylation, normal and hypomethylation groups.
Results
Clinical Data
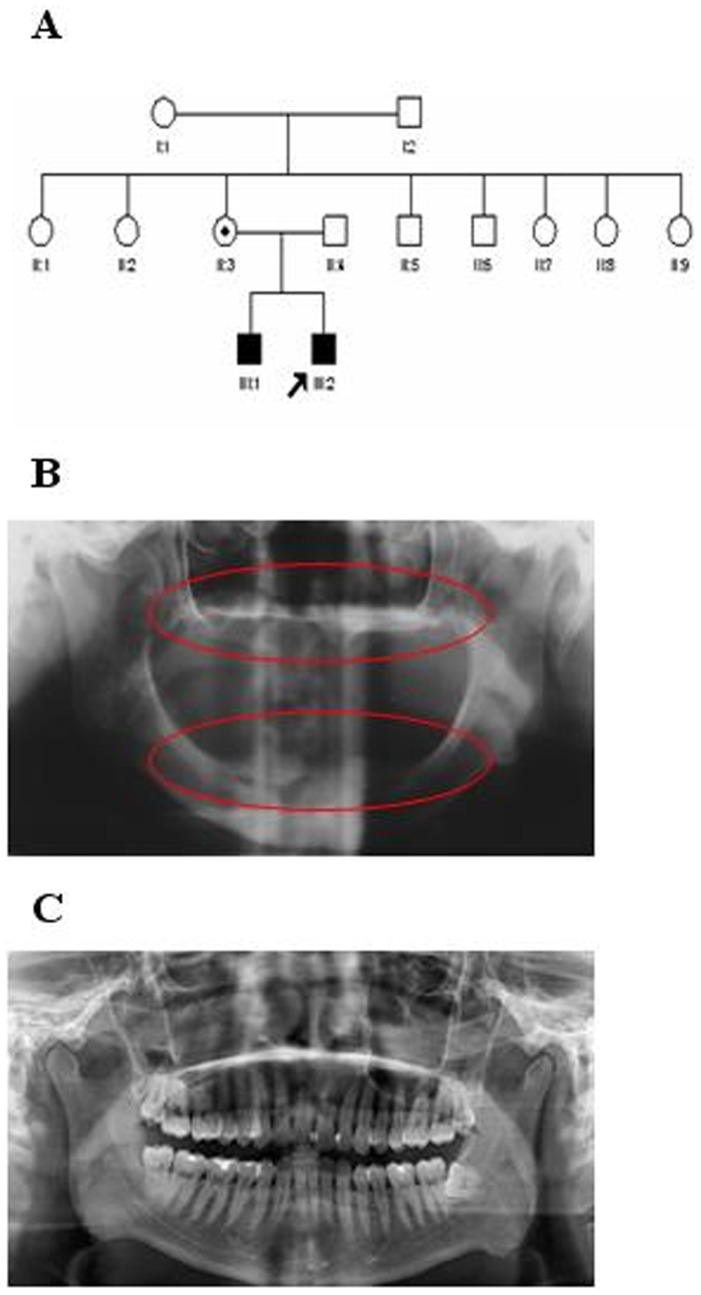
The clinical features of this Chinese family are listed in Table 1. The 2 patients experienced the classic XLHED symptoms, hypodontia, sparse hair, thin and dry skin and specific facial features (Fig. 1). Both of them were born by a normal delivery, and had normal psychomotor and intellectual development.
Table 1. Clinical phenotypes of family members in the Chinese pedigree.
| Family ID | Person ID | Gender | Affection status | Age (years) | Skin | Sparse hair | Hypohidrosis | Facial features | Eczema | Tooth existing (Hypodontia) | Nail dysplasia | Sweat glands dysplasia | Other Manifestations | |
| Hypoplastic | Thin & wrinkled | |||||||||||||
| I | II3 | F | C | 50 | – | – | + | – | – | – | NM | – | – | |
| I | III1 | M | A | 25 | ++ | ++ | ++ | + | + | + | 13, 23 | + | ++ | |
| I | III2 | M | A | 23 | ++ | ++ | ++ | ++ | + | + | anodontia | + | ++ | |
A: affected; C: carrier; F: female; M: male; NM: no missing; +: positive; number of ‘+’ symbols reflects the degree of these clinical features; -: negative; 13: Right maxillary canine; 23: Left maxillary canine.
Figure 1. Pedigree and tooth development features of the Chinese family.
(A) Males are indicated by squares, females by circles. Affected individuals are indicated by filled symbols and unaffected individuals by white symbols. Circle containing a dot refers to carrier. An arrow indicates the proband. (B) The panoramic radiographs of the proband confirmed there was no tooth germ in the alveolar bone (red circle) which was the severest symptom of tooth dysplasia. (C) The panoramic radiographs of a healthy control with normal tooth development.
The carrier (II3) had no clinical abnormity except for sparse hair. The other 22 carriers we reported before had a similar trait. The most common features were sparse hair and aberrant tooth shape. All of them were Han people which account for 90.56% of the population in China and lived in the middle of China, ranging from teens to 60s in age. They harbored 8 missense mutations, 1 frameshift mutation, 1 splicing site mutation and 2 exon deletions.
Identification of the Causative EDA Mutation
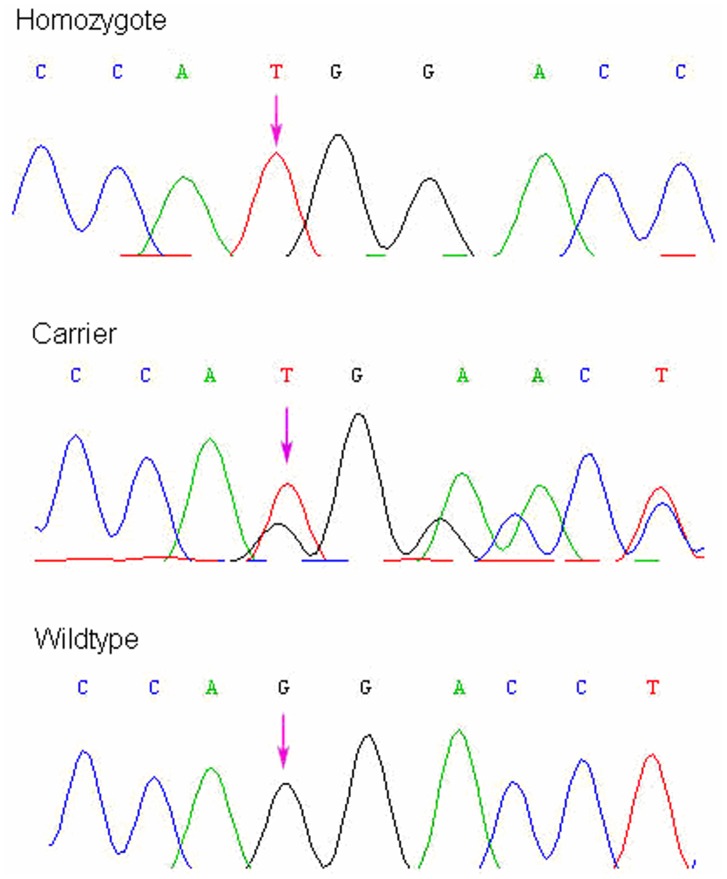
Direct sequencing was used to analyze all the coding exons and intron-exon boundaries from both directions. The proband had a nucleotide insertion in exon 5, c.573–574 insT (Fig. 2). His mother (II3) and elder brother (III1) also had the same mutation. The insertion induced a frameshift from amino acid 192 and caused the transcription to stop at amino acid 239. 200 healthy controls did not have this change.
Figure 2. Identification of the causative mutation in EDA gene.
Arrows indicate the mutation site. The affected male patient and his mother harbored a frameshift mutation c.573–574insT.
Features of EDA Promoter Methylation
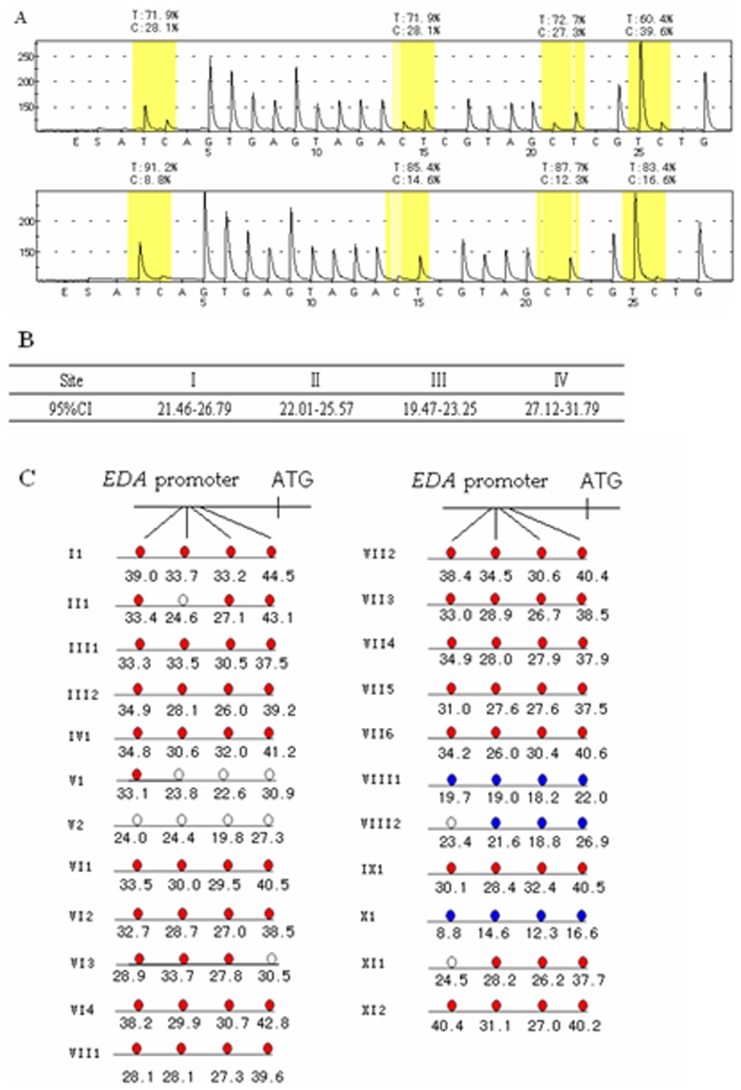
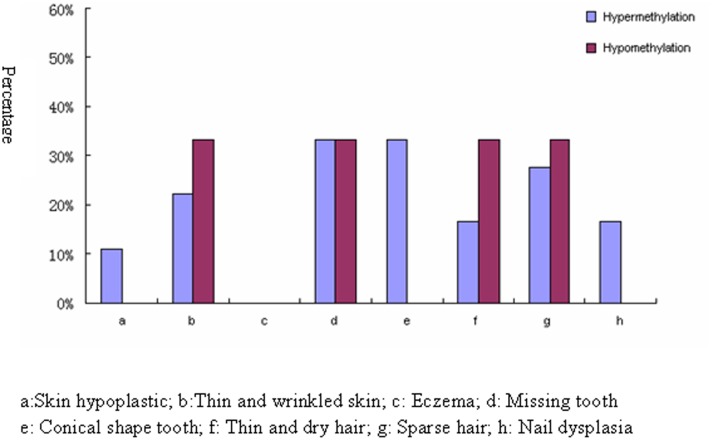
The 95% CI for methylation of each site is shown in Fig. 3B. Compared with it, 78.26% (n = 18) carriers were in hypermethylated state while 14.29% (n = 3) were hypomethylated. The other 2 samples were in normal state. The methylation state had no correlation with carriers’ mutation type and site (Table 2). Similarly, methylation state and phenotype also had no correlation. However, hypermethylated group was inclined to have more defects in nails and tooth shape when compared to hypomethylated carriers (Fig. 4).
Figure 3. EDA promoter’s methylation analysis of 23 carriers.
(A) Pyrosequencing graphs of 2 samples, a hypermethylation carrier and a hypomethylation carrier. Peak heights are proportional to the number of identical residues incorporated. Percentage in pictures means allele frequency of each site. (B) The 95% CI for the 4 sites. The figures which refer to methyl-cytosine percent at that site are calculated as described in the text. (C) The methylation state of each carriers in the 4 sites. Red, white and blue refer to hypermethylation, normal and hypomethylation respectively.
Table 2. The mutations and some selected clinical findings of twenty-three carriers.
| Familynumber | Exon/intron | Mutation type | Nucleotide changes | Amino acid change | Domaina | Sparse hair | Number of tooth missing | Conical shape tooth | Nail dysplasia | Sweat glands dysplasia | Methylation stateb (Cm value) |
| II1[12] | 1 | Missense | c.200A>T | E67V | E | – | 1 | 0 | – | – | Hyper |
| III1[12] | 3 | Missense | c.463C>T | R155C | F | + | 5 | 0 | + | NEc | Hyper |
| III2[12] | 3 | Missense | c.463C>T | R155C | F | – | NEc | NEc | – | – | Hyper |
| IV1[13] | 3 | Missense | c.467G>A | R156H | F | – | 0 | 0 | – | – | Hyper |
| V1[12] | 3 | Missense | c.491A>C | E164A | E | + | 0 | 0 | - | – | Normal |
| V2[12] | 3 | Missense | c.491A>C | E164A | E | + | 0 | 0 | + | – | Normal |
| VI1* | 3 | Deletion | – | – | E | + | 7 | 0 | + | NEc | Hyper |
| VI2* | 3 | Deletion | – | – | E | + | 7 | 1 | – | – | Hyper |
| VI3* | 3 | Deletion | – | – | E | – | 0 | 0 | – | – | Hyper |
| VI4* | 3 | Deletion | – | – | E | – | 0 | 2 | – | – | Hyper |
| VII1* | 3 | Deletion | – | – | E | – | 0 | 0 | – | – | Hyper |
| VII2* | 3 | Deletion | – | – | E | – | 0 | 0 | – | – | Hyper |
| VII3* | 3 | Deletion | – | – | E | – | 0 | 4 | – | – | Hyper |
| VII4* | 3 | Deletion | – | – | E | – | 0 | 4 | – | – | Hyper |
| VII5* | 3 | Deletion | – | – | E | – | 0 | 2 | – | – | Hyper |
| VII6* | 3 | Deletion | – | – | E | – | 0 | 2 | – | – | Hyper |
| I1 | 5 | Frameshift | 573insT | FS at 192 Term | C | – | 0 | 0 | – | – | Hyper |
| VIII1[12] | 5 | Splice donor site | IVS5+1 g>a | Altered splicing | C | – | 0 | 0 | – | – | Hypo |
| VIII2[12] | 5 | Splice donor site | IVS5+1 g>a | Altered splicing | C | – | 0 | 0 | – | – | Hypo |
| IX1[12] | 7 | Missense | c.758T>C | L253P | T | + | 9 | 0 | + | – | Hyper |
| X1[12] | 9 | Missense | c.926T>G | V309G | T | + | 3 | 0 | – | – | Hypo |
| XI1[13] | 9 | Missense | c.1045G>A | A349T | T | + | 6 | 0 | – | – | Hyper |
| XI2[13] | 9 | Missense | c.1045G>A | A349T | T | – | 0 | 0 | – | – | Hyper |
E: Extracellular domain; F: Furin domain; C: Collagen domain; T: TNF homology domain.
Hyper: hypermethylation; Hypo: hypomethylation.
not examined.
unpublished data.
Figure 4. Relationship between methylated state and phenotype of XLHED carriers.
Hypermethylated carriers are inclined to have more conical shaped tooth and nail dysplasia than hypomethylated group.
Discussion
EDA is a trimeric type II membrane protein that co-localizes with cytoskeletal structures at the lateral and apical surfaces of cells The protein includes intracellular domain, transmembrane domain, furin subdomain, collagen subdomain and TNF homology subdomain As a member of the TNF-related ligand family, EDA is involved in the early epithelial-mesenchymal interaction So far hundreds of mutations have been identified. About 80% of them are small intragenic changes, including point mutations, small deletions and insertions, and more than half of them are found in exons 1, 3 and 5. Large deletions, including entire exon loss and complete gene deletion, have also been reported But the type of mutations, the phenotype and disease severity showed no obvious correlation especially for heterozygous carriers. About 30% of them do not even have obvious symptoms, rendering accurate diagnosis of carrier status difficult [2].
In this study, we reported a known frameshift mutation in the EDA gene. The frameshift mutation, c.573–574insT, caused aberrant transcription from codon192 and a premature stop at 239. So far, at least 14 frameshift mutations have been reported in exon 5, but only 3 were insertions. Mutant EDA missed partial collagen subdomain and the whole TNF homology subdomain. The TNF homology domain consisted of 10 predicted antiparallel b-sheets linked by variable loops, in common with other members of the TNF family, which was necessary for the homotrimerization of ligands and the binding of EDA to its receptor Therefore the mutant EDA was predicted not to bind EDAR at all.
In the post-genomic era, it is becoming increasingly evident that epigenetic controlling of gene expression plays an important role in determining the phenotype. Histone modifications and DNA methylation-demethylation events are central to the epigenetic regulations of development XLHED female carriers are mosaics of functionally normal and abnormal cells. The carriers’ clinical features are likely to depend on the percentage of abnormal cells having participated to the process of ectodermal appendage formation. But it is presently unclear whether EDA promoter methylation contributes at all to the phenotype of carriers.
To obtain more precise hints, we chose the quantitative method, pyrosequencing system [22], [23], to analyze the methylation level of EDA promoter. Besides the carrier of this family, we additionally recruited 22 other carriers that we reported before. Most of them showed mild symptoms of tooth and hair impairments. All of the causative mutations were distributed in the extracellular domain. 18 of the carriers displayed hypermethylation of the EDA promoter. Some of them were even 50% higher in methylation level than normal controls. Although hypermethylated carriers were inclined to have more conical shaped tooth and nail dysplasia than that of hypomethylated group, no regular pattern seemed to exist among methylation state, mutation type, mutation site and clinical features. Some hypermethylated carriers appeared clinically normal, but some even had as many as 7 missing teeth, conical tooth and sparse hair. The 3 hypomethylated carriers came from 2 families. The 2 carriers who had a mutation in splice donor site were totally normal. The other carrier had sparse hair and missing tooth. Her missense mutation was located at the end of the transcript. The phenotypic changes may be due to the modulation of selection at another X chromosome locus or polymorphism at a locus controlling inactivation. The 2 methylation normal individuals came from the same family. They exhibited mild sparse hair. The mutation occurred between the furin cleavage site and the collagen-like domain.
In addition, the measure of promotor’s methylation by pyrosequencing is for both WT and mutated allele on both active and inactive X chromosomes, further studies are needed to explain whether this effect is specific to the EDA promoter or is generalized to the entire X chromosome.
In conclusion, a recurrent EDA missense mutation, c.573–574 insT in a Chinese patient was reported. We also performed the first study to elucidate the variation of DNA methylation patterns in EDA promoters on Chinese XLHED carriers. The results suggest most of these Chinese XLHED carriers’ have hypermethylated EDA promoter.
Acknowledgments
We appreciate the patients and their families for participating in this study.
Funding Statement
This work was supported by grants from the National Natural Scientific Foundation of China (30500562, 30772417, 30930099 (Monumental Projects) and 81120108010 (International cooperation and exchange projects)), the Chenguang Plan for Distinguished Youth of Wuhan, China (200850731374), the Open Research Fund Program of Hubei-MOST KLOS & KLOBME (201103), the Foundation for the Author of National Excellent Doctoral Dissertation (2007B69) and the Program for New Century Excellent Talents in University, Ministry of Education, China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chassaing N, Bourthoumieu S, Cossee M, Calvas P, Vincent MC (2006) Mutations in EDAR account for one-quarter of non-ED1-related hypohidrotic ectodermal dysplasia. Hum Mutat 27: 255–259. [DOI] [PubMed] [Google Scholar]
- 2. Vincent MC, Biancalana V, Ginisty D, Mandel JL, Calvas P (2001) Mutational spectrum of the ED1 gene in X-linked hypohidrotic ectodermal dysplasia. Eur J Hum Genet 9: 355–63. [DOI] [PubMed] [Google Scholar]
- 3. Rivenbark AG, Jones WD, Coleman WB (2006) DNA methylation-dependent silencing of CST6 in human breast cancer cell lines. Lab Invest 86: 1233–42. [DOI] [PubMed] [Google Scholar]
- 4. Douglas DB, Akiyama Y, Carraway H, Belinsky SA, Esteller M, et al. (2004) Hypermethylation of a small CpGuanine-rich region correlates with loss of activator protein-2alpha expression during progression of breast cancer. Cancer Res 64: 1611–1620. [DOI] [PubMed] [Google Scholar]
- 5. Prendergast GC, Ziff EB (1991) Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science 251: 186–189. [DOI] [PubMed] [Google Scholar]
- 6. Deng G, Song GA, Pong E, Sleisenger M, Kim YS (2004) Promoter methylation inhibits APC gene expression by causing changes in chromatin conformation and interfering with the binding of transcription factor CCAAT-binding factor. Cancer Res 64: 2692–2698. [DOI] [PubMed] [Google Scholar]
- 7. Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP (1989) Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell 58: 499–507. [DOI] [PubMed] [Google Scholar]
- 8. Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, et al. (1992) Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69: 905–914. [DOI] [PubMed] [Google Scholar]
- 9. Hendrich B, Bird A (1998) Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol 18: 6538–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nan X, Tate P, Li E, Bird A (1996) DNA methylation specifies chromosomal localization of MeCP2. Mol Cell Biol 16: 414–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lux W, Klobeck HG, Daniel PB, Costa M, Medcalf RL, et al. (2005) In vivo and in vitro analysis of the human tissue-type plasminogen activator gene promoter in neuroblastomal cell lines: evidence for a functional upstream kappaB element. J Thromb Haemost 3: 1009–1017. [DOI] [PubMed] [Google Scholar]
- 12. Fan H, Ye X, Shi L, Yin W, Hua B, et al. (2008) Mutations in the EDA gene are responsible for X-linked hypohidrotic ectodermal dysplasia and hypodontia in Chinese kindreds. Eur J Oral Sci 116: 412–7. [DOI] [PubMed] [Google Scholar]
- 13. Fan HL, Ye XQ, Shi B, Zhang YL, Bian Z (2007) Mutations in the ED1 gene in families with X-linked hypohidrotic ectodermal dysplasia. Zhonghua Kou Qiang Yi Xue Za Zhi 42: 272–5. [PubMed] [Google Scholar]
- 14. den Dunnen JT, Antonarakis SE (2001) Nomenclature for the description of human sequence variations. Hum Genet 109: 121–4. [DOI] [PubMed] [Google Scholar]
- 15. Povey S, Lovering R, Bruford E, Wright M, Lush M, et al. (2001) The HUGO gene nomenclature committee (HGNC). Hum Genet 109: 678–80. [DOI] [PubMed] [Google Scholar]
- 16. Yin W, Ye X, Shi L, Wang QK, J Huixi, et al. (2010) TP63 Gene Mutations in Chinese P63 Syndrome Patients. J DENT RES 89: 813–817. [DOI] [PubMed] [Google Scholar]
- 17. Monreal AW, Zonana J, Ferguson B (1998) Identification of a new splice form of the EDA1 gene permits detection of nearly all X-linked hypohidrotic ectodermal dysplasia mutations. Am J Hum Genet 63: 380–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ezer S, Bayés M, Elomaa O, Schlessinger D, Kere J (1999) Ectodysplasin is a collagenous trimeric type II membrane protein with a tumor necrosis factor-like domain and co-localizes with cytoskeletal structures at lateral and apical surfaces of cells. Hum Mol Genet 8: 2079–86. [DOI] [PubMed] [Google Scholar]
- 19. Schneider P, Street SL, Gaide O, Hertig S, Tardivel A, et al. (2001) Mutations leading to X-linked hypohidrotic ectodermal dysplasia affect three major functional domains in the tumor necrosis factor family member ectodysplasin-A. J Biol Chem 276: 18819–27. [DOI] [PubMed] [Google Scholar]
- 20. Pääkkönen K, Cambiaghi S, Novelli G, Ouzts LV, Penttinen M, et al. (2001) The mutation spectrum of the EDA gene in X-linked anhidrotic ectodermal dysplasia. Hum Mutat 17: 349. [DOI] [PubMed] [Google Scholar]
- 21. Laurikkala J, Kassai Y, Pakkasjärvi L, Thesleff I, Itoh N (2003) Identification of a secreted BMP antagonist, ectodin, integrating BMP, FGF, and SHH signals from the tooth enamel knot. Dev Biol 264: 91–105. [DOI] [PubMed] [Google Scholar]
- 22. Tao R, Jin B, Guo SZ, Qing W, Feng GY, et al. (2006) A novel missense mutation of the EDA gene in a Mongolian family with congenital hypodontia. J Hum Genet 51: 498–502. [DOI] [PubMed] [Google Scholar]
- 23. Castaldo I, Pinelli M, Monticelli A, Acquaviva F, Giacchetti M, et al. (2008) DNA methylation in intron 1 of the frataxin gene is related to GAA repeat length and age of onset in Friedreich ataxia patients. J Med Genet 45: 808–12. [DOI] [PubMed] [Google Scholar]