Abstract
Glycerol is an important osmotically compatible solute in Dunaliella. Glycerol-3-phosphate dehydrogenase (G3PDH) is a key enzyme in the pathway of glycerol synthesis, which converts dihydroxyacetone phosphate (DHAP) to glycerol-3-phosphate. Generally, the glycerol-DHAP cycle pathway, which is driven by G3PDH, is considered as the rate-limiting enzyme to regulate the glycerol level under osmotic shocks. Considering the peculiarity in osmoregulation, the cDNA of a NAD+-dependent G3PDH was isolated from D. salina using RACE and RT-PCR approaches in this study. Results indicated that the length of the cDNA sequence of G3PDH was 2,100 bp encoding a 699 amino acid deduced polypeptide whose computational molecular weight was 76.6 kDa. Conserved domain analysis revealed that the G3PDH protein has two independent functional domains, SerB and G3PDH domains. It was predicted that the G3PDH was a nonsecretory protein and may be located in the chloroplast of D. salina. Phylogenetic analysis demonstrated that the D. salina G3PDH had a closer relationship with the G3PDHs from the Dunaliella genus than with those from other species. In addition, the cDNA was subsequently subcloned in the pET-32a(+) vector and was transformed into E. coli strain BL21 (DE3), a expression protein with 100 kDa was identified, which was consistent with the theoretical value.
Introduction
Dunaliella salina, one member of the genus Dunaliella (Chlorophyceae, Volvocales), is an extremely halotolerant, unicellular, green, and motile algae. The genus Dunaliella is unique in its remarkable ability to survive in the media with a wide range of NaCl concentrations, from about 0.05 M to saturation (around 5.5 M), while maintaining a relatively low intracellular sodium concentration [1], [2]. This remarkable osmotic adaptability is mediated primarily by the massive de novo synthesis of the compatible solute, the glycerol, following salt stress [3]. These characteristics make D. salina an obvious applicable value as a model organism in studying the mechanism of osmoregulation under salt environment conditions. In addition, under high salt stress, D. salina could accumulate large amounts of β-carotene in cells, which makes it one of the best sources of natural β-carotene [4]–[7].
Glycerol is an important osmotically compatible solute in Dunaliella under salt stress [8], [9]. The extracellular osmotic pressure is released in Dunaliella by changing intracellular glycerol content. The glycerol is synthesized rapidly when the concentration of saline increases, and the glycerol transforms to starch when the concentration of saline drops [10]–[12]. At high salinity, D. salina accumulates massive amounts of glycerol and the level of intracellular glycerol is proportional and osmotically equivalent to the external NaCl concentration, reaching about 8 M or 55% (w/v) of the cell weight at saturated NaCl concentrations [13], . Moreover, the green alga Dunaliella tertiolecta could also adapt the different concentration of saline by synthesizing or eliminating the intracellular glycerol to balance the osmotic potential of intracellular and extracellular [15], . Nicotinamide adenine dinucleotide (NAD+)-dependent glycerol-3-phosphate dehydrogenase (G3PDH) plays a major role in the osmoregulation process in Dunaliella [12], [17]. In glycerol biosynthesis pathway, G3PDH catalyze dihydroxyacetone phosphate (DHAP) to form glycerol-3-phosphate, which is converted to glycerol finally by glycerol-3-phosphate phosphatase [18], [19].
It was found that there are five isozymes of G3PDH in D. salina, and these isozymes respectively take effects in different salinities and play important roles in glycerol metabolism [12]. Chen et al found that four loci produced different G3PDH isozymes functioning under different salinity conditions [12]. In this study, we isolated the cDNA of a NAD+-dependent G3PDH from D. salina, which is one isozyme with highly homology of previously isolated G3PDH in this alga [20]. Subsequently, a series of bioinformatics tools were employed for the analysis of its physical-chemical characteristic, conserved structural domain, transmembrane and signal sequence condition, secondary and spatial structure, phylogenesis, and so on. Finally, this G3PDH was subcloned in the pET-32a(+) vector and undergone prokaryotic expression to further elucidate the pathway of glycerol metabolisms in Dunaliella.
Materials and Methods
Cultivation of D. salina under Salt Stresses
Cells of D. salina strain 435 (UTEX 200) conserved in our laboratory were cultivated in the culture medium according to Chen et al [21]. Cells grown at the late log phase were harvested by centrifugation at 5,000 g for 15 min at 4°C for next experimental procedure.
Isolation of cDNA for G3PDH in D. salina
The total RNA was prepared from 10 mL of D. salina cells grown at the late log phase with using E.Z.N.A. Total RNA Kit II (OMEGA) according to the manufacture’s instruction. Subsequently, the total RNA was treated by DNase I (RNase Free) (TaKaRa) and was dissolved in 0.1% (v/v) diethyl pyrocarbonate solution (TaKaRa) [7], [22].
The first strand of cDNA was synthesized from the total RNA using PrimeScript ™ RT-PCR kit (TAKARA) according to the manufacturer’s instructions [7], [22]. Reverse transcription (RT) reaction was performed with the parameters set as follows: 42°C for 30 min, followed by 70°C for 15 min. Primers Dsgpdh1-F and Dsgpdh1-R were used to amplify the conserved fragment of the D. salina G3PDH cDNA by using Premix Ex Taq (TaKaRa) following the manufacturer’s instructions. The PCR procedure is as the following: 1 cycle of 94°C, 5 min; 30 cycles of 94°C, 30 s, 51°C, 30 s, and 72°C, 1 min; and 1 cycle of 72°C, 10 min; used primers listed in Table 1.
Table 1. Primers used in this study (5′-3′).
| Procedure | Primer | Primer sequence (5→3) |
| ESTisolation | Dsgpdh1-F | CAACGAGAACCATGAGAACC |
| Dsgpdh1-R | CACTGAGGGGGAGATGAACTTGC | |
| 3'RACE | Dsgpdh3'F | CAATGTCGCCAGCAATGTTA |
| 5'RACE | Dsgpdh5'F1 | AACTGAACTTCACCCCCACAGACAT |
| Dsgpdh5’F2 | GGCTCGTGGAGTGGAGGTGT | |
| cDNAisolation | Dsgpdh-F | TTAGTAGTAGTCGTTCACTACACGG |
| Dsgpdh-R | ATGCTTCTCCAGAAAGGAAACATTG | |
| ORFsubclone | ORF-F | CGGGATCCATGCTTCTCCAGAAAGGAAAC |
| ORF-R | CCCTCGAGTTAGTAGTAGTCGTTCACTACACG |
Based on the obtained conserved cDNA fragment sequence, gene specific primer Dsgpdh3’F was designed and 3′ RACE was conducted with oligo dT-Adaptor primers using RNA PCR Kit (AMV) Ver.3.0 (TaKaRa). The first-strand cDNA was amplified by LA Taq (TaKaRa) with the parameters set as follows: 94°C, 4 min; 30 cycles of 94°C, 30 s, 46°C, 30 s, and 72°C, 1 min with a final extension at 72°C for 10 min.
The 5′ RACE operation was accomplished with SMART™ MMLV Reverse Transcriptase (Clontech) and synthesized primers SMARTAO and 5′-RACE CDS. The second 5′RACE was conducted using the Dsgpdh5’F2primer designed according to the fragment obtained from the first 5′RACE reaction with Dsgpdh5’F1 primer. Other handlings including touchdown PCR were employed according to the SMARTer™ RACE cDNA Amplifcation Kit User Manual, with the exception of LA Taq DNA polymerase (TaKaRa) for touchdown PCR, rather than Advantage 2 Polymerase Mix.
The full-length G3PDH cDNA was obtained with specific primes (Dsgpdh-F and Dsgpdh-R, Table 1) corresponding to the 5′ and 3′ ends of the G3PDH gene. The PCR procedure to amplify the G3PDH cDNA fragment is as follows: 1 cycle of 94°C, 5 min; 30 cycles of 94°C, 30 s, 55°C, 30 s, and 72°C, 1 min; and 1 cycle of 72°C, 10 min. All amplified fragments were cloned into pCR2.1 vector (Invitrogen) and undergone Sanger sequencing.
All PCR products were separated by electrophoresis in 1.5% (w/v) agar gels, cloned in the pMD18-T vector (TaKaRa) and sequenced before the further experiments. Plasmid preparations, transformations, and other standard molecular biology techniques were carried out as described previously [23].
Bioinformatics Analysis and Phylogenetic Construction
Sequence analysis was performed using Blast Software (http://blast.ncbi.nlm.nih.gov/). Component analysis of G3PDH was calculated using DNAStar software 7.1.0 (Lasergene). Physical and chemical characteristics of G3PDH were analyzed by ProtParam tool (http://www.expasy.ch/tools/protparam.html). Multiple alignments among similar enzymes were conducted using Clustal X 1.83 (NCBI, Bethesda, MD). Phylogenetic and molecular evolutionary analysis of the amino acid sequences of different G3PDHs were conducted using the Neighbor Joining method and the molecular evolution genetics analysis (MEGA) software, version 4.0.2. The conserved structural domain of G3PDH was predicted via National Center for Biotechnology Information conserved domain database (CDD) in NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), and the protein conserved module of G3PDH was analyzed via the Pfam database (http://pfam.sanger.ac.uk/) to search its domain combinations. The amino acid sequence was subjected to TMHMM server (http://www.cbs.dtu.dk/services/TMHMM/) for transmembrane analysis and SignalP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP/) for the prediction of protein signal sequence. Subcellular localization presumption was performed using WoLF PSORT (http://wolfpsort.org/). Secondary structure was predicted via the NPS@ service (http://npsa-pbil.ibcp.fr/) and PredictProtein (https://www.predictprotein.org/); 3D structure was constructed using 3D-JIGSAW (http://bmm.cancerresearchuk.org/~3djigsaw/).
Plasmid Construction and Protein Expression
Plasmid pET-32a-G3pdh was constructed by insertion of the D. salina G3PDH open reading frame (ORF) into the BamH I and Xho I (all restriction endonucleases are products of TaKaRa, Japan) restriction sites of expression vector pET-32a(+) (Novagen, Darmstadt, Germany). The G3PDH cDNA fragment was used as templates to synthesize the G3PDH ORF by using PrimSTAR HS DNA Taq (TaKaRa, Japan) with forward primer ORF-F and reverse primer ORF-R, which will introduce the BamH I and Xho I site into the 5′ and 3′ end of the ORF, respectively. The PCR procedure is as the following: 1 cycle of 94°C, 30 s; 30 cycles of 94°C, 30 s, 60°C, 30 s, and 72°C, 2 min; and 1 cycle of 72°C, 10 min. PCR product was separated by electrophoresis in 1.5% (w/v) agar gels, cloned in the pMD18-T vector and transformed into E. coli JM109, which was verified by double digestion of BamH I and Xho I. Both plasmids pET-32a and pMD18-T-JM109-G3pdh were prepared from E. coli BL21 and JM109, then digested by BamH I and Xho I at 30°C for 45 min or 2 h. The digested products were separated by electrophoresis in 1% (w/v) and 1.5% (w/v) agar gels, and the ORF and pET-32a fragments were recycled using E.Z.N.A. kit (OMEGA, USA). T4 DNA ligase (0.5 µL containing 200 NEB units; New England Biolabs, USA) was then added, and samples were incubated at 16°C for 12–16 h. 5 µL of the ligation mixture was used to transform electronically competent E. coli (100 µL of BL21(DE3)). The correct strain of E. coli BL21 (DE3)/pET-32a-G3pdh was verified by double digestion of BamH I and Xho I.
E. coli strains BL21 (DE3) were grown in LB medium at 37°C in darkness on a platform shaker at 230 cycles min−1. Ampicillin (100 µg mL−1) was used for selection or maintenance of plasmids. To induce the expression of the D. salina G3PDH, a final concentration of 1.0 mmol L−1 isopropyl-β-D-thiogalactopyranoside (IPTG) was added to the E. coli culture when the optical density (OD) value reached 0.4–0.6, and the culture was allowed to continue growing for 3–4 h before harvesting by centrifugation.
SDS-PAGE Electrophoresis Analysis
The transformed cells with plasmid pET-32a-G3pdh or pET-32a was disrupted by ultrasonication in PBS buffer containing 1 mmol L−1 phenylmethysulfonyl fluoride (PMSF). The supernatants were collected by centrifugation at 12,000 g for 30 min at 4°C. Protein concentration was estimated as described [24]. Then supernatant in each samples were boiled for 5 min after adding 5×SDS loading butter (4:1, v:v); protein molecular weight marker was also boiled for 3 min before loading samples. 10 µg of each samples were subjected to SDS-PAGE in a Bio-Rad protein electrophoresis system (Bio-Rad, Hercules, CA, USA), which was carried out at 80 V, and increased to 120 V after 1 h. The concentration of separating gel and stacking gel were 12% (w/v) and 5% (w/v). Protein electrophoretic profiles in gels were visualized through Coomassie Blue R-250 staining procedure.
Results
Isolation of cDNA for G3PDH from D. salina
A pair of specific primers were designed to obtain G3PDH cDNA conserved fragment from D. salina on the basis of the G3PDH gene sequences of D. salina, Chlamydomonas reinhardii, Arabidopsis thaliana, Pichia stipitis, zygosaccha and four predicted G3PDH gene sequences. When using total RNA from D. salina cells as RT substrate, an expected 583 bp fragment amplified with primers Dsgdph1-F/R was cloned and sequenced (Fig. 1a).
Figure 1. Isolation of D.salina G3PDH gene fragments.
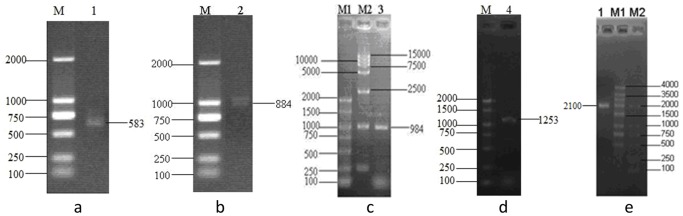
(a)EST fragment of 583 bp. M, DNA marker; lane 1, PCR product. (b)3′ RACE fragment of the 884 bp. M, DNA marker; lane 2, PCR product; (c)First 5′ RACE fragment of 984 bp. M1, M2, DNA marker; lane 3, PCR product. (d)Second 5′ RACE fragment of 1253 bp. M, DNA marker; lane 4, PCR product. (e) The full-length cDNA of D. salina.
On the basis of this cDNA conserved fragment, the 3′ end fragment was amplified by 3′ RACE reaction, which was 884 bp in length (Fig. 1b); and then two 5′ RACE reactions were fulfilled resulting two fragments with 984 bp in the first step (Fig. 1c) and 1253 bp in the second step (Fig. 1d), respectively. Then, based on the sequence assembly, full-length G3PDH cDNA was amplified using specific primers Dsgpdh-F/R, which was 2100 bp (Fig. 2).
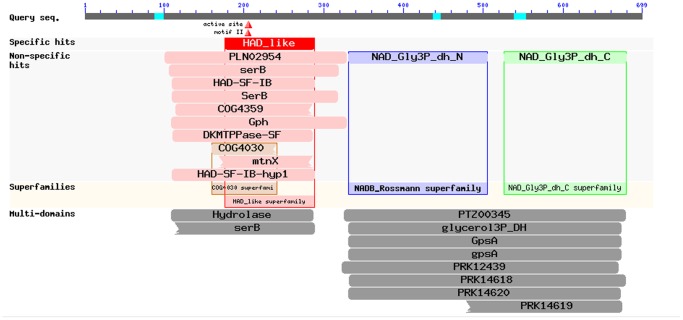
Figure 2. Conserved domains in G3PDH detected by NCBI Conserved Domains Search.
The deduced 699-amino-acid sequence is used to search. Three conserved regions including phosphoserine phosphatase (SerB) domain, N- and C- terminals binding to NAD+ were predicted.
Bioinformatics Analysis of G3PDH cDNA and Amino Acid Sequence
Nucleotide sequence analysis showed that the D. salina G3PDH cDNA contained 2100 bp nucleotides with an ORF of 2100 bp, which contained 19.29% A, 31.48% G, 29.05% C and 20.18% T. The ORF encoded a 699-amino-acid-long peptide including 78 basic amino acids (lysine, arginine), 83 acidic amino acids (aspartic acid, glumatic acid), 266 hydrophobic amino acids (isoleucine, leucine, phenylalanine, tryptophan, valine) and 144 polar amino acids (Asparagine, cysteine, serine, threonine, tyrosine). Analysis by ProtParam tool revealed that the molecular weight of this peptide was 76.6 kDa, the isoelectric point was 6.49.
A complete homologous search by BLAST demonstrated that the nucleotide and putative protein sequence had, respectively, sequence identities of 91% and 95% with the published D. salina G3PDH with NAD+ as coenzyme (AY845323.1), 83% and 78% with D. viridis G3PDH1 (EU624406.1), 76% and 72% with D. viridis G3PDH2 (EU624407.1), which indicated that the protein encoded by the obtained cDNA in this study might belong to the G3PDH family with NAD+ as coenzyme.
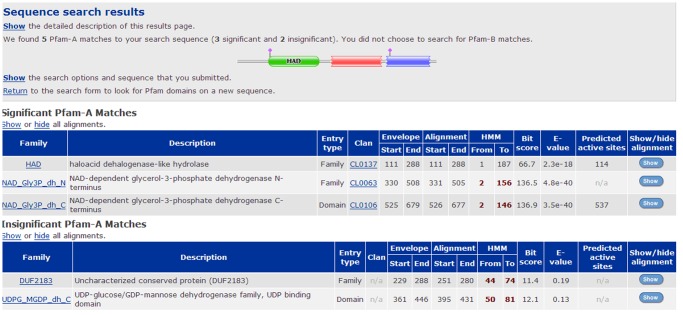
The Conserved Domain Database (CDD) provided by NCBI was employed to predict the structural and functional region (Fig. 2), which manifested that the putative polypeptide from D. salina contained three conserved regions including phosphoserine phosphatase (SerB) domain, N- and C- terminals binding to NAD+ (Fig. 2). Using Pfam database to search and predict the structural and functional domain of this putative polypeptide gained a similar result as shown by Fig. 3. The difference from CDD conservative structure speculated by NCBI was that SerB domain was affiliated to similar hydrolase domain family. The point also elucidated the first 330 amino acids should possess hydrolase function, and the latter amino acids played the main role of glycerol-3-phosphate dehydrogenase. Consequently, the results predicted that this deduced protein was a NAD+-dependent G3PDH (EC 1.1.1.8). Eukaryotic neural network (NN) search by SignalP 3.0 showed that this polypeptide had no signal peptide. It was also predicted to be non-secretory protein by Markov models (HMM) of SignalP 3.0. TMHMM Server v. 2.0 predicted that the G3PDH had no transmembrane domain. Presumption of subcellular localization performed using WoLF PSORT showed that D. salina G3PDH may be situated in the chloroplast.
Figure 3. Conservative module analysis of G3PDH by Pfam.
Similar with CDD search, three conserved domains were found; with the exception that SerB domain was predicted to be affiliated to similar hydrolase domain family.
Prediction of Protein Structure
The secondary structure of the protein was deduced by NPS@service PHD, GOR1, SOPMA and PREDATOR methods. On account of the distinct emphases of various methods, the predicted results were also different. So these methods were made a comparison and PredictProtein was adopted to analyze online (Table 2). As shown by Table 2, the G3PDH protein had abundant α-helixes, some extended strands, many random coils and a few β-turns, but had no 310-helix, π-helix and other rare secondary structure.
Table 2. Prediction of protein secondary structure of D. salina G3PDH.
| α-helix | Extendedstrand | Randomcoil | β-turn | |
| PHD | 45.21% | 8.15% | 46.64% | 0% |
| GOR1 | 52.79% | 32.76% | 7.73% | 6.72% |
| SOPMA | 46.35% | 13.45% | 32.05% | 8.15% |
| PREDATOR | 36.48% | 10.73% | 52.79% | 0% |
| PredictProtein | 45.21% | 12.45% | 42.35% | 0% |
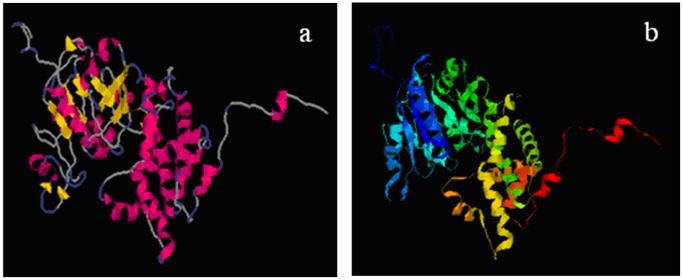
Three-dimensional structure of the D. salina G3PDH was predicted by 3D-JIGSAW comparative modeling program based on homologues of known structures automatically, and was visualized using RasMol software (Fig. 4). A big gap could be observed in the center of the protein by Fig. 4 using both the cartoon and ribbon models, which was regarded as the possible active site where G3PDH reacted with substrate and avoided external interference due to the protection of substrate.
Figure 4. Schematic representation of three-dimensional models of the D.salina G3PDH.
(a) Comparative modeling was performed using 3D-JIGSAW. (b) Comparative modeling was performed using CPH models. The α-helix and β-sheet regions of the putative protein are indicated with ribbons and arrows, respectively. The loop regions are also designated in the schematics.
Multiple Sequence Alignment and Phylogenetic Analysis
The deduced amino acid sequence of G3PDH was multiple sequences aligned with those from different species by Clustal X1.8. These sequences and Genbank accession numbers are as the following: D. salina G3PDH (GI 61816942), D. viridis G3PDH (GI 189187651), D. viridis G3PDH (GI 189187649), Chlamydomonas reinhardtii predicted G3PDH (GI 159463132), Chlorella variabilis hypothetical G3PDH (GI 307107298), Volvox carteri hypothetical G3PDH (GI 302833661), Hordeum vulgare predicted G3PDH (GI 326525148), Sorghum bicolor hypothetical G3PDH (GI 242060059), Zea mays G3PDH (GI 226494897), Oryza sativa Japonica Group G3PDH (GI 115442511), Cuphea lanceolata G3PDH (GI 840731), Vitis vinifera hypothetical G3PDH (GI 147796339), Arabidopsis thaliana putative G3PDH (GI 19424089), Populus trichocarpa predicted G3PDH (GI 224140865), Ricinus communis putative G3PDH (GI 255572030), Selaginella moellendorffii hypothetical G3PDH (GI 302820075), Physcomitrella patens subsp. patens predicted G3PDH (GI 168031121). The results indicated the general conservatism of botanical and algal G3PDH genes (Fig. 5). Consequently, according to the location of the conservative region, the functional region of the G3PDH presumably started at the 331st amino acid of the sequence and the 330 amino acids prior to it possibly involved in other properties of protein, which had explanation in structural and functional predication section above.
Figure 5. Alignment of D.salina G3PDH with G3PDHs sequences from other species.
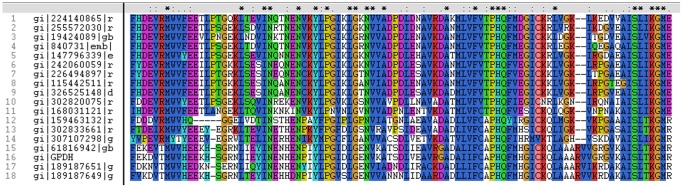
GPDH: G3PDH in the present study. Other G3PDHs are shown as GenBank accession number: D. salina G3PDH (GI 61816942), D. viridis G3PDH (GI 189187651), D. viridis G3PDH (GI 189187649), Chlamydomonas reinhardtii predicted G3PDH (GI 159463132), Chlorella variabilis hypothetical G3PDH (GI 307107298), Volvox carteri hypothetical G3PDH (GI 302833661), Hordeum vulgare predicted G3PDH (GI 326525148), Sorghum bicolor hypothetical G3PDH (GI 242060059), Zea mays G3PDH (GI 226494897), Oryza sativa Japonica Group G3PDH (GI 115442511), Cuphea lanceolata G3PDH (GI 840731), Vitis vinifera hypothetical G3PDH (GI 147796339), Arabidopsis thaliana putative G3PDH (GI 19424089), Populus trichocarpa predicted G3PDH (GI 224140865), Ricinus communis putative G3PDH (GI 255572030), Selaginella moellendorffii hypothetical G3PDH (GI 302820075), Physcomitrella patens subsp. patens predicted G3PDH (GI 168031121).
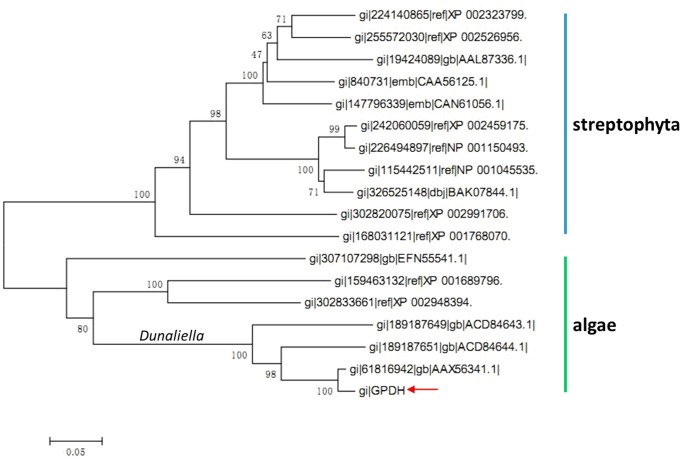
The phylogenetic tree for the complete homologous G3PDHs was constructed using neighbor-joining method by MEGA 4.0.2 software. As Figure 6 shown, the G3PDH obtained in this study (indicated by red arrow in Fig. 6) share highest evolutionary position with other homologues from the Dunaliella genus, they all clustered into the green algae group with Chlamydomonas reinhardtii and Chlorella variabilis.
Figure 6. Phylogenetic tree (neighbor-joining) for G3PDH of 18 species.
D. salina G3PDH (GI 61816942), D. viridis G3PDH (GI 189187651), D. viridis G3PDH (GI 189187649), Chlamydomonas reinhardtii predicted G3PDH (GI 159463132), Chlorella variabilis hypothetical G3PDH (GI 307107298), Volvox carteri hypothetical G3PDH (GI 302833661), Hordeum vulgare predicted G3PDH (GI 326525148), Sorghum bicolor hypothetical G3PDH (GI 242060059), Zea mays G3PDH (GI 226494897), Oryza sativa Japonica Group G3PDH (GI 115442511), Cuphea lanceolata G3PDH (GI 840731), Vitis vinifera hypothetical G3PDH (GI 147796339), Arabidopsis thaliana putative G3PDH (GI 19424089), Populus trichocarpa predicted G3PDH (GI 224140865), Ricinus communis putative G3PDH (GI 255572030), Selaginella moellendorffii hypothetical G3PDH (GI 302820075), Physcomitrella patens subsp. patens predicted G3PDH (GI 168031121).
Prokaryotic Expression, Protein Purification and Enzymatic Assay
The G3PDH ORF sequence of 2100 bp was amplified by PCR with D. salina cDNA as templates. The cDNA was subsequently subcloned in the pET-32a(+) expression vector in the BamH I/Xho I sites. The constructed prokaryotic expression vector pET-32a-G3pdh was transformed into E. coli strain BL21 (DE3) and IPTG was used for induction. The G3PDH in transgenic strains were analyzed by SDS-PAGE. As shown by Fig. 7, a clear protein expression band could be observed at the position of 100 kDa (the addition of target gene 76.6 kDa and histidine marker 23 kDa), which was consistent with the theoretical value. Whereas, an obvious protein expression band appeared at the location of 20 kDa in positive control and no band was found in the samples due to the inhibition of the protein expression by the introduced target gene (Fig. 7).
Figure 7. 10% (w/v) SDS-PAGE of expression of interest protein in E.coli.
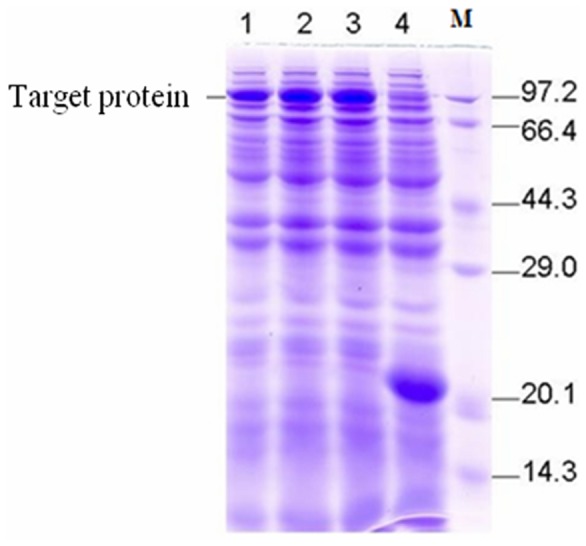
M, protein marker; lane 1, 2 and 3, protein sample with loading volume of 10, 15 and 20 µl, respectively; lane 4, negative control (empty vector control, pET-32a).
Discussion
As is known, the osmotic adjustment response of Dunaliella (especially D. salina) functioned by varying the intracellular concentration of a compatible solute glycerol to balance the osmotic pressure inside and outside of cells. When subjected to hyperosmotic shock D. salina cells rapidly shrink followed by synthesis of glycerol to increases the internal osmolarity for resuming original volume of cells. Under hypoosmotic shock, it was found rapid swells followed by a decrease in internal glycerol and volume resumption [21]. The biosynthesis of glycerol in D. salina involved the key enzymes G3PDH, which convert DHAP to glycerol-3-phosphate. Some studies have been performed to investigate the G3PDH for elucidating the mechanism of osmotic stress tolerance by glycerol. In a study, a novel G3PDH (NAD+) (EC1.1.1.8) gene (PfGPD) was cloned from halotolerant yeast Pichia farinosa, and the PfGPD gene was induced by salt stress [25]. In another study, He et al cloned the cDNA encoding a NAD+-dependent G3PDH from D. salina, and the cDNA may encode an osmoregulated isoform primarily involved in glycerol synthesis [20]. In addition, He et al have cloned two novel chloroplastic G3PDH cDNAs (DvGPDH1 and DvGPDH2) from Dunaliella viridis, which encode two polypeptides of 695 and 701 amino acids, respectively [26]. Q-PCR analysis revealed that both genes exhibited transient transcriptional induction of gene expression upon hypersalinity shock, followed by a negative feedback of gene expression.
In the present study, the cloned 2100 bp G3PDH cDNA from D. salina acts with NAD+ as coenzyme, and the comparative study of conservative regions discovered NAD+ and 3-phosphoglycerate binding sites in the G3PDH protein, theoretically testifying the cloned cDNA encodes G3PDH of osmosis-adjusting type. The G3PDH protein deduced from G3PDH cDNA contains 699 amino acids, of which the molecular weight is 76.6 kDa and the isoelectric point is 6.49, with −3.33 charge in pH 7.0. This G3PDH gene and protein had 91% and 95% identity with the putative G3PDH gene sequence (AY845323.1) and protein (AAX56341.1) [20]. The putative G3PDH protein contained 701 amino acids, with the molecular weight of 76.9 kDa, isoelectric point of 6.1 and −6.08 charge in pH 7.0 environment. So, it was showed some certain structural differences between them, and also reflected the diversity and complicity of G3PDH isoenzyme with NAD+ as coenzyme. It was reported that three isoforms of G3PDH have been separated from D. tertiolecta [27], [28]. The chloroplasts contained the two major isoforms, and the third, minor form was in the cytosol. The first chloroplast form was the major form when the cells were grown on high NaCl, and it has been a form for glycerol production for osmoregulation. The second form increased in specific activity when inorganic phosphate was increased and played roles in stimulating cell growth and glyceride synthesis. The presumption of subcellular localization by WoLF PSORT showed that G3PDH in the present study may situate in the chloroplast as the osmoregulation form in glycerol production. Similarly, it was thought that the G3PDH in the study by He et al was also an osmoregulation form in chloroplast [20]. However, another study cloned and sequenced a cDNA encoding the D. salina FAD-G3PDH, which situated in the mitochondrial. The expression of FAD-G3PDH was enhanced by salt treatment. Its catalytic site facing toward the cytosol, combined action of this enzyme with the cytosolic NAD+-dependent G3PDH forms the glycerol-3-phosphate shuttle [29]. In this shuttle, cytosolic NAD+-dependent G3PDH oxidizes cytosolic NADH to NAD+, and catalyzes the reduction of DHAP to glycerol-3-phosphate. Subsequently glycerol-3-phosphate passes the outer mitochondrial membrane and is oxidized to DHAP by FAD-G3PDH, simultaneously delivers its electrons to the respiratory chain [30]. Due to the essential role in glycolytic pathway, G3PDH is one of the typically constitutive housekeeping genes in living organisms [31], [32]. Consistently, results of sequence alignment showed that G3PDH genes were conservative between plants and algae. Phylogenetic analysis indicated that G3PDHs of green algae clustered into one group. Difference of G3PDHs functions between plant and green algae will be interesting in coming study considering the unicellular green algae maybe more sensitive to the salinity of environmental conditions.
According to the analysis of conservative region of the G3PDH in the present study, it could be speculated that the G3PDH functional domain originate at the 331 amino acid of the amino sequence and the first 330 amino acids are potentially correlated with other properties of the protein. Namely, G3PDH protein has two independent functional domains, SerB and G3PDH domains. SerB (EC 3.1.3.3) and glycerol-3-phosphate phosphatase (EC 3.1.3.21) are attributed to the same type of hydrolase, and two enzymes have similar functions due to their active centers of similar size. Therefore, it was speculated the G3PDH might also have glycerol-3-phosphate phosphatase activity and can catalyze DHAP to glycerol directly without glycerol-3-phosphate phosphatase. Similarly, in the studies by He et al and He et al, protein domain analysis revealed that DsGPDH2 in D. salina and DvGPDH1 and DvGPDH2 in D. viridis all encoded unique bi-domain proteins with C-terminal G3PDH domains and additional N-terminal SerB domains [20], [26]. It has been reported that such bi-domain G3PDHs only exist in green alga, but not in higher plants or other species, such as yeasts and animals [26]. For example, only one catalytic domain has been found exist in polypeptide chain of G3PDHs in yeasts Debaryomyces hansenii, Candida glycerinogenes and Candidamagnoliae [33]–[35]. The existence of unique bi-domain G3PDHs in these green algae might be the evolutionary consequence, which maintained a unique osmoregulation mechanism in green algae for survival in severe environments [26].
Some key enzyme genes related to glycerol metabolism, such as the cDNA of fructose-1, 6-diphosphate aldolase (DsALDP) and NAD+-G3PDH are cloned from D. salina. These genes have been transferred into bacteria or plants to increase the salt-tolerance of these species. Zhang et al transferred the DsALDP gene into E. coli cultured in media with different NaCl concentration to analyze its expression [36]. As a result, the bacteria expressing DsALDP exhibited a higher salt tolerance with increasing NaCl concentration than bacteria with no DsALDP expression. Moreover, Zhang et al transferred the DsALDP gene into tobacco by Agrobacterium tumefaciens, and DsALDP gene was expressed effectively in transgenic tobacco, which exhibited a higher salt tolerance [37]. In another report, a G3PDH gene from D. salina has been transferred into led discs cells of tobacco. RT-PCR analysis showed that G3PDH gene integrated into tobacco genome has produced mRNA [38]. In the present study, the prokaryotic expression vector pET-32a-G3pdh was constructed and transferred into E. coli strain BL21 (DE3). The analysis by SDS-PAGE showed that the G3PDH protein was expressed successfully in transgenic strains, and the further work would emphasize on transforming this G3PDH gene into other higher plants to improve their salt tolerance.
In conclusion, in the present research the cDNA of a NAD+-G3PDH was successfully isolated from D. salina. The cDNA was 2100 bp long, which encoded a deduced protein sequence of 699 amino acids with an estimated molecular weight of 76.6 kDa. Protein domain analysis revealed that G3PDH protein has two independent functional domains, SerB and G3PDH domains. The D. salina G3PDH was a nonsecretory protein that may be located in the chloroplast. The D. salina G3PDH had a closer relationship with Dunaliella G3PDHs than with those of other species in the phylogenetic analysis. The secondary and three-dimensional structure of the D. salina G3PDH is predicted. In addition, the prokaryotic expression vector pET-32a-G3pdh was constructed and transferred into E. coli, in which G3PDH protein was expressed successfully. To fully understand glycerol metabolism and osmotic adjustment based on glycerol in Dunaliella, future investigation should focus on the gene clone of some enzymes related to glycerol metabolism and the application of transgenic technology to increase salt-tolerance of other plants by transferring these cloned genes.
Funding Statement
The authors have no support or funding to report.
References
- 1. Fraser PD, Bramley PM (2004) The biosynthesis and nutritional uses of carotenoids. Prog Lipid Res 43: 228–265. [DOI] [PubMed] [Google Scholar]
- 2. Chen H, Jiang JG (2009) Osmotic responses of Dunaliella to the changes of salinity. J Cell Physiol 219: 251–258. [DOI] [PubMed] [Google Scholar]
- 3. Alkayal F, Albion RL, Tillett RL, Hathwaik LT, Lemos MS, et al. (2010) Expressed sequence tag (EST) profiling in hyper saline shocked Dunaliella salina reveals high expression of protein synthetic apparatus components. Plant Sci 179: 437–449. [DOI] [PubMed] [Google Scholar]
- 4. Yan Y, Zhu YH, Jiang JG, Song DL (2005) Cloning and sequence analysis of the phytoene synthase gene from a unicellular chlorophyte, Dunaliella salina . J Agric Food Chem 53: 1466–1469. [DOI] [PubMed] [Google Scholar]
- 5. Ye ZW, Jiang JG, Wu GH (2008) Biosynthesis and regulation of carotenoids in Dunaliella: progresses and prospects. Biotechnol Adv 26: 352–360. [DOI] [PubMed] [Google Scholar]
- 6. Zhu YH, Jiang JG, Yan Y, Chen XW (2005) Isolation and characterization of phytoene desaturase cDNA involved in the beta-carotene biosynthetic pathway in Dunaliella salina . J Agric Food Chem 53: 5593–5597. [DOI] [PubMed] [Google Scholar]
- 7. Zhu YH, Jiang JG, Chen Q (2008) Characterization of cDNA of lycopene beta-cyclase responsible for a high level of beta-carotene accumulation in Dunaliella salina . Biochem Cell Biol 86: 285–292. [DOI] [PubMed] [Google Scholar]
- 8. Ahmed AM, Zidan MA (1987) Glycerol production by Dunaliella bioculata . J Basic Microbiol 27: 419–425. [Google Scholar]
- 9. Ramos AA, Polle J, Tran D, Cushman JC, Jin E-S, et al. (2011) The unicellular green alga Dunaliella salina Teod. as a model for abiotic stress tolerance: genetic advances and future perspectives. Algae 26: 3–20. [Google Scholar]
- 10. Gimmler H, MÖLler E-M (1981) Salinity-dependent regulation of starch and glycerol metabolism in Dunaliella parva . Plant Cell Environ 4: 367–375. [Google Scholar]
- 11. Goyal A, Brown AD, Gimmler H (1987) Regulation of salt-induced starch degradation in Dunaliella tertiolecta . J Plant Physiol 127: 77–96. [Google Scholar]
- 12. Chen H, Jiang JG, Wu GH (2009) Effects of salinity changes on the growth of Dunaliella salina and its isozyme activities of glycerol-3-phosphate dehydrogenase. J Agric Food Chem 57: 6178–6182. [DOI] [PubMed] [Google Scholar]
- 13. Ben-Amotz A, Avron M (1973) The role of glycerol in the osmotic regulation of the halophilic alga Dunaliella parva . Plant Physiol 51: 875–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sadka A, Lers A, Zamir A, Avron M (1989) A critical examination of the role of de novo protein synthesis in the osmotic adaptation of the halotolerant alga Dunaliella . FEBS Letters 244: 93–98. [Google Scholar]
- 15. Goyal A (2007) Osmoregulation in Dunaliella, Part I: Effects of osmotic stress on photosynthesis, dark respiration and glycerol metabolism in Dunaliella tertiolecta and its salt-sensitive mutant (HL 25/8). Plant Physiol Biochem 45: 696–704. [DOI] [PubMed] [Google Scholar]
- 16. Goyal A (2007) Osmoregulation in Dunaliella, Part II: Photosynthesis and starch contribute carbon for glycerol synthesis during a salt stress in Dunaliella tertiolecta . Plant Physiol Biochem 45: 705–710. [DOI] [PubMed] [Google Scholar]
- 17. Chen H, Lu Y, Jiang JG (2012) Comparative analysis on the key enzymes of the glycerol cycle metabolic pathway in Dunaliella salina under osmotic stresses. PLoS One 7: e37578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang ZX, Zhuge J, Fang H, Prior BA (2001) Glycerol production by microbial fermentation: a review. Biotechnol Adv 19: 201–223. [DOI] [PubMed] [Google Scholar]
- 19. Cui L, Chai Y, Li J, Liu H, Zhang L, et al. (2010) Identification of a glucose-6-phosphate isomerase involved in adaptation to salt stress of Dunaliella salina . J Appl Phycol 22: 563–568. [Google Scholar]
- 20. He Q, Qiao D, Bai L, Zhang Q, Yang W, et al. (2007) Cloning and characterization of a plastidic glycerol 3-phosphate dehydrogenase cDNA from Dunaliella salina . J Plant Physiol 164: 214–220. [DOI] [PubMed] [Google Scholar]
- 21. Chen H, Lao YM, Jiang JG (2011) Effects of salinities on the gene expression of a (NAD+)-dependent glycerol-3-phosphate dehydrogenase in Dunaliella salina . Sci Total Environ 409: 1291–1297. [DOI] [PubMed] [Google Scholar]
- 22. Ye ZW, Jiang JG (2010) Analysis of an essential carotenogenic enzyme: zeta-carotene desaturase from unicellular alga Dunaliella salina . J Agric Food Chem 58: 11477–11482. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J., D.W R, editors (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY.
- 24. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 25. Peng F, Li G, Wang X, Jiang Y, Yang Y (2010) Cloning and characterization of a glycerol-3-phosphate dehydrogenase (NAD+) gene from the halotolerant yeast Pichia farinosa . Yeast 27: 115–121. [DOI] [PubMed] [Google Scholar]
- 26. He Y, Meng X, Fan Q, Sun X, Xu Z, et al. (2009) Cloning and characterization of two novel chloroplastic glycerol-3-phosphate dehydrogenases from Dunaliella viridis . Plant Mol Biol 71: 193–205. [DOI] [PubMed] [Google Scholar]
- 27. Gee R, Goyal A, Byerrum RU, Tolbert NE (1993) Two isoforms of dihydroxyacetone phosphate reductase from the chloroplasts of Dunaliella tertiolecta . Plant Physiol 103: 243–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ghoshal D, Mach D, Agarwal M, Goyal A (2002) Osmoregulatory isoform of dihydroxyacetone phosphate reductase from Dunaliella tertiolecta: purification and characterization. Protein Expr Purif 24: 404–411. [DOI] [PubMed] [Google Scholar]
- 29. Yang W, Cao Y, Sun X, Huang F, He Q, et al. (2007) Isolation of a FAD-GPDH gene encoding a mitochondrial FAD-dependent glycerol-3-phosphate dehydrogenase from Dunaliella salina . J Basic Microbiol 47: 266–274. [DOI] [PubMed] [Google Scholar]
- 30. Ronnow B, Kielland-Brandt MC (1993) GUT2, a gene for mitochondrial glycerol 3-phosphate dehydrogenase of Saccharomyces cerevisiae . Yeast 9: 1121–1130. [DOI] [PubMed] [Google Scholar]
- 31. Yamada H, Chen D, Monstein HJ, Hakanson R (1997) Effects of fasting on the expression of gastrin, cholecystokinin, and somatostatin genes and of various housekeeping genes in the pancreas and upper digestive tract of rats. Biochem Biophys Res Commun 231: 835–838. [DOI] [PubMed] [Google Scholar]
- 32. Foss DL, Baarsch MJ, Murtaugh MP (1998) Regulation of hypoxanthine phosphoribosyltransferase, glyceraldehyde-3-phosphate dehydrogenase and beta-actin mRNA expression in porcine immune cells and tissues. Anim Biotechnol 9: 67–78. [DOI] [PubMed] [Google Scholar]
- 33. Chen X, Fang H, Rao Z, Shen W, Zhuge B, et al. (2008) Cloning and characterization of a NAD+-dependent glycerol-3-phosphate dehydrogenase gene from Candida glycerinogenes, an industrial glycerol producer. FEMS Yeast Res 8: 725–734. [DOI] [PubMed] [Google Scholar]
- 34. Lee DH, Kim MD, Ryu YW, Seo JH (2008) Cloning and characterization of CmGPD1, the Candida magnoliae homologue of glycerol-3-phosphate dehydrogenase. FEMS Yeast Res 8: 1324–1333. [DOI] [PubMed] [Google Scholar]
- 35. Thome PE (2004) Isolation of a GPD gene from Debaryomyces hansenii encoding a glycerol 3-phosphate dehydrogenase (NAD+). Yeast 21: 119–126. [DOI] [PubMed] [Google Scholar]
- 36. Zhang XN, Wang H, Qu ZC, Ye MM, Shen DL (2002) Cloning and prokaryotic expression of a salt-induced cDNA encoding a chloroplastic fructose-1,6-diphosphate aldolase in Dunaliella salina (Chlorophyta). DNA Seq 13: 195–202. [DOI] [PubMed] [Google Scholar]
- 37. Zhang X, Lin C, Chen H, Wang H, Qu Z, et al. (2003) Cloning of a NaCl-induced fructose-1, 6-diphosphate aldolase cDNA from Dunaliella salina and its expression in tobacco. Sci China C Life Sci 46: 49–57. [DOI] [PubMed] [Google Scholar]
- 38. Zhang H, Qiao D, Bai L, Zhang Q, Deng X, et al. (2004) Vector construction and expression in tobacco of GPDH Gene from Dunaliella salina . J Sichuan Univ 41: 439–444. [Google Scholar]