Abstract
As a vital antioxidant, L-ascorbic acid (AsA) affects diverse biological processes in higher plants. Lack of AsA in cell impairs plant development. In the present study, we manipulated a gene of GDP-mannose pyrophosphorylase which catalyzes the conversion of D-mannose-1-P to GDP-D-mannose in AsA biosynthetic pathway and found out the phenotype alteration of tomato. In the tomato genome, there are four members of GMP gene family and they constitutively expressed in various tissues in distinct expression patterns. As expected, over-expression of SlGMP3 increased total AsA contents and enhanced the tolerance to oxidative stress in tomato. On the contrary, knock-down of SlGMP3 significantly decreased AsA contents below the threshold level and altered the phenotype of tomato plants with lesions and further senescence. Further analysis indicated the causes for this symptom could result from failing to instantly deplete the reactive oxygen species (ROS) as decline of free radical scavenging activity. More ROS accumulated in the leaves and then triggered expressions of defence-related genes and mimic symptom occurred on the leaves similar to hypersensitive responses against pathogens. Consequently, the photosynthesis of leaves was dramatically fallen. These results suggested the vital roles of AsA as an antioxidant in leaf function and defence response of tomato.
Introduction
In higher plants, reactive oxygen species (ROS) are produced as by-products in most energy-generating processes, such as photosynthesis and respiration. An electron reduction of O2 leads to the formation of superoxide radical (O2 −), which is then disproportionated by superoxide dismutase (SOD) to O2 and hydrogen peroxide (H2O2). ROS production in plant cells is low under optimal growth conditions, but increases dramatically when plants are subjected to abiotic stresses and pathogen attack. Unfavourable environmental conditions, such as cold, heat, drought, and salt, limit the rate of carbon fixation, which results in an increase in photoinhibition and overproduction of superoxide radicals and H2O2 [1]. Furthermore, oxidative burst is one of the most rapid defence reactions to pathogen attack, which changes the production of superoxide and H2O2 at the infection site [2].
Excessive ROS can induce programmed cell death and necrosis [3]. In higher plants, the levels of ROS are strictly regulated by an efficient battery of enzymatic and non-enzymatic antioxidants [2]. Among them, ascorbic acid (AsA) acts as one of the most abundant antioxidants against oxidative stress [4]. In plants, chloroplasts are potentially the major site for the generation of ROS, in which AsA is present at a high level, at concentration of 20 mM or more. Thus, AsA plays a central function in photoprotection, including scavenger of ROS generated by photosynthesis and respiration, cofactor for violaxanthin deepoxidase, and photosystem II electron donor [5].
Although alternative biosynthetic pathways have been proposed [6]–[8], Smirnoff-Wheeler’s pathway [9] (Fig. 1) has been proved to be the major functional pathway by biochemical and genetic approach. GDP-D-mannose pyrophosphorylase (GMP) catalyzes the conversion of D-Mannose-1-P to GDP-D-Mannose, an initial step in the Smirnoff-Wheeler’s pathway [9]. The importance of GMP in the control of AsA biosynthesis has been confirmed in some plants. The significant reduction of AsA in the vtc1 mutant of Arabidopsis was caused by a point mutation in GMP gene [10]. Antisense inhibition of GMP gene in the transgenic potato (Solanum tuberosum) plants reduced AsA contents both in leaves and tubers [11]. In acerola (Malpighia glabra), the MgGMP gene expression displayed a strong correlation with the AsA contents in the ripening fruit [12]. In addition, over-expression of MgGMP gene increased AsA content by approximately two-fold in tobacco (Nicotiana tabacum) [13].
Figure 1. Major AsA biosynthetic pathway in higher plants (Smirnoff-Wheeler.
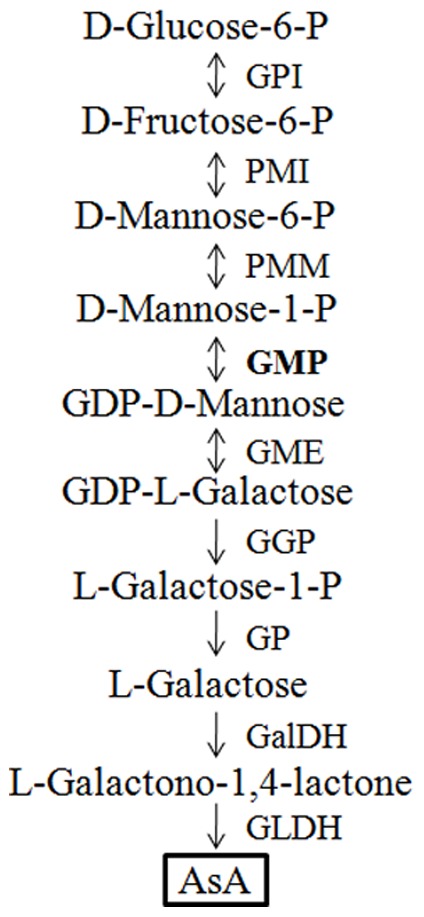
’s pathway). GPI, glucose-6-phosphate isomerase; PMI, phosphomannose isomerase; PMM, phosphomannomutase; GMP (marked in bold), GDP-D-mannose pyrophosphorylae; GME, GDP-D-mannose-3′,5′-epimerase; GGP, GDP-L-galactose-1-phosphate phosphorylase; GP, L-galactose-1-phosphate phosphatase; GalDH, L-galactose dehydrogenase; GLDH, L-galactono-1,4-lactone dehydrogenase.
Tomato is a representative of edible fruit plants with an abundance of AsA. Recently, several progresses about AsA metabolism in tomato have been achieved [14]–[19]. Genes involved in tomato AsA biosynthesis have been largely identified [20]–[24], and some of them have been functionally characterized, such as L-galactono-1,4-lactone dehydrogenase (GLDH) [20] and GDP-D-mannose 3′, 5′-epimerase (GME) [21], [22]. In tomato genome, GMP has four isoforms (GMP1-GMP4). Currently, some progresses have been made in understanding the biological function of tomato GMP genes, especially GMP3. Expression profiling analysis revealed that SlGMP3 mRNA accumulated during the early stages of tomato fruit development and was down-regulated in mature green fruits subjected to ethylene and various stresses [16]. In addition, over-expression of SlGMP3 gene in tobacco significantly increased AsA content in leaf and improved the tolerance to high and low temperature stress by enhancing antioxidation capacity [25]. However, whether SlGMP3 affects the AsA biosynthesis and accumulation in tomato remains unclear.
To better understand the regulation of AsA biosynthesis in tomato, we generated SlGMP3 over-expressing and knocked-down transgenic tomato plants, and analyzed the subsequent effects on AsA metabolism and plant growth. Results indicated that elevated GMP3 expression increased total AsA levels in tomato leaves and fruits, and then enhanced photo-oxidative stress tolerance, whereas RNAi silencing of GMP in tomato could effectively reduce AsA contents, leading to ROS accumulation with leaf lesion formation and senescence, and activation of defence responses. These imply the vital role of AsA as an antioxidant in tomato.
Materials and Methods
Plant Materials
Tomato (Solanum lycopersicum cv. Ailsa Craig, AC) plants were grown in a naturally illuminated glasshouse. Tissues from roots, stems, leaves, flowers, and fruits at various developmental stages of AC plants were collected, immediately frozen in liquid nitrogen, and stored at −80°C until use.
Gene Cloning, Vector Construction and Tomato Transformation
The 1236 bp fragment, including the full open reading frame of SlGMP3 (SGN-U568547), was amplified using the primer sets of GMP3F and GMP3R (Table S1), and then cloned into pMD18-T vector (Takara, Japan). Subsequently, over-expression and RNAi vectors were constructed by subcloning the full length cDNA of SlGMP3 into pMV and pHGRV vectors, respectively, under the control of CaMV35S promoter. The constructs were introduced into Agrobacterium tumefaciens strain EHA105 by electroporation. Tomato seeds of Ailsa Craig were used for transformation, performed according to the methods described by Fillatti et al. [26]. Independent transgenic plants were confirmed by polymerase chain reaction (PCR) using primer combinations between CaMV35S promoter specific primers CaMV35SF or gate35SF and SlGMP3 gene specific primer GMP3R (Table S1) and Southern blot. The transcript levels of SlGMP3 as well as other three members of SlGMP gene family in the transgenic lines were analyzed via semi-quantitative reverse transcriptase (RT)-PCR and real-time RT-PCR.
Semi-quantitative and Real-time RT-PCR Analysis
Total RNA was isolated using TRIzol® reagent (Introvigen, USA), and DNase I was used to clean out DNA before reverse transcription. The reverse transcript reaction was performed with MMLV reverse transcriptase (Toyobo, Osaka, Japan) following the manufacturer’s protocol. Reverse transcript products were used as template for semi-quantitative RT-PCR and real-time RT-PCR. All reaction was assayed in triplicates. Semi-quantitative RT-PCR was performed to preliminarily analyze the expression levels of SlGMP3 in the transgenic lines. It was carried out using the following program: an initial denaturation of 94°C for 3 min, followed by 24–28 circles of 94°C for 30 s, 56°C for 30 s, and 72°C for 1 min, and a final extension at 72°C for 10 min. PCR products were detected by 1% agarose gel in 1×TAE with EtBr. Real-time RT-PCR was performed using LightCycler 480 SYBR Green I Master kit (Roche, Germany) according to the supplier’s instruction to analyze the expression levels of the genes involved in AsA biosynthesis, photosynthesis, antioxidant system, and defence response in the transgenic tomato plants. Real-time RT-PCR amplification step consisted of a pre-incubation at 95°C for 5 min, followed by 40 circles of 95°C for 10 s, 60°C for 15 s and 72°C for 20 s. PCR products were monitored by the LightCycler 480 Real-Time PCR Detection System. Tomato β-actin gene was used as a reference gene to optimize semi-quantitative RT-PCR and as internal standard in real-time RT-PCR. The primers used are listed in Tables S1, S2, S3 and S4.
Oxidative Stress Treatment
One-month-old plantlets of SlGMP3 over-expressing lines and wild type grown in compost plastic pots in a naturally illuminated glasshouse were sprayed with 75 µM methyl viologen (MV) solution or distilled water (control) for 2 d. The MV dissolved in 0.1% Tween-20 solution (100 mL) was applied to six plants. The fourth leaf samples were taken at 7 d post-treatment. The effect of oxidative stress on SlGMP3 over-expressing transgenic plants was assessed by measuring the chlorophyll and malondialdehyde (MDA) contents in the leaves. The experiment was replicated three times.
Measurement of Total Ascorbate
The AsA content was determined using high-performance liquid chromatography as described by Rizzolo et al. [27]. Briefly, samples were ground under liquid nitrogen and homogenised in 5 mL of cold 0.1% (w/v) metaphosphoric acid. The homogenate was centrifuged at 12,000 g for 10 min at 4°C. Then the supernatant was filtered through a Millipore membrane (0.22 µm). An aliquot of 300 µL was incubated with 300 µL 50 mM dithiothreitol for 15 min at room temperature. Then, the extracts were analysed by HPLC using an SB-aq column (Agilent) eluted with acetate buffer (0.2 mol/L, pH 4.5) at a flow rate of 1.0 mL/min to measure total ascorbate. Elutes were detected at 254 nm, and a standard curve from 2 to 40 µg/mL AsA was obtained.
Determination of Chlorophyll, MDA, and Net Photosynthesis Rate
Chlorophyll content was determined by grinding leaf tissues under liquid nitrogen and extracting with 80% (v/v) acetone under low light intensity using the procedure described by Wellburn’s [28]. MDA was assayed for indirect evaluation of lipid peroxidation using trichloroacetic acid, as described previously by Heath and Parker [29]. Net photosynthesis rate was measured using a portable photosynthetic system (CIRAS-2, PP System, USA) according to the supplier’s manual.
Histochemistry
Leaf samples taken from two-month-old tomato plants were stained with trypan blue and 3,-3′-diaminobenzidine (DAB) solution to visualize dead cells and detect the presence of H2O2, respectively. Trypan blue staining was performed as previously described [30]. Leaves were submerged in a 70°C LPTB solution [2.5 mg/mL trypan blue, 25% (w/v) lactic acid, 23% water-saturated phenol, 25% glycerol, and H2O], vacuum infiltrated for 5 min, and then repeated one time. Subsequently, the samples were heated over boiling water for 2 min and cooled for 1 h. The LPTB solution was then replaced with a chloral hydrate solution (25 g in 10 mL H2O) for destaining. After multiple exchanges, the samples were equilibrated in 70% glycerol and photographed. H2O2 was visually detected in the leaves of tomato plants using DAB as the substrate [31]. Briefly, the leaves were cleaned and placed in 1 mg/mL DAB, pH 3.8, under light at 25°C for 8 h. The experiment was terminated by immersing the leaves in boiling 96% ethanol for 10 min. After cooling, the leaves were placed in fresh 96% ethanol for 4 h at room temperature and photographed. The deep brown polymerization product was produced via the reaction of DAB with H2O2.
Statistic Analysis
Data analysis was performed using SAS software, and significant differences were calculated using the Student’s t-test at 95% confidence limit.
Results
Expression Patterns of GMP Genes in Tomato
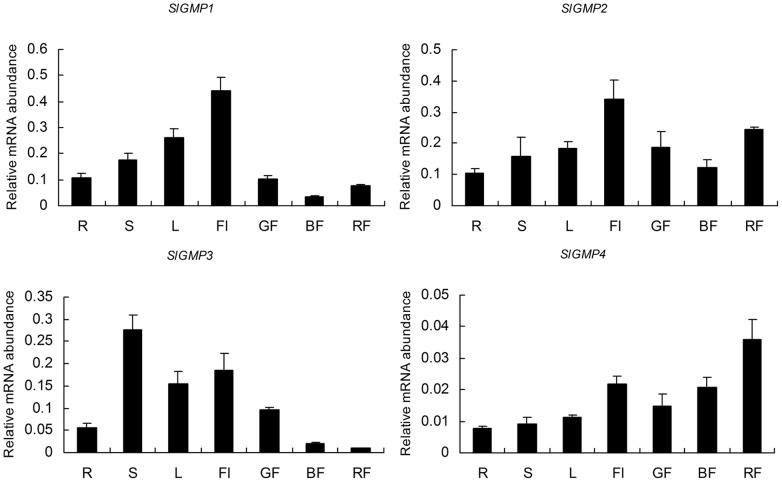
Four unigenes corresponding to the amino acid sequences of GMP (GMP1, SGN-U563807; GMP2, SGN-U568548; GMP3, SGN-U568547; and GMP4, SGN-U584300) exist in tomato genome as reported by Massot et al. [17]. Blast results showed that GMP1 and GMP3 are both located on chromosome 3, and GMP2 and GMP4 are located on chromosome 6 and 9, respectively. The full-length cDNA of SlGMP3 was isolated previously by our group [23]. We investigated the spatial and temporal expression patterns of four GMP members via real-time RT-PCR analysis. All of them were expressed constitutively in the tissues tested though distinct expression patterns (Fig. 2). For SlGMP3, the transcript levels were high in stems, flowers, and young leaves, whereas low in roots, and fruits at breaker and red ripe stages (Fig. 2). The expressions of the other members, SlGMP1 and SlGMP2, followed in a similar pattern, which were high in flowers, low in roots and breaker fruits, and slightly increased at the red ripe stage (Fig. 2). For SlGMP4, the expression was extremely distinct at low in most tissues, especially in vegetative tissues (Fig. 2).
Figure 2. Relative expression analysis of four members of SlGMP gene family in various tissues of tomato variety Ailsa Craig.
R: root, S: stem, L: leaf, Fl: flower, GF: green fruit, BF: breaker fruit, and RF: red fruit. Data obtained by real-time RT-PCR were normalized against Actin.
Identification of Tomato Transgenic Plants
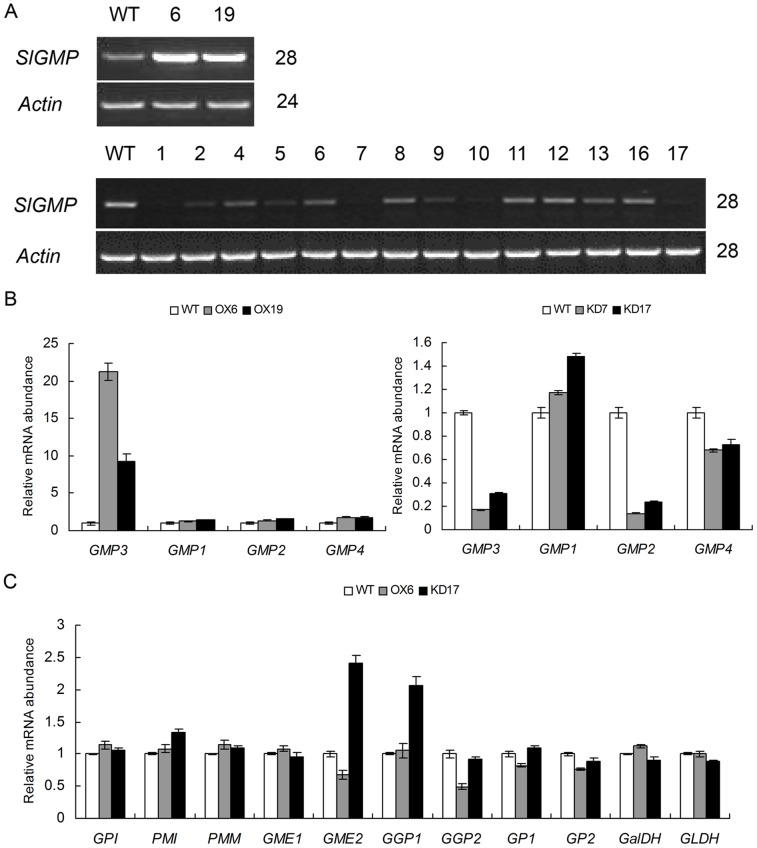
Thirty-eight SlGMP3 over-expressing (OX) and seventeen RNAi (KD) transgenic plants were obtained and confirmed by PCR using genomic DNA as template and 35S forward and gene-specific reverse primers. The expression level of SlGMP3 gene in young leaves of transgenic as well as wild-type plants was examined by semi-quantitative RT-PCR (Fig. 3A) and real-time RT-PCR (Fig. 3B). Two over-expressing lines (OX6 and OX19) and two RNAi lines (KD7 and KD17) with significant changes were selected for further study.
Figure 3. Expression analysis of four SlGMPs and other AsA biosynthesis-related genes in SlGMP3 transgenic plants.
(A) RT-PCR analysis of SlGMP3 expression in the young leaves of two SlGMP3-OX lines (upper panel) and 17 SlGMP3 RNAi lines (bottom panel). The PCR circle numbers are indicated on the right. (B) Relative expression analysis of four members of SlGMP gene family in the young leaves of lines OX6 and OX19 (left) and lines KD7 and KD17 (right) via real-time RT-PCR. (C) Relative expression analysis of AsA biosynthesis-related genes in the young leaves of lines OX6 and KD17 via real-time RT-PCR. Data were obtained by normalizing against Actin and shown as a percentage of wild-type plants.
In order to make clear whether other three members of SlGMP gene family were affected in the SlGMP3 transgenic plants, we investigated their expressions in young leaves (Fig. 3B). In two SlGMP3 over-expressing lines, the expression of SlGMP1, SlGMP2 and SlGMP4 was not significantly affected. However, in SlGMP3 RNAi lines, only SlGMP2 was markedly down-regulated. This is due to the fact that SlGMP2 has 86% homology to SlGMP3 with ten > = 20 base pair length identity (Fig. S1), while SlGMP1 and SlGMP4 share low identity (only 23% and 40%, respectively) with SlGMP3. Due to repression of both SlGMP2 and SlGMP3, SlGMP3 RNAi lines were designated as SlGMP2/3-KD lines in the following.
Alteration of AsA Pool Size in Transgenic Tomato Plants
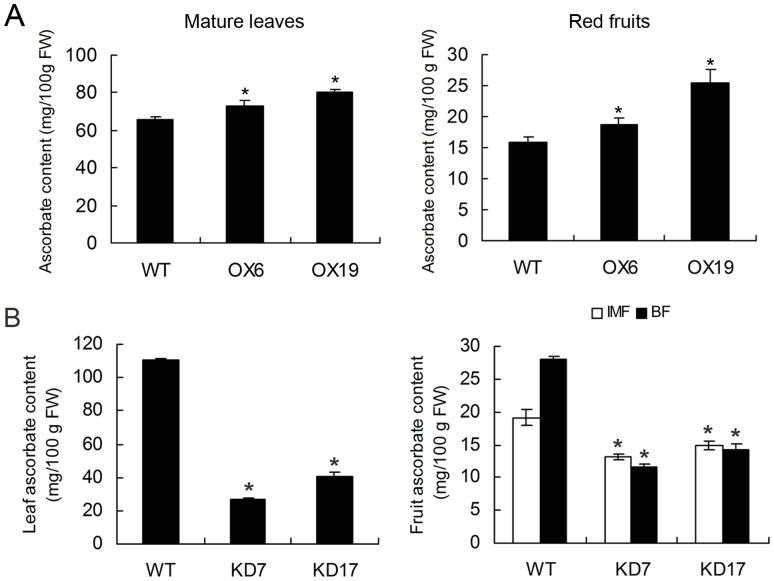
The total AsA contents of both SlGMP3-OX and SlGMP2/3-KD lines were altered compared to the non-transgenic plants. Constitutive over-expression of SlGMP3 increased the total AsA contents in the fully expanded leaves and red ripe fruits (Fig. 4A). However, in SlGMP2/3-KD lines, the total AsA contents in leaves, immature green fruits, and breaker fruits decreased dramatically (Fig. 4B). With the developments of fruit, the AsA levels increased up to 46% in breaker stage compared with immature green stage in wild-type plants, whereas no major difference was observed between two stages of SlGMP2/3-KD lines (Fig. 4B). Moreover, a correlation between AsA content and the suppression extent of SlGMP genes was observed in KD7 and KD17 lines. These results indicate that over-expressing SlGMP3 could enhance AsA accumulation, and knock-down of SlGMP3 with SlGMP2 significantly affects the total AsA pool size in tomato leaves and fruits.
Figure 4. AsA content analysis in SlGMP3 transgenic plants.
(A) Total AsA contents in the mature leaves (left) and red fruits (right) of lines OX6 and OX19. (B) Total AsA contents in the mature leaves (left) and immature green fruits (IMF) and breaker fruits (BF) (right) of lines KD7 and KD17. As the experiments of (A) and (B) were carried at two separate times, there were some differences in AsA contents in leaves of the wild-type tomato plants. Data are presented as mean ± SD of four independent plants per line. Asterisk indicates significant differences from the control (P>0.95).
The expression levels of other genes in the Smirnoff-Wheeler’s pathway were also examined in transgenic and wild-type plants. The results showed the regulation of AsA biosynthesis altered in leaves. The transcript abundances of biosynthetic genes GME and GDP-L-galactose-1-phosphate phosphorylase (GGP) were remarkably affected. Over-expression of SlGMP3 down-regulated SlGME2 and SlGGP2 (reduced nearly 50%) (Fig. 3C). On the contrary, in SlGMP2/3-KD transgenic line KD17, SlGME2 and SlGGP1 were significantly up-regulated (over one fold) compared with the wild-type plants (Fig. 3C). Most other genes of AsA biosynthetic pathway remained unchanged in transgenic plants.
Enhanced Tolerance to Oxidative Stress through Over-expressing SlGMP3 in Tomato
To evaluate whether the over-expression of SlGMP3 could protect transgenic plants against oxidative damage, MV was used to induce oxidative stress and sprayed on one-month-old plantlets of line OX6 and wild type. After the treatment, we found less chlorotic spots on the leaves of line OX6 than wild type. The oxidative damage caused by MV was examined through the extents of leaf chlorophyll loss and MDA (membrane-lipid peroxidation product) increase. Under normal conditions, chlorophyll content was not significantly different between wild-type and transgenic plants. However, after MV treatment, the chlorophyll content in the wild-type plants decreased by 34%, whereas no significant change in line OX6 (Fig. 5A). And the same as the MDA content in wild-type plants increased up to 70%, whereas only 21% increase was found in line OX6 (Fig. 5B). These results indicate that over-expressing SlGMP3 improves the tolerance of the plants to oxidative stress.
Figure 5. Improvement of the photo-oxidative stress tolerance in tomato plants via over-expressing SlGMP3.

Chlorophyll content (A) and MDA content (B) in the fourth leaf of the MV-treated and untreated plants measured at 7 d post-treatment. Data are presented as mean ± SD (N = 6) from triplicate independent measurements. Asterisk indicates significant differences from the control (P>0.95).
Phenotypic Alteration in SlGMP2/3-KD Plants
The SlGMP3-OX plants grew normally during the whole life circle. On the contrary, the SlGMP2/3-KD plants exhibited a lesion-mimic phenotype from the very early stage of seedlings, and the lesions spontaneously developed on leaves. Some lesions initially appeared on the cotyledon surfaces at three weeks post germination; the cotyledons turned yellow (Fig. 6A) and then dropped off the plants. Subsequently, lesions occurred on the lower true leaves (Fig. 6B), and spread from the bottom to the upper leaves (Fig. 6C). The necrotic lesions started from the leaflet tips of the compound leaves (Fig. 6D), and developed in a typical circular pattern and spread until the entire leaves turned chlorotic, and finally fell. The cell deaths of the lesioned leaves were observed via trypan blue staining. The dead cells were consistent with macroscopic lesions before staining in the leaves of lines KD7 and KD17, whereas no staining was observed in wild-type leaves (Fig. 6D). The mature leaves of lines KD7 and KD17 withered faster than the wild type. With plants growing up to the reproductive stage, most leaves of line KD7 were dried out, whereas the wild-type plants remained green (Fig. 6C). We noticed the occurrence of lesions was more severe in line KD7 than line KD17. It indicates that phenotypic changes are correlated with the inhibition extent of SlGMP genes (Fig. 3B) in tomato.
Figure 6. Phenotype comparisons of SlGMP2/3-KD and wild-type plants.
(A) Altered cotyledon morphology of SlGMP2/3-KD vs wild-type plants. KD7 and KD17 cotyledons developed lesions (left) at three weeks post germination and accelerated senescence (right) compared with the wild type. (B) Seedlings of wild-type (left) and KD7 (right) plants. The bottom leaves of KD7 plant started to wilt. (C) Plant morphology of the three-month-old wild-type (left) and KD7 (right) plants. Middle and bottom leaves of KD7 plants became dry wilted. (D) Lesion formation on KD7 and KD17 leaves. The leaf lesion areas on the two-month-old plants of KD7 and KD17 (upper panel) are consistent with the areas of dead cells revealed through trypan blue staining (bottom panel). (E) H2O2 accumulation in the leaves of KD7 and KD17. H2O2 accumulation was revealed via DAB staining in leaves without (upper panel) and with (lower panel) visible lesions from the two-month-old plants of KD7 and KD17. (F) Tomato fruits harvested from four-month-old wild-type and transgenic plants.
Although the flowering time and the yield of SlGMP2/3-KD plants were not influenced, the fruit weight was significantly decreased in line KD7 (Fig. 6F and Table S5).
Oxidative Burst in the Leaves of SlGMP2/3-KD Plants
The spontaneous lesions on the leaves of SlGMP2/3-KD plants developed in a similar manner as a hypersensitive response after pathogen attack. Oxidative burst, wherein large quantities of ROS are generated, is an early plant response to microbial pathogen attack. Both types of leaves with and without lesions were analyzed via DAB staining to check whether the lesions observed in the leaves of SlGMP2/3-KD plants were caused by H2O2 accumulation. The lesioned leaves stained more, whereas the wild type did not (Fig. 6E), showing a close relationship between lesion and H2O2 levels in tomato leaves.
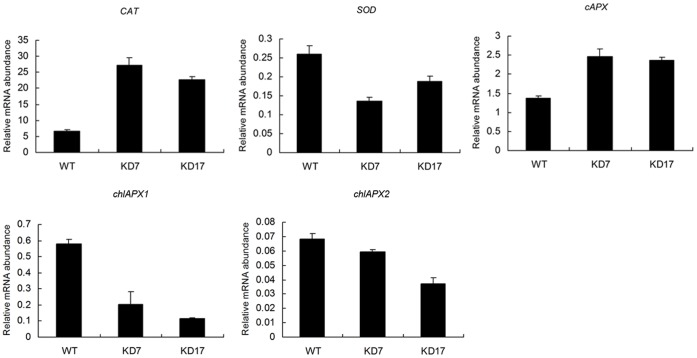
To examine whether the accumulation of high ROS concentrations affected the antioxidant defence system of plants, the transcript levels of several genes involved in detoxifying H2O2 or O2− were measured, including catalase (CAT), SOD, and ascorbate peroxidase (APX) using real-time RT-PCR. We found that SOD and chloroplastic APX genes were down-regulated in lesioned leaves of lines KD7 and KD17, whereas both their CAT and cytosolic APX genes were significantly up-regulated, especially for CAT, which increased over two-fold compared with the wild type (Fig. 7). These results imply that the partial antioxidant defence system is triggered in response to the oxidative burst in SlGMP2/3-KD leaves.
Figure 7. Relative transcript levels of the oxidative stress-related genes in the tomato lesioned leaves.
The expression levels of oxidative related genes CAT, SOD, cAPX, and chlAPXs in the tomato lesioned leaves were measured by real-time RT-PCR. Data were normalized against Actin.
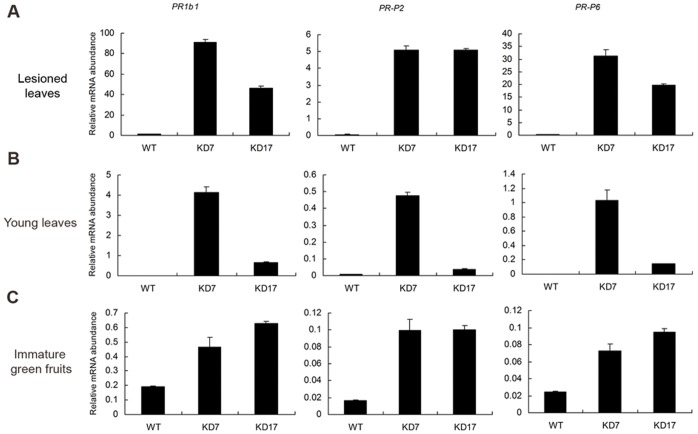
Activation of Defence Response in SlGMP2/3-KD Plants
Many mutants with such lesions in plants have shown an enhanced systemic resistance to microbial pathogens. To reveal whether SlGMP2/3-KD plants activated pathogen defence response, the expressions of three pathogenesis-related (PR) genes, including PR1b1 [32], PR-P2 [33], and PR-P6 [34], [35], were analyzed by real-time RT-PCR. These genes are known to be strongly induced locally in tomato-pathogen interactions. The results showed that the transcripts of PR1b1, PR-P2, and PR-P6 were more abundant in lesioned leaves of lines KD7 and KD17 than those in the wild type (Fig. 8A). Moreover, three defence genes were also activated in young leaves without any lesion (Fig. 8B) and in immature green fruits without any occurrence of lesions (Fig. 8C) though the activation extent was less than that in lesioned leaves. In addition, as shown in Fig. 8, more transcripts of PR1b1, PR-P2, and PR-P6 were induced in leaves than immature fruits. All these results seem that systemic acquired resistance (SAR) is activated in SlGMP2/3-KD plants.
Figure 8. Relative transcript levels of the pathogenesis-related genes in the tomato lesioned and normal leaves and fruits.
Expression analysis of the pathogenesis-related genes PR1b1, PR-P2, and PR-P6 was performed in necrotic mature leaves (A), non-necrotic normal young leaves (B) and immature green fruits (C) of lines KD7 and KD17 by real-time RT-PCR. Data were normalized against Actin.
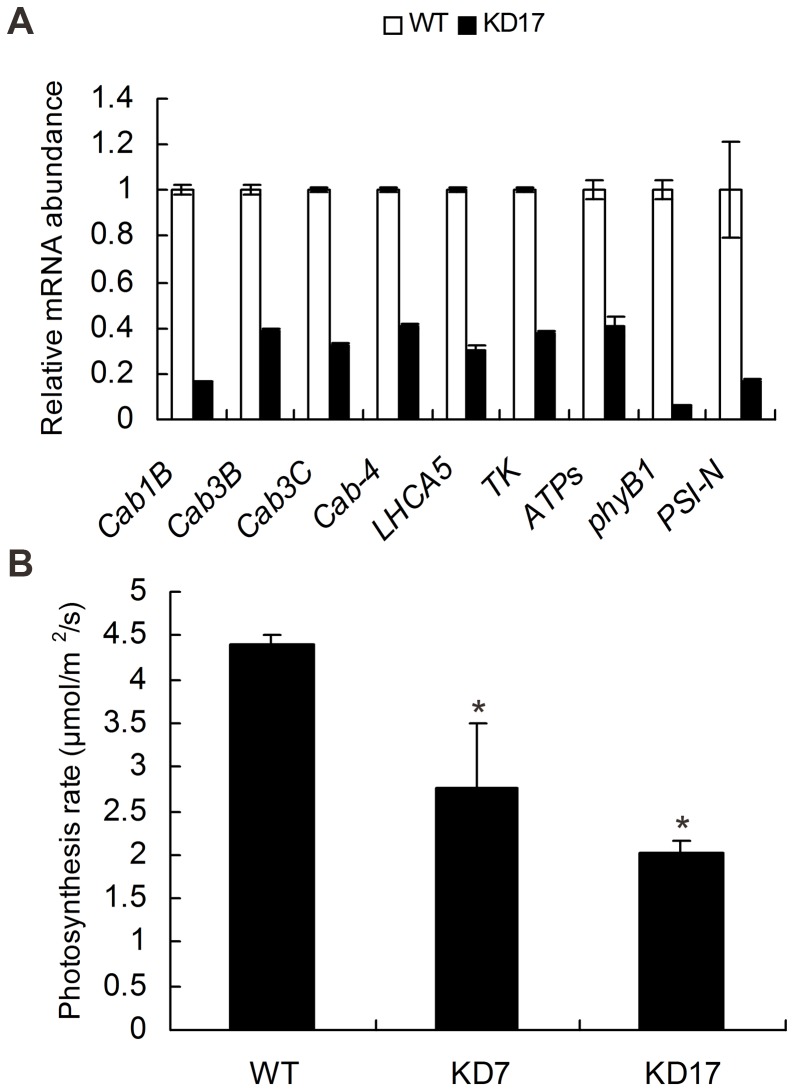
Impairment of the Photosynthetic System in SlGMP2/3-KD Plants
Primary results obtained through TOM2 Oligo chip microarray showed that some photosynthesis-related genes were up-regulated in the breaker fruits of SlGMP3-OX line OX19, and further confirmed via real-time RT-PCR analysis. These genes are involved in chlorophyll a-b binding, light harvesting processes in photosystem II, as well as ATP and phytochrome biosyntheses. Therefore, these genes were also analyzed in SlGMP2/3-KD plants. The nine selected photosynthesis-related genes were significantly down-regulated, and the net photosynthesis rates drastically declined in the slightly lesioned leaves of lines KD7 and KD17 (Fig. 9A, B).
Figure 9. Photosynthetic system was impaired in leaves of SlGMP2/3-KD tomato plants.
(A) Relative transcript levels of the photosynthesis-related genes in wild-type leaves and KD17 necrotic leaves were assayed via real-time RT-PCR. Data were normalized against Actin and shown as a percentage of wild-type plants. (B) Net photosynthesis rates of KD7 and KD17 necrotic leaves and wild-type leaves were measured using a portable photosynthetic system (CIRAS-2, PP System, USA). Data are presented as mean ± SD (N = 4) from triplicate independent measurements. Asterisk indicates significant differences from the control (P>0.95).
Discussion
GMP has been shown to affect AsA biosynthesis in Arabidopsis [10], potato [11], and acerola [12], [13]. Recently, expression of yeast-derived GMP gene in tomato was found to enhance AsA levels in leaves, green and red fruits [36], indicating biotechnological manipulation of AsA biosynthesis in tomato can be achieved through increasing GMP activity. In recent years, four GMP isoforms are found to exist in tomato genome [17]. However, until now, whether AsA biosynthesis could be regulated by manipulation of the four SlGMP genes in tomato is not clear yet. In 2006, due to the limited mRNA/EST sequence information in public databases, only one GMP gene (SlGMP3) was obtained from tomato by our group. The putative amino acid sequence of SlGMP3 has high similarity with AtGMP and StGMP which have been proved to play important roles in ascorbate biosynthesis in Arabidopsis and potato, respectively. So we focused our work on SlGMP3 gene to make clear its function in tomato. In this study, we generated transgenic tomato plants using constitutive over-expression and RNAi strategies for SlGMP3 gene and studied the plant response to stress regarding the role of GMP in the Smirnoff-Wheeler’s pathway and AsA as a major antioxidant of the plant cell.
Over-expression of SlGMP3 increased the total AsA content and enhanced antioxidant capacity, which was consistent with the results in tobacco expressing SlGMP3 [25]. Since we constructed RNAi vector using the full length cDNA of SlGMP3, SlGMP2 as well as SlGMP3 were simultaneously suppressed, leading to significant reduction in AsA contents in leaves and fruits of tomato. This confirmed the importance of the Smirnoff-Wheeler’s pathway and the vital role of GMP activity in AsA biosynthesis in tomato plant. Plant AsA efflux is regulated by environmental factors and especially the light environment. Massot et al. [17] found that among the four GMP genes, SlGMP1 and SlGMP3 were regulated by light, whereas SlGMP2 and SlGMP4 were not in tomato leaves. It implies SlGMP1 may also play an important role in controlling AsA biosynthesis in tomato. It should be mentioned that SlGMP2 was severely suppressed in SlGMP2/3-KD lines as SlGMP3, then we don’t exclude the possibility that SlGMP2 participates in controlling ascorbate content and causing phenotypic alteration of KD lines. Further investigation is needed to study on SlGMP1 and SlGMP2.
Significant increase in transcript abundances of SlGME2 and SlGGP1 downstream of SlGMP was found in the SlGMP2/3-KD plants, possibly because of a biological process to maintain a relative, stable ascorbate output. Similarly, up-regulation of SlGMP genes and SlGGP1 were observed in SlGME RNAi transgenic tomato plants [21]. The step controlled by GME was considered as an important control point in tomato AsA synthesis because it lies at the intersection of AsA synthesis and cell wall polysaccharides [21]. RNAi silencing of SlGME1 and SlGME2 in tomato effectively reduces AsA content, resulting in ROS accumulation, leaf bleaching and developmental defects [21], and over-expression of SlGME2 could improve AsA accumulation in tomato leaves and red ripe fruits in our group [22]. For GGP gene, it encodes an enzyme that catalyzes the first committed step in AsA biosynthesis in plants and has been identified as rate limiting in Arabidopsis [37] and tobacco [38] leaves. It has been confirmed that over-expression of GGP gene from kiwifruit increased AsA up to six-fold in tomato fruits [39]. Recently, the vital role of SlGGP1 in light-regulated AsA biosynthesis in tomato has been found [17]. We propose except SlGME1, SlGME2 and SlGMP3, SlGGP1 may be another good candidate for enhancing AsA accumulation in tomato. All these above underscore a complex regulation of the AsA pool size in tomato.
Lesions were formed on leaves throughout the whole life of SlGMP2/3-KD tomato plants. Cell death phenotype was previously found in the Arabidopsis mutant vtc1 and GMP antisense transgenic potato plants lacking AsA [11], [40]. However, the cell death pattern of SlGMP2/3-KD tomato plants is more similar to the GMP antisense potato plants [11] than vtc1 [40]. Interestingly, the cell death phenotype started earlier in SlGMP2/3-KD tomato plants than GMP antisense potato plants. Lesions were formed on tomato leaves even from seedling stage (Fig. 6A), while GMP antisense potato plants started to develop this phenotype 10 weeks after transfer to soil, which was considered that the imbalance between ROS and antioxidants due to the reduction in AsA contents changed the phenotype in the GMP antisense transgenic potato plants [11]. In this study, DAB staining revealed that higher concentration of H2O2 was involved in activating cell death program, which is distinct from the vtc1 mutant, wherein the H2O2 content is not changed [41]. This is consistent with the case of lesion formation on tobacco leaves due to reduced levels of the H2O2-detoxifying enzyme CAT [42]. In our study, the increased AsA content improved the tolerance to photo-oxidative stress in SlGMP3-OX plants, which further supported the results of gene knocked-down.
CAT is the key enzyme that directly detoxifies H2O2 in plant antioxidant system. CAT mRNA abundance was significantly up-regulated in the lesioned leaves of SlGMP2/3-KD plants. The increased expression of CAT appeared to be a compensatory mechanism for the reduced levels of AsA under oxidative stress. However, the capacity of the antioxidant system is not largely changed in the Arabidopsis mutant vtc1-1 [41], which might suggest that the low intracellular level of AsA is sufficient for scavenging ROS in Arabidopsis.
Salicylic acid (SA) is known to play an important role in the activation of defence responses in plants against microbial infection. In several identified lesion mimic mutants, the activation of PR genes was correlated with the accumulation of high SA levels [43], [44]. PR1b1 and PR-P2, the marker genes in tomato SA-dependent defence pathway were activated in SlGMP2/3-KD plants. In previous study, H2O2 is considered to act upstream of SA in the signal transduction pathway [45]. We speculate that the enhanced production of H2O2 might induce defence response partly through SA response pathway in SlGMP2/3-KD plants. More studies are needed to further confirm this hypothesis.
To determine whether the activated defence response could enhance resistance to pathogens in SlGMP2/3-KD plants, five-week-old plantlets were challenged with the virulent bacterium Pseudomonas syringae pv. tomato DC3000 (Pst) according to the method described by Anderson [46]. However, the resistance of lines KD7 and KD17 to Pst DC3000 at 24/20°C day/night temperature (16 h/8 h) and 70% relative humidity was slightly improved but not statistically significant (data not shown). This result may be attributed to the low light intensity of the artificial climate chamber, in which the transgenic plants showed weaker phenotype of spontaneous cell death than that in greenhouse with a high light intensity.
Multiple functions for AsA in photosynthesis have been proposed [5]. AsA-deficient plants show zeaxanthin depletion and an inhibition of photosynthesis when exposed to high light [47]. Rubisco levels are significantly reduced in the vtc1 mutant [48]. It is suggested that genes encoding the components of the electron transport system could be regulated by AsA [49]. In the chloroplast, AsA-GSH (reduced glutathione) cycle is the key pathway to protect the photosynthetic machinery by removing ROS. In SlGMP2/3-KD tomato lines, the expressions of several genes encoding photosynthetic enzymes were very significantly reduced as well as overall photosynthetic activity. This implies that the impairment of photosynthetic system may be due to the lower AsA level.
In conclusion, we have studied one of the AsA biosynthetic pathway steps using over-expression and RNAi strategies for SlGMP3 gene in tomato plants. In doing so, we found the vital roles of AsA as an antioxidant in leaf senescence and defence response in tomato. The increased H2O2 level caused by low AsA levels might be responsible for the induction of the SA signal transduction pathway, which led to necrosis development and defence response activation. In addition to AsA biosynthesis, GMP is also need for synthesis of mannose, fucose and L-galactose containing polysaccharides in the cell wall. We do not exclude the possibility that cell wall defects participate in causing some of the phenotypes seen in our experiment and this will be investigated in our further study.
Supporting Information
Nucleotide sequence alignment of SlGMP2 and SlGMP3 genes.
(TIF)
Primers used for SlGMP3 amplification and identification of the transformants and RT-PCR.
(DOC)
Primers used for real-time RT-PCR of the AsA biosynthesis-related genes.
(DOC)
Primers used for real-time RT-PCR of the photosynthesis-related genes.
(DOC)
Primers used for real-time RT-PCR of the pathogenesis-related genes and antioxidant enzyme genes.
(DOC)
The fruit weights and yields of SlGMP2/3 -KD lines and wild-type plants.
(DOC)
Acknowledgments
The authors thank Dr. K. Ziaf for critically reading this manuscript and Dr. F. Xiao for technical assistance.
Funding Statement
This work was supported by the National Basic Research Program of China (No. 2011CB100600), and the National Natural Science Foundation of China (No. 31230064). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Foyer CH, Noctor G (2005) Oxidant and antioxidant signalling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ 28: 1056–1071. [Google Scholar]
- 2. Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399. [DOI] [PubMed] [Google Scholar]
- 3. Vranová E, Inzé D, Van Breusegem F (2002) Signal transduction during oxidative stress. J Exp Bot 53: 1227–1236. [PubMed] [Google Scholar]
- 4. Smirnoff N (2000) Ascorbic acid: metabolism and functions of a multi-facetted molecule. Curr Opin Plant Biol 3: 229–235. [PubMed] [Google Scholar]
- 5. Smirnoff N (2000) Ascorbate biosynthesis and function in photoprotection. Philos Trans R Soc Lond B Bio Sci 355: 1455–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Agius F, González-Lamothe R, Caballero JL, Muñoz-Blanco J, Botella MA, et al. (2003) Engineering increased vitamin C levels in plants by overexpression of a D-galacturonic acid reductase. Nat Biotechnol 21: 177–181. [DOI] [PubMed] [Google Scholar]
- 7. Lorence A, Chevone BI, Mendes P, Nessler CL (2004) myo-Inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol 134: 1200–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wolucka BA, Van Montagu M (2003) GDP-mannose 3′, 5′-epimerase forms GDP-L-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. J Biol Chem 278: 47483–47490. [DOI] [PubMed] [Google Scholar]
- 9. Wheeler GL, Jones MA, Smirnoff N (1998) The biosynthetic pathway of vitamin C in higher plants. Nature 393: 365–369. [DOI] [PubMed] [Google Scholar]
- 10. Conklin PL, Norris SR, Wheeler GL, Williams EH, Smirnoff N, et al. (1999) Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc Natl Acad Sci USA 96: 4198–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Keller R, Renz FS, Kossmann J (1999) Antisense inhibition of the GDP-mannose pyrophosphorylase reduces the ascorbate content in transgenic plants leading to developmental changes during senescence. Plant J 19: 131–141. [DOI] [PubMed] [Google Scholar]
- 12. Badejo AA, Jeong ST, Goto-Yamamoto N, Esaka M (2007) Cloning and expression of GDP-D-mannose pyrophosphorylase gene and ascorbic acid content of acerola (Malpighia glabra L.) fruit at ripening stages. Plant Physiol Biochem 45: 665–672. [DOI] [PubMed] [Google Scholar]
- 13. Badejo AA, Tanaka N, Esaka M (2008) Analysis of GDP-D-mannose pyrophosphorylase gene promoter from acerola (Malpighia glabra) and increase in ascorbate content of transgenic tobacco expressing the acerola gene. Plant Cell Physiol 49: 126–132. [DOI] [PubMed] [Google Scholar]
- 14. Badejo AA, Wada K, Gao Y, Maruta T, Sawa Y, et al. (2011) Translocation and the alternative D-galacturonate pathway contribute to increasing the ascorbate level in ripening tomato fruits together with the D-mannose/L-galactose pathway. J Exp Bot 63: 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Di Matteo A, Sacco A, Anacleria M, Pezzotti M, Delledonne M, et al. (2010) The ascorbic acid content of tomato fruits is associated with the expression of genes involved in pectin degradation. BMC Plant Biol 10: 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ioannidi E, Kalamaki MS, Engineer C, Pateraki I, Alexandrou D, et al. (2009) Expression profiling of ascorbic acid-related genes during tomato fruit development and ripening and in response to stress conditions. J Exp Bot 60: 663–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Massot C, Stevens R, Génard M, Longuenesse JJ, Gautier H (2012) Light affects ascorbate content and ascorbate-related gene expression in tomato leaves more than in fruits. Planta 235: 153–163. [DOI] [PubMed] [Google Scholar]
- 18. Stevens R, Buret M, Duffé P, Garchery C, Baldet P, et al. (2007) Candidate genes and quantitative trait loci affecting fruit ascorbic acid content in three tomato populations. Plant Physiol 143: 1943–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stevens R, Page D, Gouble B, Garchery C, Zamir D, et al. (2008) Tomato fruit ascorbic acid content is linked with monodehydroascorbate reductase activity and tolerance to chilling stress. Plant Cell Environ 31: 1086–1096. [DOI] [PubMed] [Google Scholar]
- 20. Alhagdow M, Mounet F, Gilbert L, Nunes-Nesi A, Garcia V, et al. (2007) Silencing of the mitochondrial ascorbate synthesizing enzyme L-galactono-1, 4-lactone dehydrogenase affects plant and fruit development in tomato. Plant Physiol 145: 1408–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gilbert L, Alhagdow M, Nunes-Nesi A, Quemener B, Guillon F, et al. (2009) GDP-D-mannose 3, 5-epimerase (GME) plays a key role at the intersection of ascorbate and non cellulosic cell wall biosynthesis in tomato. Plant J 60: 499–508. [DOI] [PubMed] [Google Scholar]
- 22. Zhang C, Liu J, Zhang Y, Cai X, Gong P, et al. (2011) Overexpression of SlGMEs leads to ascorbate accumulation with enhanced oxidative stress, cold, and salt tolerance in tomato. Plant Cell Rep 30: 389–398. [DOI] [PubMed] [Google Scholar]
- 23. Zou L, Li H, Ouyang B, Zhang J, Ye Z (2006) Cloning, expression, and mapping of GDP-D-mannose pyrophosphorylase cDNA from tomato (Lycopersicon esculentum). Acta Genet Sinica 33: 757–764. [DOI] [PubMed] [Google Scholar]
- 24. Zou L, Li H, Ouyang B, Zhang J, Ye Z (2006) Cloning and mapping of genes involved in tomato ascorbic acid biosynthesis and metabolism. Plant Sci 170: 120–127. [Google Scholar]
- 25. Wang HS, Yu C, Zhu ZJ, Yu XC (2011) Overexpression in tobacco of a tomato GMPase gene improves tolerance to both low and high temperature stress by enhancing antioxidation capacity. Plant Cell Rep 30: 1029–1040. [DOI] [PubMed] [Google Scholar]
- 26. Fillatti JAJ, Kiser J, Rose R, Comai L (1987) Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Nat Biotechnol 5: 726–730. [Google Scholar]
- 27. Rizzolo A, Forni E, Polesello A (1984) HPLC assay of ascorbic acid in fresh and processed fruit and vegetables. Food Chem 14: 189–199. [Google Scholar]
- 28. Wellburn R (1994) The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J Plant Physiol 144: 307–313. [Google Scholar]
- 29. Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts.I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125: 189–198. [DOI] [PubMed] [Google Scholar]
- 30. Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11: 1187–1194. [Google Scholar]
- 32. Tornero P, Gadea J, Conejero V, Vera P (1997) Two PR-1 genes from tomato are differentially regulated and reveal a novel mode of expression for a pathogenesis-related gene during the hypersensitive response and development. Mol Plant Microbe Interact 10: 624–634. [DOI] [PubMed] [Google Scholar]
- 33. Linthorst HJ, Danhash N, Brederode FT, Van Kan JA, De Wit PJ, et al. (1991) Tobacco and tomato PR proteins homologous to win and pro-hevein lack the “hevein” domain. Mol Plant Microbe Interact 4: 586–592. [DOI] [PubMed] [Google Scholar]
- 34. Joosten MH, Bergmans CJ, Meulenhoff EJ, Cornelissen BJ, De Wit PJ (1990) Purification and serological characterization of three basic 15-kilodalton pathogenesis-related proteins from tomato. Plant Physiol 94: 585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Kan JA, Joosten MH, Wagemakers CA, van den Berg-Velthuis GC, de Wit PJ (1992) Differential accumulation of mRNAs encoding extracellular and intracellular PR proteins in tomato induced by virulent and avirulent races of Cladosporium fulvum. Plant Mol Biol 20: 513–527. [DOI] [PubMed] [Google Scholar]
- 36. Cronje C, George GM, Fernie AR, Bekker J, Kossmann J, et al. (2012) Manipulation of L-ascorbic acid biosynthesis pathways in Solanum lycopersicum: elevated GDP-mannose pyrophosphorylase activity enhances L-ascorbate levels in red fruit. Planta 235: 553–564. [DOI] [PubMed] [Google Scholar]
- 37. Bulley SM, Rassam M, Hoser D, Otto W, Schünemann N, et al. (2009) Gene expression studies in kiwifruit and gene over-expression in Arabidopsis indicates that GDP-L-galactose guanyltransferase is a major control point of vitamin C biosynthesis. J Exp Bot 60: 765–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Laing WA, Wright MA, Cooney J, Bulley SM (2007) The missing step of the L-galactose pathway of ascorbate biosynthesis in plants, an L-galactose guanyltransferase, increases leaf ascorbate content. Proc Natl Acad Sci USA 104: 9534–9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bulley S, Wright M, Rommens C, Yan H, Rassam M, et al. (2012) Enhancing ascorbate in fruits and tubers through over-expression of the L-galactose pathway gene GDP-L-galactose phosphorylase. Plant Biotechnol J 10: 390–397. [DOI] [PubMed] [Google Scholar]
- 40. Pavet V, Olmos E, Kiddle G, Mowla S, Kumar S, et al. (2005) Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiol 139: 1291–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Veljovic-Jovanovic SD, Pignocchi C, Noctor G, Foyer CH (2001) Low ascorbic acid in the vtc-1 mutant of Arabidopsis is associated with decreased growth and intracellular redistribution of the antioxidant system. Plant Physiol 127: 426–435. [PMC free article] [PubMed] [Google Scholar]
- 42. Takahashi H, Chen Z, Du H, Liu Y, Klessig DF (1997) Development of necrosis and activation of disease resistance in transgenic tobacco plants with severely reduced catalase levels. Plant J 11: 993–1005. [DOI] [PubMed] [Google Scholar]
- 43. Weymann K, Hunt M, Uknes S, Neuenschwander U, Lawton K, et al. (1995) Suppression and restoration of lesion formation in Arabidopsis lsd mutants. Plant Cell 7: 2013–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Greenberg JT, Guo A, Klessig DF, Ausubel FM (1994) Programmed cell death in plants: a pathogen-triggered response activated coordinately with multiple defense functions. Cell 77: 551–563. [DOI] [PubMed] [Google Scholar]
- 45. Chamnongpol S, Willekens H, Moeder W, Langebartels C, Sandermann H, et al. (1998) Defense activation and enhanced pathogen tolerance induced by H2O2 in transgenic tobacco. Proc Natl Acad Sci USA 95: 5818–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Anderson JC, Pascuzzi PE, Xiao FM, Sessa G, Martin GB (2006) Host-mediated phosphorylation of type ? effector AvrPto promotes Pseudomonas virulence and avirulence in tomato. Plant Cell 18: 502–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Müller-Moulé P, Golan T, Niyogi KK (2004) Ascorbate-deficient mutants of Arabidopsis grow in high light despite chronic photooxidative stress. Plant Physiol 134: 1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pastori GM, Kiddle G, Antoniw J, Bernard S, Veljovic-Jovanovic S, et al. (2003) Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling. Plant Cell 15: 939–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kiddle G, Pastori GM, Bernard S, Pignocchi C, Antoniw J, et al. (2003) Effects of leaf ascorbate content on defense and photosynthesis gene expression in Arabidopsis thaliana . Antioxid Redox Signal 5: 23–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide sequence alignment of SlGMP2 and SlGMP3 genes.
(TIF)
Primers used for SlGMP3 amplification and identification of the transformants and RT-PCR.
(DOC)
Primers used for real-time RT-PCR of the AsA biosynthesis-related genes.
(DOC)
Primers used for real-time RT-PCR of the photosynthesis-related genes.
(DOC)
Primers used for real-time RT-PCR of the pathogenesis-related genes and antioxidant enzyme genes.
(DOC)
The fruit weights and yields of SlGMP2/3 -KD lines and wild-type plants.
(DOC)