Abstract
The discovery of chromosomal translocations in prostate cancer has greatly enhanced our understanding of prostate cancer biology. Genomic rearrangements involving the ETS family of transcription factors are estimated to be present in 50-70% of prostate cancer cases. These rearrangements fuse the ETS factors with promoters of genes that are androgen regulated. Thus, the expression of ETS factors, such as ERG, ETV1, ETV4 and ETV5, is mediated by androgen. In-vitro and in-vivo studies suggest that overexpression of ETS proteins increase cell proliferation and confer an invasive phenotype to prostate cancer cells. Epidemiological studies demonstrate that ETS-fusion positive patients exhibit tumors corresponding to a more advanced disease. The ability of ETS factors to serve as markers for screening and diagnosing prostate cancer patients is being investigated, and the results have been largely positive to date. Additionally, ETS factors present an excellent opportunity as therapeutic targets and several strategies have been devised to directly target ETS proteins or their binding partners and downstream effectors.
Keywords: Prostate cancer, chromosomal translocation, transcription factor, ETS, TMPRSS2-ERG, ETV1
Introduction
Prostate cancer is the second most leading cause of cancer death in men in the United States, behind only lung cancer. It accounts for 29% of all cancer cases and is responsible for 9% of all cancer deaths (American Cancer Society). Prostate cancer is associated with significant molecular heterogeneity. The mechanisms responsible for disease initiation, progression and metastasis are not yet fully understood. Thus, a better understanding of prostate cancer biology is important for improving prostate cancer screening, developing specific diagnostic tools and ultimately treating the disease.
The discovery of chromosomal translocations in prostate cancer has greatly enhanced our understanding of prostate cancer biology. Chromosomal rearrangements are a common mechanism driving oncogenesis in sarcomas and hematologic malignancies [1]. Recently, fusions involving the erythroblastosis virus E26 transforming sequences (ETS) family of transcription factors has been shown to play an important role in prostate cancer pathogenesis [2]. The ETS family of transcription factors consists of a highly conserved group of genes that play important roles in cellular proliferation, differentiation, migration, invasion and angiogenesis [3]. ETS proteins share significant homology with each other and contain a C-terminal ETS domain that is involved in DNA-binding [4]. Chromosomal rearrangements involving the ETS family of transcription factors result in truncated ETS proteins that are fused to androgen regulated gene promoters. Thus, the expression of these genes is regulated by androgen in prostate cells harboring ETS fusions. The most common fusion product involves the 5’ promoter region of TMPRSS2, a trans-membrane serine protease that is expressed exclusively in the prostate, fused to the 3’ region of ERG [2]. This fusion results in the over-expression of a nearly full-length ERG protein. Different studies have shown that the fusion protein is present in approximately 50% of prostate cancer cases. Prostate cells demonstrating androgen dependent ERG over-expression have a molecular signature indicative of an aggressive phenotype [5]. Moreover, the presence of the fusion transcript has been largely associated with a poor prognosis, lower incidence of recurrence free survival and higher Gleason scores, indicative of a more advanced disease [6]. Fusions involving other ETS factors, such as ETV1 (10% cases), ETV4 (<1%), ETV5 (<1%) and ELK4 (<1%) have also been implicated in prostate cancer [7,8]. To date, 14 different genes (TMPRSS2, SLC45A3, C15orf21 CANT1, EST14, FOXP1, HERVK17, FLJ35294, HERV-K, ACSL3 and NDRG1, DDX5, HNRPA2B1, KLK2) have been identified as fusion partners for ERG, ETV1, ETV4, ETV5 and ELK4 [9-12]. Thus, the ETS family of transcription factors represents a novel class of macromolecules that can be exploited for their usefulness as diagnostic tools and therapeutic targets in the treatment of prostate cancer.
Initial discovery
The presence of ETS translocations in human prostate cancer was initially reported in a landmark paper published in 2005 [2]. The authors used a bioinformatics approach to probe genes over-expressed across several prostate cancer microarray data sets. They identified ERG and ETV1, two genes that encode ETS transcription factors and are involved in oncogenic transformations in myeloid leukemia and Ewing’s sarcoma [13]. Although the over-expression of ERG in prostate cancer had been previously reported [14], Tomlins et al now proposed a mechanism to account for this over-expression. The authors noticed that ERG and ETV1 expression was mutually exclusive and over-expression products included exons 4-7 of the ETS factors more commonly than exons 1 and 2. This led them to believe that the over-expression mechanism involved chromosomal rearrangements. 5’ RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) to obtain the complete transcript of the overexpressed product revealed TMPRSS2 as the 5’ fusion partner. TMPRSS2 is a serine protease expressed in the normal prostate tissue and is strongly induced by androgen in androgen-sensitive prostate cells. Thus, the fusion of 5’-unstranslated region of TMPRSS2 with 3’-regions of ERG or ETV1 placed these protooncogenes under direct regulation by androgen stimulation.
Incidence of ETS fusions
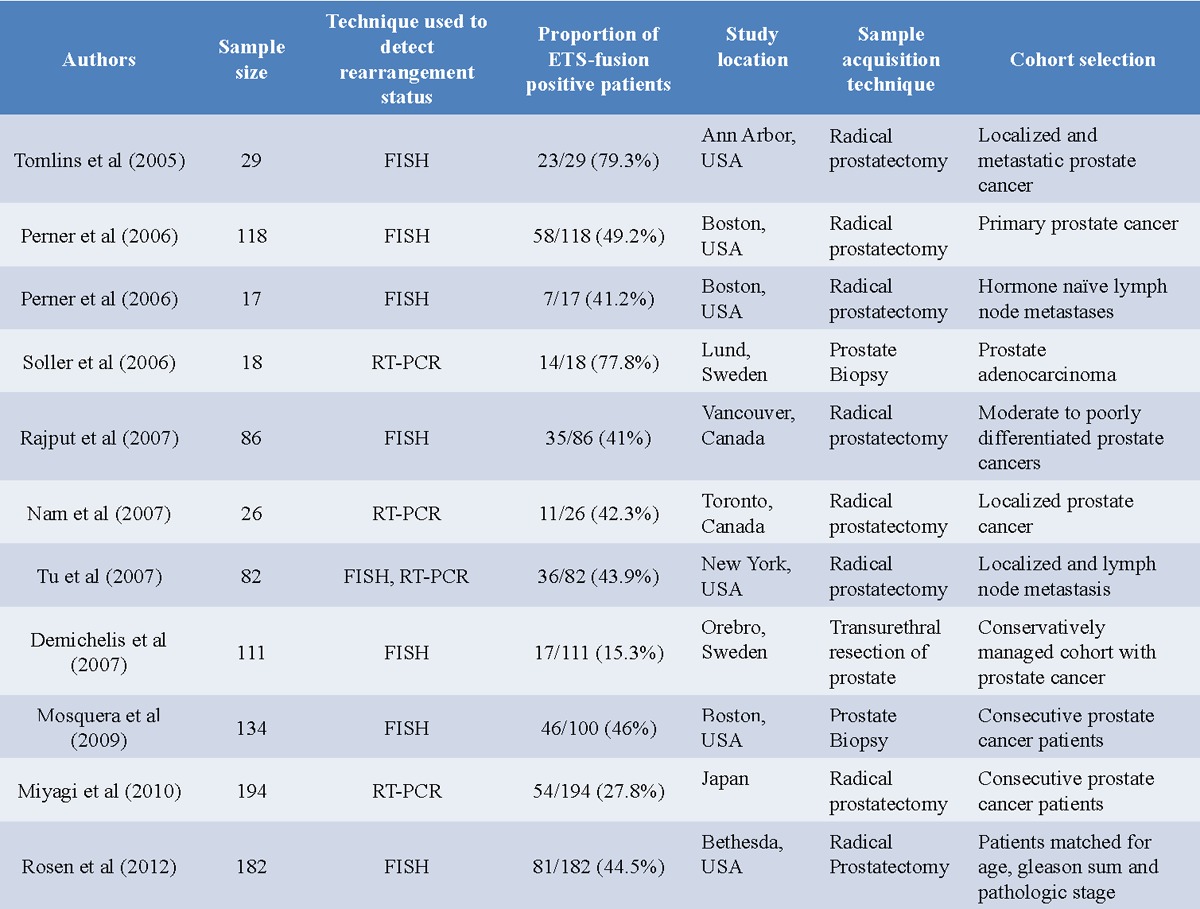
The discovery of chromosomal fusions as a mechanism for ETV1 and ERG over-expression marked a new era in the understanding of prostate cancer biology. Several other groups have, since then, confirmed the original findings of Tomlins et al. However, the frequency of gene recombination varies between different studies (15%-78%), probably owing to the sensitivity of the technique used, the number of samples included in the study, the number of fusion variants probed and the tumor stage [15-23]. Earlier studies used fluorescence in-situ hybridization or RT-PCR to probe for recombination between ERG and TMPRSS2. More recently, ERG expression has been detected by immunohistochemistry using a monoclonal ERG antibody [24].
Using FISH technique, Perner et al [15], identified TMPRSS2-ERG rearrangements in 49.2% of 118 primary prostate cancers and 41.2% of 18 hormone-naive lymph node metastases. Soon after, Soller et al [16] used RT-PCR to detect TMPRSS2-ERG in 14 out of 18 cases of prostate adenocarcinoma (78%). Using FISH on prostate cancer tissue microarrays, Rajput et al [17] found that 41% of moderate to poorly differentiated tumors (35/86 cases) exhibited TMPRSS2-ERG fusion status. However, only 6.7% of well differentiated tumors (1/15 cases) and none of the prostatic hyperplasia cases contained these gene rearrangements (0/5). Other studies by Nam et al [18] and Tu et al [19] point to a 40%-50% occurrence of gene rearrangement, whereas a study by Demichiles et al [20] reported only a 15% occurrence (17/111) of TMPRSS2-ERG in a Swedish cohort of men with localized prostate cancer who underwent expectant management. Most of the studies probe for the rearranged product in men who underwent surgery. To determine whether the high frequency observed in these prostatectomy specimens (40-78%) reflects selection bias, Mosquera et al [21] determined the prevalence of TMPRSS2-ERG fusion status among PSA screened men undergoing prostate biopsy and found that 46% of men with prostate cancer harbored the fusion gene, consistent with the frequency observed in the surgical cohorts. Efforts have also been made to correlate the frequency of gene alterations with ethnic backgrounds. A study of 194 Japanese prostate cancer patients revealed the presence of TMPRSS2-ERG rearrangement in only 28% of cases [25]. Another study analyzed the frequency of ERG oncoprotein expression between 91 African-American and 91 Caucasian patients who were matched for age, Gleason score and pathologic state. A markedly greater frequency of ERG expression was found between the index tumors of the Caucasians Americans (63.3%) and those of the African Americans (28.6%) [22].
The correct estimation of TMPRSS2-ERG incidence is also complicated by the fact that significant molecular heterogeneity exists between independent tumor foci in multifocal prostate cancer [26,27]. Minner et al performed a study to analyze the extent of heterogeneity for TMPRSS2-ERG fusion [23]. They developed a heterogeneity tissue microarray containing samples from 10 different tumor blocks of 190 large prostate cancers. ERG immunostaining was homogeneously positive in 29 prostate cancers (16%), whereas heterogeneous ERG positivity was seen in 74 cancers (42%). Furthermore, ERG heterogeneity was within one tumor focus (intrafocal heterogeneity) in 69 cases (93% of heterogeneous cases) and between different tumor foci (interfocal heterogeneity) in 5 cases (7%). This study shows that ERG heterogeneity exists in a significant portion of prostate cancer cases, and ERG rearrangement status may not be accurately determined by analyzing a single tissue biopsy per patient (Table 1).
Table 1.
Summary of published studies demonstrating the frequency of TMPRSS2-ERG fusion in prostate cancer
|
Although, the individual estimates of fusion occurrence vary widely between different studies, overall the data clearly suggests that the TMPRSS2-ERG rearrangement is present in a majority of prostate cancer cases. Further increase in the sensitivity of techniques used and the discovery of new splice variants may lead to an even higher proportion of prostate cancers detected to carry the fusion product.
Mechanism of genomic rearrangement
TMPRSS2 and ERG are located 3Mb apart on chromosome 21. The rearrangement between these two genes can occur either through balanced or unbalanced translocation between two chromosome 21s, or through interstitial deletion (Edel). The literature, to date, suggests that intronic deletion is the more common mechanism for the rearrangement, being responsible for 60%-80% of TMPRSS2-ERG cases [5,15,19,28,29]. The deletion of the intervening region between TMPRSS2 and ERG on chromosome 21, in addition to ERG activation, may also lead to loss of intervening genes [30]. However, whether this deletion leads to any additional biological significance resulting in a more aggressive phenotype, is yet to be determined.
Two independent studies have elucidated the mechanism underlying the non-random juxtaposition of TMPRSS2 and ERG in prostate cells [31,32]. Both studies probe the synergistic ability of androgen signaling coupled with genotoxic stress to induce chromosomal rearrangements. Mani et al [31] stimulated the androgen responsive LNCaP prostate cell-line, which does not harbor the TMPRSS2-ERG rearrangement, with increasing amounts of dihydrotestosterone (DHT). Using FISH, they observed that the treatment induced proximity between TMPRSS2 and ERG regions on chromosome 21. To determine whether this induced proximity facilitated gene fusions, the authors used radiation as a surrogate for genotoxic stress and observed that DHT stimulated LNCaP cells that were exposed to radiation to harbor the TMPRSS2-ERG fusion. Thus, androgen signaling brings the 5’ and 3’ fusion partners in close proximity, increasing the likelihood of fusions in the presence of agents that cause DNA double stranded breaks.
Lin et al [32] used a similar strategy to determine the specific machinery required for the site-specific DNA double-stranded breaks (DSBs). They identified activation-induced cytidine deaminase (AID)/GADD45 and the LINE-1 repeat-encoded ORF2 endonuclease as the enzymes responsible for mediating double-stranded breaks at translocation loci brought in close proximity by ligand bound androgen receptor. Ligation of the DSBs via non-homologous end joining pathway finally results in the generation of rearranged product.
Alternatively spliced forms
The study of TMPRSS2-ERG fusion product in prostate cancer is rendered complex by the fact that more than 20 alternatively spliced TMPRSS2-ERG isoforms have been identified in prostate cancer [33-36]. These isoforms differ in the location of TMPRSS2-ERG junction and are heterogeneous in the presence of various TMPRSS2 and ERG exons. The transcripts range from those encoding full-length ERG protein, N-terminal truncated ERG proteins, ERG isoforms expressing only the DNA binding domain (ETS) and finally those encoding truncated ERG proteins lacking the ETS domain but expressing pointed/SAM domains [36]. The isoforms have been shown to promote proliferation, invasion and motility with variable activities depending on the structure of the 5’ region encoding the fusion proteins [34]. Interestingly, the splice variants lacking the ETS domain are more abundantly expressed in prostate cancer patients than those encoding full-length ERG protein [35]. However, recent data suggest increased ratio of full-length over truncated splice forms correlates with a less favorable prognosis. In particular, the fusion isoform involving exon 1-2 of TMPRSS2 and exon 4-7 of ERG is strongly associated with aggressive prostate cancer. The presence of this fusion product is correlated with early recurrence and seminal vesicle invasion [33]. However, the reason for association of particular isoforms with aggressive disease is not known and further investigation is required to answer this phenomena.
Biological function of TMPRSS2-ERG protein
In a physiological setting, ERG expression is mainly restricted to endothelial and hematopoietic cells [37-39]. Its expression pattern underlies the roles of ERG in angiogenesis, endothelial cell differentiation, platelet development, stem-cell function and hematopoiesis. ERG is not expressed in epithelial cells, including the prostatic epithelium [40-42].
In-vitro over-expression of truncated ERG in the same proportion as that found in the majority of prostate cancer patients leads to an increase in migration and cell invasion in primary as well as benign immortalized prostatic epithelial cells [6]. ERG over-expression in these cell lines leads to the activation of several matrix metalloproteinases (MMP3, MMP9, ADAM19) and genes involved in the plasminogen activator pathway (PLAT, PLAU). Both classes of gene products have been directly implicated in several cancers [43,44]. Consequently, ERG knock-down in the TMPRSS2-ERG positive prostate cancer cell line VCaP results in decreased expression of these target genes. This selective decrease in gene expression is accompanied by decreased invasion and migration.
The effects of ERG on survival can be attributed partly to its effects on PIM-1 expression [45]. PIM-1 is an oncogene with pro-survival attributes. It is found in several tumors of epithelial and hematological origin, including prostate cancer, and favors genomic instability [46]. ERG also upregulates expression of the chemokine receptor CXCR4, the extracellular matrix protein CRISP3 and the extracellular matrix glycophosphoprotein osteopontin [47-49]. ERG induces the expression of these proteins through binding to ETS binding sequences on the promoter regions of these genes. CXCR4, CRISP3 and osteopontin are key regulators of tumor invasion and metastasis in a variety of cancers [50-52]. ERG also regulates the expression of c-MYC oncogene, which consequently affects cellular morphology and expression of prostate differentiation related genes [53]. Recent data suggests that ERG is implicated in the epithelial-to-mesenchymal transition, epigenetic regulation, inflammation and DNA damage repair pathways. EMT occurs at the invasive front of tumors and produces single migratory cells that are able to escape the primary tumor site. This is concomitant with deregulation of the Wnt pathway [54]. Increased ERG activity is associated with EMT via repression of epithelial-specific genes, such as E-cadherin and cytokeratins, and increased expression of mesenchymal-specific genes, such as CDH2 and CDH11 [55]. Loss of E-cadherin is a hallmark of EMT [54]. ERG also upregulates EMT facilitators ZEB1 and ZEB2, which are involved in regulation of E-cadherin, perhaps via SNAIL factors [56]. A genome-wide screen revealed several WNT genes, such as WNT11, WNT2, WNT9A, CCND1, FZD7 and FZD4, as direct targets of ERG [57]. Notably, FZD4 is a direct mediator of WNT signaling and EMT [55]. Wnt-11 promotes neuroendocrine-like differentiation, survival and migration of prostate cancer cells [58].
ERG is implicated in epigenetic regulation through its effects on the transcription of polycomb group protein EZH2. EZH2 is a key element of the Polycomb Repressive Complex 2 (PRC2) and is responsible for the establishment of the repressive H3K27 methylation marker. ERG concomitantly represses expression of the prostate-specific tumor suppressor NKX3.1 via EZH2 [59]. NKX3.1 is important for prostate cancer initiation and progression as it integrates multiple signaling pathways such as PI3K, Akt, p53 and androgen receptor signaling [60]. NKX3.1 silencing by ERG and EZH2 leads to increased proliferation and invasion. ERG also positively influences the prostaglandin pathway by modulating levels of 15-hydroxyprostaglandin dehydrogenase (HPDG) and prostaglandin-E2 (PGE2), directly implicating it in the inflammatory response pathway [61]. Furthermore, ERG activates Nf-kB pathway through toll-like receptor 4, further implicating it in the inflammatory pathway [62].
In-vivo studies with animal models have been equally rewarding in furthering our understanding of ERG activity. Transgenic over-expression of ERG in the mouse prostate results in the development of prostatic intraepithelial neoplasia (PIN). However, it does not lead to carcinoma, thus indicating that ERG is not sufficient for the development of prostate cancer in mice [6]. However, TMPRSS2-ERG knock-down in VCaP xenograft models in SCID mice results in decreased tumor growth in-vivo [53]. These results suggest that ETS fusions may occur in the context of other genetic lesions that drive the transition of PIN to prostate cancer or from primary to metastatic disease. To this end, studies by two groups show that TMPRSS2-ERG rearrangement co-operates with PTEN loss and activation of PI3K pathway to mediate progression of PIN to prostatic adenocarcinoma [63,64]. Similar to transgenic ERG mouse models, Pten-deficient mice develop high-grade PIN without progression to invasive cancer. Moreover, Pten-loss is observed in approximately 70% of prostate tumors. Tumors expressing the TMPRSS2-ERG genomic rearrangement demonstrate significantly reduced expression of PTEN. Also, ERG over-expression and PTEN loss is correlated across a majority of tumor specimens. Moreover, when PTEN heterozygous mice are crossed with mice that over-express truncated ERG in their prostate, the offspring consistently demonstrated lesions indicative of prostate carcinoma. These findings further stress the fact that ETS fusions function concomitantly in the presence of other genomic alterations to induce malignancy.
Clinical relevance of ERG expression
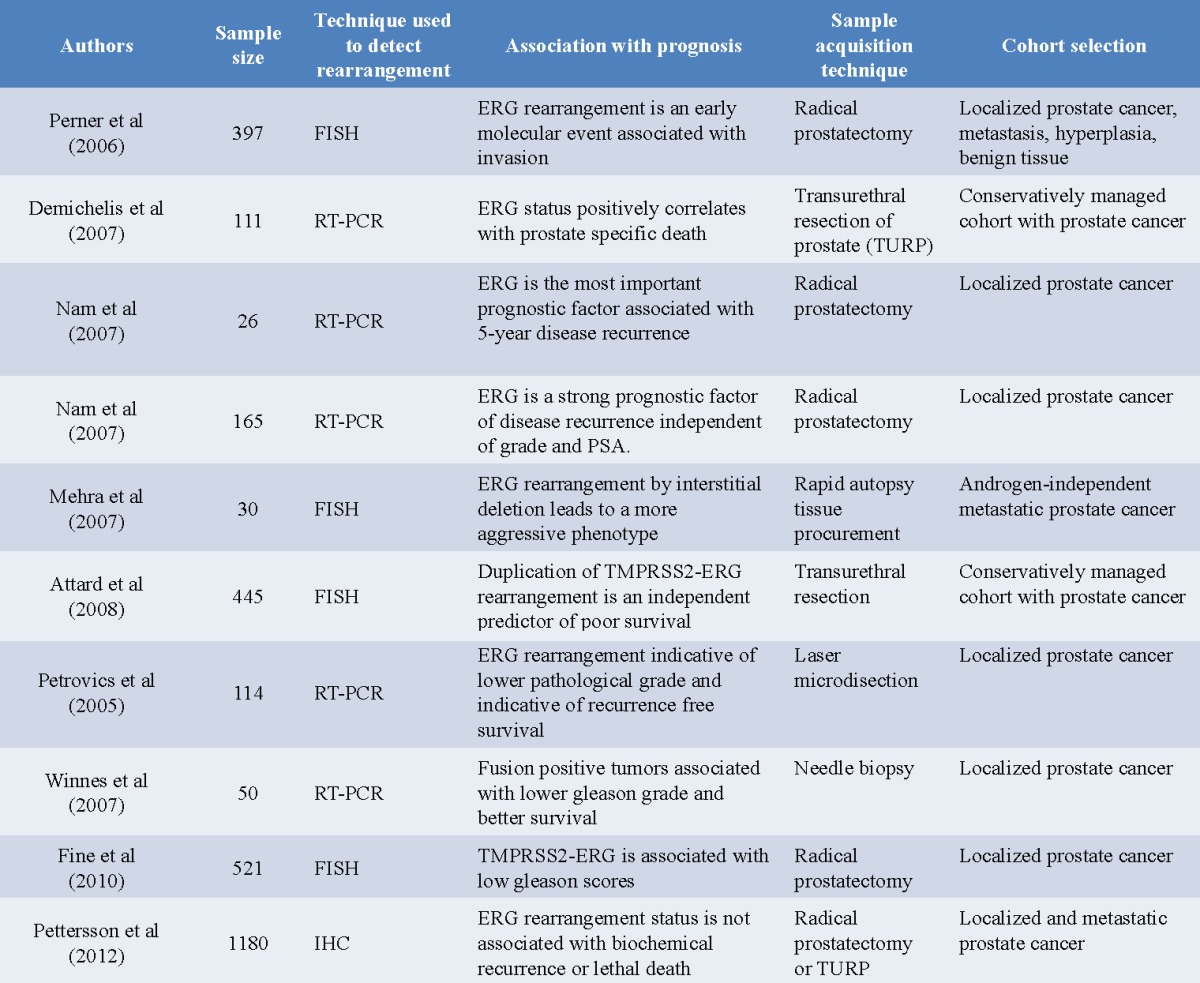
Recently, the influence of fusion status on clinical outcome has become an area of intense investigation. Several groups have attempted to correlate the presence of ETS fusion products with cancer stage and clinical outcomes [15,18,20,65]. Although most studies associate TMPRSS2-ERG fusion status with higher Gleason scores, aggressive disease, poor prognosis and decreased incidence of disease free survival, there are differences in the methods, sample size and selection of the prostate patient cohort. Hence, conflicting reports have emerged as a result of these studies.
One of the first studies in this area was performed on a group of prostate cancer patients with localized prostate cancers that were subjected to expectant (watchful waiting) therapy [20]. The study revealed a positive correlation between the presence of the fusion protein and metastasis and prostate cancer specific death. A similar study conducted on patients undergoing radical prostatectomy for localized prostate cancer showed a higher rate of disease recurrence (58.4%) among patients exhibiting the fusion product as compared to patients that were fusion negative (8.1%) [66]. Using a multivariate analysis approach, this group also found that the presence of TMPRSS2-ERG is an important prognostic factor associated with disease recurrence [18]. In another study, TMPRSS2-ERG fusion was determined to be an early molecular event associated with invasion and metastasis [67]. Perhaps the most significant association between fusion status and clinical outcome is provided by a FISH based study of 445 prostate cancers from patients who had been conservatively managed [65]. Absence of ERG alterations was found to be a favorable indicator of cause specific survival (90% survival at 8 years). A new category of prostate tumors was also identified, characterized by a duplication of TMPRSS2-ERG fusion along with deletion of sequences 5’ to ERG. This class of tumors was termed as 2+Edel and patients harboring this mutation exhibited extremely poor cause specific survival (25% survival at 8 years). Moreover, the presence of 2+Edel fusion status as a prognostic factor was found to be independent of Gleason scores as well as serum PSA levels.
Other independent studies have demonstrated either no association or negative correlation between ERG fusion status and clinical outcome. In a multifocal prostate cancer tissue microarray based study, no correlation was found between TMPRSS2-ERG rearrangement and pathologic stage, Gleason score or recurrence free survival [28]. Similarly, another study ruled out any correlation between fusion status and seminal vesicle invasion or lymph node metastasis. Other groups, on the other hand have reported that fusion-positive tumors are associated with lower Gleason grades and better recurrence free survival [69,70]. These reports mirror observations made earlier by Petrovics et al [14] in a report preceding the discovery of TMPRSS2-ERG: ERG rearrangement, where ERG expressing prostate tumors were found to be moderately to well differentiated, while exhibiting a lower pathological grade and indicative of recurrence free survival after radical prostatectomy.
Recently, a large scale study was completed on a cohort of 1,180 men treated with radical prostatectomy. A meta-analysis of prior-research was also conducted as part of this study and it included 5,074 men followed for biochemical recurrence and 2,049 men followed for lethal disease. The results of this study suggest that TMPRSS2-ERG, or ERG overexpression, is associated with tumor stage but does not strongly predict recurrence or mortality among men treated with radical prostatectomy [71] (Table 2).
Table 2.
Summary of published studies demonstrating the association between ERG rearrangement status and clinical outcome
|
As of yet there is no equivocal consensus regarding the presence of fusion products and association with a particular phenotype, although most studies point towards a more aggressive disease state in the presence of genomic rearrangements. Also, it is difficult to compare different studies due to variability in the method of sample selection (watchful waiting/radical prostatectomy), end-point of studies (higher Gleason score/metastasis/cancer specific death), detection method (FISH/RT-PCR/IHC) and sample size. Interpretation of these studies is further complicated by the significant molecular heterogeneity in prostate cancer. Gene fusions may be present only in certain foci within the prostate and there is no way to determine whether the metastatic node is a direct result of the foci used to assign TMPRSS2-ERG rearrangement status.
TMPRSS2-ERG as a biomarker
Monitoring circulating levels of prostate specific antigen (PSA) is the most common form of screening for prostate cancer cases [72]. However, using circulating PSA levels as a biomarker for prostate cancer is complicated by the fact that PSA is also produced by benign prostate epithelia, benign prostatic hypertrophy and infection in the prostate [73]. Hence, PSA has limitations in both sensitivity and specificity. Recently, much interest has been attributed to the detection of TMPRSS2-ERG fusion transcripts to supplement or replace PSA testing as the benchmark standard for detecting prostate cancer. Unlike PSA, there is no evidence to suggest that truncated ERG protein is shed from the prostate. Instead, researchers have focused their efforts on detecting ERG fusion transcripts in the urine. As prostate cancer cells are routinely shed in the urine, TMPRSS2-ERG fusion transcripts can be easily detected by a non-invasive method [74,75]. The technique usually involves collecting urine immediately after a digital rectal exam (DRE) as the examination process results in shedding of prostate cells. The urine is sedimented by centrifugation, RNA is extracted and RT-PCR is used to measure the amount of fusion transcript present.
The technique was first used to quantify the amount of TMPRSS2-ERG transcripts present in the urine of 19 prostate cancer patients [74]. The presence of fusion transcript in the urine was indicative of fusion status in the prostate of patients. Patients whose urine did not contain the fusion product tested negative for TMPRSS2-ERG rearrangement when prostate tissue biopsies were probed by FISH. Another study attempted to quantify ERG mRNA in the urine of 237 patients scheduled to undergo needle biopsy of the prostate [76]. When normalized to PSA mRNA, a higher urine ERG score was found to be associated significantly with prostate cancer on biopsy. Importantly, urine ERG score performed best in men demonstrating PSA levels less than 4ng/ml. A diagnostic marker for this segment of screening populations is particularly important in order to determine which patients should be recommended for biopsy.
Other studies sought to develop a biomarker analysis profile consisting of TMPRSS2-ERG and multiple other biomarkers, which would provide superior results compared to conventional PSA testing. Van Gils et al [77] studied the diagnostic usefulness of a combination of TMPRSS2-ERG transcripts and prostate cancer antigen 3 (PCA3) RNA in urinary sediments after DRE. PCA3 is a non-coding RNA that is specific to the prostate and is over-expressed in prostate cancer [75]. The researchers found that although TMPRSS2-ERG fusion transcripts and PCA3 RNA could be detected in the urine with a sensitivity of 37% and 62% respectively, the combination of both markers increased the sensitivity to 73%. Laxman et al [78] probed an even wider multiplex of biomarkers as diagnostic tools for the early detection of prostate cancer in a cohort of 234 patients. They found that increased GOLPH2, SPINK1, PCA3 expression and TMPRSS2-ERG fusion status were significant predictors of prostate cancer with a sensitivity of 66% and specificity of 76% [76,78]. TMPRSS2-ERG is only found in about 50% of prostate cancer, which renders it highly specific but with low sensitivity. Hence, combining it with more sensitive markers such as PCA3 is a step forward in the right direction. Indeed, a recent multi-center study of 1312 men found that stratification by urine TMPRSS2-ERG and PCA3 score is associated with the presence of cancer, tumor volume, and clinically significant cancer in prostatectomy and biopsy patients [79]. Salami et al have since developed a multivariable algorithm which combines serum PSA, PCA3, and TMPRSS2-ERG and improves prostate cancer prediction with 90% specificity and 80% sensitivity [80].
Other ETS fusions in prostate cancer
Although TMPRSS2-ERG rearrangements form the dominant class of fusions found in prostate cancers, rearrangements involving other ETS family members have also been implicated [81]. ETV1 fusion products are estimated to be found in approximately 10% of prostate cancer cases. Unlike the ERG proto-oncogene, where the 3’-fusion partner is exclusively TMPRSS2, ETV1 can be rearranged with the 5’ untranslated region of several genes. In fact, TMPRSS2-ETV1 is found in less than 1% of prostate cancer cases. An added level of complexity is introduced by the fact that the 5’ ETV1 partners may or may not be androgen regulated.
In addition, the commonly used prostate cell-lines LNCaP and MDA-PCa 2B, which show the outlier ETV1 over-expression, only express the wild-type full-length ETV1 transcript. However, in these cells, the ETV1 is translocated from chromosome 7 to an intronic region on chromosome 14. This intronic region is prostate specific and is regulated by androgens in its entirety [82].
Similar to ERG, over-expression of ETV1 in benign prostatic epithelial cell-lines results in the induction of a subset of genes involved in migration and invasion [82]. However, unlike ERG, full-length and N-terminal truncated ETV1 seem to have differential activities [10]. Although, both gene products can confer invasiveness when over-expressed in benign prostate epithelia cell-lines, full-length ETV1 appears to be more oncogenic and is able to induce anchorage-independent growth. The mechanism for this difference in activity is not known and further studies are required to investigate the role of full-length and truncated ETV1 proteins on prostate cancer growth and invasion.
Transgenic mice that over-express ETV1 in the prostate develop prostatic intraepithelial neoplasia (PIN), without progression to invasive prostatic adenocarcinoma [83]. Thus, it is likely that, similar to ERG over-expression in mouse prostate, ETV1 fusion products function in the context of pre-existing genomic lesions.
Other ETS family members, such as ETV4, ETV5 and ELK4 are involved in chromosomal translocations in prostate cancer at a much lesser frequency (<1%). The mechanism driving the expression of these genes is similar, in that, the 5’ partner is usually the untranslated region of an androgen-regulated gene. Both ETV4 and ETV5 have been shown to fuse with the 5’ untranslated region of TMPRSS2 in human prostate cancer tissue samples [7,8]. Additionally, the androgen regulated gene SLC45A3 can be rearranged with both ETV5 and ELK4 [11]. Moreover, ETV4 can also fuse with KLK2 and CANT1, two prostate specific and androgen regulated genes [10]. In all cases, the fusion leads to an androgen-regulated over-expression of the 3’ ETS partner. Similar to ERG and ETV1, in-vitro studies have shown that the over-expression of these proteins confer an aggressive phenotype to prostate cancer cells, concomitant with a molecular signature that results in the over-expression of a subset of genes involved in invasion.
Unlike other ETS family members, ELK4 is an endogenously androgen regulated gene. It is expressed in both benign prostate epithelia, as well as prostate cancer samples, although the expression is higher in the latter case [11]. It is yet to be determined what advantages are conferred to the tumor by placing ELK4 under the regulation of a different androgen regulated promoter, namely SLC45A3. ELK and SLC45A3 are both located on chromosome 1 and it is believed that chromosomal translocation is not the major mechanism responsible for the fusion product. Whether the fusion product is a result of interstitial deletion or trans-splicing is yet to be determined.
Therapeutic applications
Inhibition of ERG protein and ERG pathway provides a promising therapeutic target for the treatment of prostate cancer and inhibition of prostate cancer metastasis. Transcription factors have been historically considered difficult targets due to the complex regulation of their target genes, lack of enzymatic activity and the widespread network of protein binding partners required for their function. However, the successful modulation of transcription factor function in several cancers has now revealed that this large and important class of proteins is indeed “druggable” (Figure 1).
Figure 1.
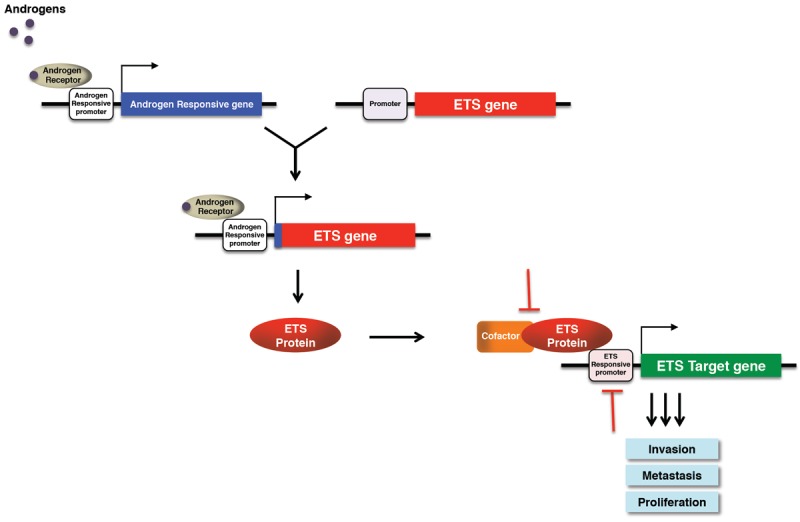
Chromosomal rearrangements involving the ETS family of transcription factors result in truncated ETS proteins that are fused to androgen regulated gene promoters. This lead to androgen regulation of ETS protein expression. ETS proteins regulate the expression of a subset of genes that increase cell proliferation, invasion and metastasis. Inhibition of ETS proteins provide a promising therapeutic target for the treatment of prostate cancer and inhibition of prostate cancer metastasis. These proteins can be targeted by blocking their binding to DNA or inhibiting their interaction with associated cofactors.
It should be noted that ETS proteins contain several regions of intrinsic disorder. Intrinsic disorder accords flexibility and movement to proteins, thus allowing them to participate in multi-protein complexes. These interactions are rapid, yet specific. In the case of transcription factors, disordered regions can implement and co-ordinate multiple functions, thus allowing the cell to regulate DNA binding, protein interactions and context-dependent gene regulation. A quick analysis of ERG amino acid sequence using disorder prediction softwares such as PONDR and DisEMBL reveals that whereas the ETS and SAM domains of ERG are relatively ordered, the remainder of the protein is intrinsically disordered. These intrinsically disordered regions are likely to be critical for ERG function.
We recently showed that YK-4-279, a small molecule inhibitor of EWS-FLI1 in Ewing’s sarcoma, robustly inhibits transcription by ERG and ETV1 in prostate cancer cells [84]. Inhibition of ERG and ETV1 activity resulted in reduced migration and invasion of rearrangement-positive prostate cancer cells. Cells lacking rearranged ERG or ETV1 were unresponsive to YK-4-279 treatment. Recently it was demonstrated that ERG transcriptional activity can also be modulated by preventing its binding to the promoter region of target genes. The heterocyclic dithiophene diamidine DB1255 specifically targets a portion of the ERG DNA recognition site by binding to the minor groove of the DNA as a dimer [85]. This leads to reduced ERG transcriptional activity.
Targeting downstream effectors or binding partners of ERG is also an optimal strategy for inhibiting ERG mediated effects on the cell. This can be a rational strategy with a fast track to clinical use, especially in cases where direct inhibitors are already available. A recent study identified phospholipase 2 group VII (PLA2G7) as a potential drug target in ERG oncogene positive prostate cancers [86]. PLA2G7 is involved in the cell’s response to oxidative stress and promotes cell migration and invasion. Silencing PLA2G7 by lipid-lowering statins sensitizes ERG-positive cells to oxidative stress and reduces proliferation [87]. Recent findings have identified Poly (ADP-Ribose) Polymerase (PARP), a key DNA repair protein, to interact with ERG in a DNA independent manner [88]. PARP1 is shown to be important for ERG protein function and inhibition of PARP1 impairs ERG mediated tumorigenesis and cell invasion. An important clinical advantage of this finding is that several PARP inhibitors are at an advanced stage of clinical testing, and this discovery has opened a wide spectrum of new strategies to target ERG.
Finally, in a recent study, Shao et al targeted TMPRSSS2-ERG mRNA transcripts using liposomal nanovectors packaged with siRNAs [89]. The targeting siRNAs significantly inhibited tumor growth in a mouse model. The degree of growth inhibition directly correlated with the extent of fusion gene knockdown. Growth inhibition was accompanied with inhibition of angiogenesis, reduced proliferation and an increase in apoptosis of tumor cells.
Concluding remarks
The discovery of ETS rearrangements in prostate cancer has greatly enhanced our understanding of prostate cancer pathology and changed our perception regarding the role of chromosomal rearrangements in solid tumor biology. As a diagnostic tool, it has the potential to complement, or even supersede PSA screening as a biomarker for prostate cancer detection. As a therapeutic target, it can be exploited to target primary tumors and prevent metastatic dissemination of tumor cells.
Acknowledgments
This review focused on articles published up to January 15, 2013. We sincerely apologize to the authors of many scientific studies that were published during this period and not discussed in this article due to space constraints.
References
- 1.Ordonez JL, Osuna D, Garcia-Dominguez DJ, Amaral AT, Otero-Motta AP, Mackintosh C, Sevillano MV, Barbado MV, Hernandez T, de Alava E. The clinical relevance of molecular genetics in soft tissue sarcomas. Adv Anat Pathol. 2010;17:162–181. doi: 10.1097/PAP.0b013e3181d98cbf. [DOI] [PubMed] [Google Scholar]
- 2.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 3.Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem. 2011;80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, Yan J, Talukder S, Turunen M, Taipale M, Stunnenberg HG, Ukkonen E, Hughes TR, Bulyk ML, Taipale J. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehra R, Tomlins SA, Yu J, Cao X, Wang L, Menon A, Rubin MA, Pienta KJ, Shah RB, Chinnaiyan AM. Characterization of TMPRSS2-ETS gene aberrations in androgen-independent metastatic prostate cancer. Cancer Res. 2008;68:3584–3590. doi: 10.1158/0008-5472.CAN-07-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, Mehra R, Chinnaiyan AM. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helgeson BE, Tomlins SA, Shah N, Laxman B, Cao Q, Prensner JR, Cao X, Singla N, Montie JE, Varambally S, Mehra R, Chinnaiyan AM. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68:73–80. doi: 10.1158/0008-5472.CAN-07-5352. [DOI] [PubMed] [Google Scholar]
- 8.Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, Cao X, Wei JT, Rubin MA, Shah RB, Chinnaiyan AM. TMPRSS2: ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 9.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermans KG, van der Korput HA, van Marion R, van de Wijngaart DJ, Ziel-van der Made A, Dits NF, Boormans JL, van der Kwast TH, van Dekken H, Bangma CH, Korsten H, Kraaij R, Jenster G, Trapman J. Truncated ETV1, fused to novel tissue-specific genes, and full-length ETV1 in prostate cancer. Cancer Res. 2008;68:7541–7549. doi: 10.1158/0008-5472.CAN-07-5930. [DOI] [PubMed] [Google Scholar]
- 11.Rickman DS, Pflueger D, Moss B, VanDoren VE, Chen CX, de la Taille A, Kuefer R, Tewari AK, Setlur SR, Demichelis F, Rubin MA. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res. 2009;69:2734–2738. doi: 10.1158/0008-5472.CAN-08-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pflueger D, Rickman DS, Sboner A, Perner S, LaFargue CJ, Svensson MA, Moss BJ, Kitabayashi N, Pan Y, de la Taille A, Kuefer R, Tewari AK, Demichelis F, Chee MS, Gerstein MB, Rubin MA. N-myc downstream regulated gene 1 (NDRG1) is fused to ERG in prostate cancer. Neoplasia. 2009;11:804–811. doi: 10.1593/neo.09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oikawa H, Utsugisawa T, Murai K, Narigasawa Y, Miyairi Y, Shimosegawa K, Suzuki T, Kuriya S. Tetrasomy of Philadelphia chromosome in myeloblastic crisis of chronic myelogenous leukemia. Int J Hematol. 1995;61:229–230. [PubMed] [Google Scholar]
- 14.Petrovics G, Liu A, Shaheduzzaman S, Furusato B, Sun C, Chen Y, Nau M, Ravindranath L, Dobi A, Srikantan V, Sesterhenn IA, McLeod DG, Vahey M, Moul JW, Srivastava S. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24:3847–3852. doi: 10.1038/sj.onc.1208518. [DOI] [PubMed] [Google Scholar]
- 15.Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, Tchinda J, Tomlins SA, Hofer MD, Pienta KG, Kuefer R, Vessella R, Sun XW, Meyerson M, Lee C, Sellers WR, Chinnaiyan AM, Rubin MA. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 16.Soller MJ, Isaksson M, Elfving P, Soller W, Lundgren R, Panagopoulos I. Confirmation of the high frequency of the TMPRSS2/ERG fusion gene in prostate cancer. Genes Chromosomes Cancer. 2006;45:717–719. doi: 10.1002/gcc.20329. [DOI] [PubMed] [Google Scholar]
- 17.Rajput AB, Miller MA, De Luca A, Boyd N, Leung S, Hurtado-Coll A, Fazli L, Jones EC, Palmer JB, Gleave ME, Cox ME, Huntsman DG. Frequency of the TMPRSS2:ERG gene fusion is increased in moderate to poorly differentiated prostate cancers. J Clin Pathol. 2007;60:1238–1243. doi: 10.1136/jcp.2006.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nam RK, Sugar L, Wang Z, Yang W, Kitching R, Klotz LH, Venkateswaran V, Narod SA, Seth A. Expression of TMPRSS2:ERG gene fusion in prostate cancer cells is an important prognostic factor for cancer progression. Cancer Biol Ther. 2007;6:40–45. doi: 10.4161/cbt.6.1.3489. [DOI] [PubMed] [Google Scholar]
- 19.Tu JJ, Rohan S, Kao J, Kitabayashi N, Mathew S, Chen YT. Gene fusions between TMPRSS2 and ETS family genes in prostate cancer: frequency and transcript variant analysis by RT-PCR and FISH on paraffin-embedded tissues. Mod Pathol. 2007;20:921–928. doi: 10.1038/modpathol.3800903. [DOI] [PubMed] [Google Scholar]
- 20.Demichelis F, Fall K, Perner S, Andren O, Schmidt F, Setlur SR, Hoshida Y, Mosquera JM, Pawitan Y, Lee C, Adami HO, Mucci LA, Kantoff PW, Andersson SO, Chinnaiyan AM, Johansson JE, Rubin MA. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 21.Mosquera JM, Mehra R, Regan MM, Perner S, Genega EM, Bueti G, Shah RB, Gaston S, Tomlins SA, Wei JT, Kearney MC, Johnson LA, Tang JM, Chinnaiyan AM, Rubin MA, Sanda MG. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res. 2009;15:4706–4711. doi: 10.1158/1078-0432.CCR-08-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosen P, Pfister D, Young D, Petrovics G, Chen Y, Cullen J, Bohm D, Perner S, Dobi A, McLeod DG, Sesterhenn IA, Srivastava S. Differences in Frequency of ERG Oncoprotein Expression Between Index Tumors of Caucasian and African American Patients With Prostate Cancer. Urology. 2012;80:749–753. doi: 10.1016/j.urology.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minner S, Gartner M, Freudenthaler F, Bauer M, Kluth M, Salomon G, Heinzer H, Graefen M, Bokemeyer C, Simon R, Sauter G, Schlomm T, Wilczak W. Marked heterogeneity of ERG expression in large primary prostate cancers. Mod Pathol. 2013;26:106–16. doi: 10.1038/modpathol.2012.130. [DOI] [PubMed] [Google Scholar]
- 24.Park K, Tomlins SA, Mudaliar KM, Chiu YL, Esgueva R, Mehra R, Suleman K, Varambally S, Brenner JC, MacDonald T, Srivastava A, Tewari AK, Sathyanarayana U, Nagy D, Pestano G, Kunju LP, Demichelis F, Chinnaiyan AM, Rubin MA. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12:590–598. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyagi Y, Sasaki T, Fujinami K, Sano J, Senga Y, Miura T, Kameda Y, Sakuma Y, Nakamura Y, Harada M, Tsuchiya E. ETS family-associated gene fusions in Japanese prostate cancer: analysis of 194 radical prostatectomy samples. Mod Pathol. 2010;23:1492–1498. doi: 10.1038/modpathol.2010.149. [DOI] [PubMed] [Google Scholar]
- 26.Furusato B, Tan SH, Young D, Dobi A, Sun C, Mohamed AA, Thangapazham R, Chen Y, McMaster G, Sreenath T, Petrovics G, McLeod DG, Srivastava S, Sesterhenn IA. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis. 2010;13:228–237. doi: 10.1038/pcan.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barry M, Perner S, Demichelis F, Rubin MA. TMPRSS2-ERG fusion heterogeneity in multifocal prostate cancer: clinical and biologic implications. Urology. 2007;70:630–633. doi: 10.1016/j.urology.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lapointe J, Kim YH, Miller MA, Li C, Kaygusuz G, van de Rijn M, Huntsman DG, Brooks JD, Pollack JR. A variant TMPRSS2 isoform and ERG fusion product in prostate cancer with implications for molecular diagnosis. Mod Pathol. 2007;20:467–473. doi: 10.1038/modpathol.3800759. [DOI] [PubMed] [Google Scholar]
- 29.Mehra R, Han B, Tomlins SA, Wang L, Menon A, Wasco MJ, Shen R, Montie JE, Chinnaiyan AM, Shah RB. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res. 2007;67:7991–7995. doi: 10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]
- 30.Mao X, Boyd LK, Yanez-Munoz RJ, Chaplin T, Xue L, Lin D, Shan L, Berney DM, Young BD, Lu YJ. Chromosome rearrangement associated inactivation of tumour suppressor genes in prostate cancer. Am J Cancer Res. 2011;1:604–617. [PMC free article] [PubMed] [Google Scholar]
- 31.Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, Palanisamy N, Chinnaiyan AM. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, Rosenfeld MG. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Cai Y, Ren C, Ittmann M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66:8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Cai Y, Yu W, Ren C, Spencer DM, Ittmann M. Pleiotropic biological activities of alternatively spliced TMPRSS2/ERG fusion gene transcripts. Cancer Res. 2008;68:8516–8524. doi: 10.1158/0008-5472.CAN-08-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Y, Dobi A, Sreenath T, Cook C, Tadase AY, Ravindranath L, Cullen J, Furusato B, Chen Y, Thangapazham RL, Mohamed A, Sun C, Sesterhenn IA, McLeod DG, Petrovics G, Srivastava S. Delineation of TMPRSS2-ERG splice variants in prostate cancer. Clin Cancer Res. 2008;14:4719–4725. doi: 10.1158/1078-0432.CCR-08-0531. [DOI] [PubMed] [Google Scholar]
- 36.Clark J, Merson S, Jhavar S, Flohr P, Edwards S, Foster CS, Eeles R, Martin FL, Phillips DH, Crundwell M, Christmas T, Thompson A, Fisher C, Kovacs G, Cooper CS. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene. 2007;26:2667–2673. doi: 10.1038/sj.onc.1210070. [DOI] [PubMed] [Google Scholar]
- 37.Mohamed AA, Tan SH, Mikhalkevich N, Ponniah S, Vasioukhin V, Bieberich CJ, Sesterhenn IA, Dobi A, Srivastava S, Sreenath TL. Ets family protein, erg expression in developing and adult mouse tissues by a highly specific monoclonal antibody. J Cancer. 2010;1:197–208. doi: 10.7150/jca.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLaughlin F, Ludbrook VJ, Cox J, von Carlowitz I, Brown S, Randi AM. Combined genomic and antisense analysis reveals that the transcription factor Erg is implicated in endothelial cell differentiation. Blood. 2001;98:3332–3339. doi: 10.1182/blood.v98.12.3332. [DOI] [PubMed] [Google Scholar]
- 39.Birdsey GM, Dryden NH, Amsellem V, Gebhardt F, Sahnan K, Haskard DO, Dejana E, Mason JC, Randi AM. Transcription factor Erg regulates angiogenesis and endothelial apoptosis through VE-cadherin. Blood. 2008;111:3498–3506. doi: 10.1182/blood-2007-08-105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loughran SJ, Kruse EA, Hacking DF, de Graaf CA, Hyland CD, Willson TA, Henley KJ, Ellis S, Voss AK, Metcalf D, Hilton DJ, Alexander WS, Kile BT. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat Immunol. 2008;9:810–819. doi: 10.1038/ni.1617. [DOI] [PubMed] [Google Scholar]
- 41.Taoudi S, Bee T, Hilton A, Knezevic K, Scott J, Willson TA, Collin C, Thomas T, Voss AK, Kile BT, Alexander WS, Pimanda JE, Hilton DJ. ERG dependence distinguishes developmental control of hematopoietic stem cell maintenance from hematopoietic specification. Genes Dev. 2011;25:251–262. doi: 10.1101/gad.2009211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng AP, Loughran SJ, Metcalf D, Hyland CD, de Graaf CA, Hu Y, Smyth GK, Hilton DJ, Kile BT, Alexander WS. Erg is required for self-renewal of hematopoietic stem cells during stress hematopoiesis in mice. Blood. 2011;118:2454–2461. doi: 10.1182/blood-2011-03-344739. [DOI] [PubMed] [Google Scholar]
- 43.Fingleton B. Matrix metalloproteinases: roles in cancer and metastasis. Front Biosci. 2006;11:479–491. doi: 10.2741/1811. [DOI] [PubMed] [Google Scholar]
- 44.de Launoit Y, Baert JL, Chotteau-Lelievre A, Monte D, Coutte L, Mauen S, Firlej V, Degerny C, Verreman K. The Ets transcription factors of the PEA3 group: transcriptional regulators in metastasis. Biochim Biophys Acta. 2006;1766:79–87. doi: 10.1016/j.bbcan.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 45.Magistroni V, Mologni L, Sanselicio S, Reid JF, Redaelli S, Piazza R, Viltadi M, Bovo G, Strada G, Grasso M, Gariboldi M, Gambacorti-Passerini C. ERG deregulation induces PIM1 overexpression and aneuploidy in prostate epithelial cells. PLoS One. 2011;6:e28162. doi: 10.1371/journal.pone.0028162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roh M, Gary B, Song C, Said-Al-Naief N, Tousson A, Kraft A, Eltoum IE, Abdulkadir SA. Overexpression of the oncogenic kinase Pim-1 leads to genomic instability. Cancer Res. 2003;63:8079–8084. [PubMed] [Google Scholar]
- 47.Cai J, Kandagatla P, Singareddy R, Kropinski A, Sheng S, Cher ML, Chinni SR. Androgens Induce Functional CXCR4 through ERG Factor Expression in TMPRSS2-ERG Fusion-Positive Prostate Cancer Cells. Transl Oncol. 2010;3:195–203. doi: 10.1593/tlo.09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ribeiro FR, Paulo P, Costa VL, Barros-Silva JD, Ramalho-Carvalho J, Jeronimo C, Henrique R, Lind GE, Skotheim RI, Lothe RA, Teixeira MR. Cysteine-rich secretory protein-3 (CRISP3) is strongly up-regulated in prostate carcinomas with the TMPRSS2-ERG fusion gene. PLoS One. 2011;6:e22317. doi: 10.1371/journal.pone.0022317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flajollet S, Tian TV, Flourens A, Tomavo N, Villers A, Bonnelye E, Aubert S, Leroy X, Duterque-Coquillaud M. Abnormal expression of the ERG transcription factor in prostate cancer cells activates osteopontin. Mol Cancer Res. 2011;9:914–924. doi: 10.1158/1541-7786.MCR-10-0537. [DOI] [PubMed] [Google Scholar]
- 50.Wai PY, Kuo PC. The role of Osteopontin in tumor metastasis. J Surg Res. 2004;121:228–241. doi: 10.1016/j.jss.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 51.Akashi T, Koizumi K, Tsuneyama K, Saiki I, Takano Y, Fuse H. Chemokine receptor CXCR4 expression and prognosis in patients with metastatic prostate cancer. Cancer Sci. 2008;99:539–542. doi: 10.1111/j.1349-7006.2007.00712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roh M, Franco OE, Hayward SW, van der Meer R, Abdulkadir SA. A role for polyploidy in the tumorigenicity of Pim-1-expressing human prostate and mammary epithelial cells. PLoS One. 2008;3:e2572. doi: 10.1371/journal.pone.0002572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun C, Dobi A, Mohamed A, Li H, Thangapazham RL, Furusato B, Shaheduzzaman S, Tan SH, Vaidyanathan G, Whitman E, Hawksworth DJ, Chen Y, Nau M, Patel V, Vahey M, Gutkind JS, Sreenath T, Petrovics G, Sesterhenn IA, McLeod DG, Srivastava S. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008;27:5348–5353. doi: 10.1038/onc.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 55.Gupta S, Iljin K, Sara H, Mpindi JP, Mirtti T, Vainio P, Rantala J, Alanen K, Nees M, Kallioniemi O. FZD4 as a mediator of ERG oncogene-induced WNT signaling and epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2010;70:6735–6745. doi: 10.1158/0008-5472.CAN-10-0244. [DOI] [PubMed] [Google Scholar]
- 56.Leshem O, Madar S, Kogan-Sakin I, Kamer I, Goldstein I, Brosh R, Cohen Y, Jacob-Hirsch J, Ehrlich M, Ben-Sasson S, Goldfinger N, Loewenthal R, Gazit E, Rotter V, Berger R. TMPRSS2/ERG promotes epithelial to mesenchymal transition through the ZEB1/ZEB2 axis in a prostate cancer model. PLoS One. 2011;6:e21650. doi: 10.1371/journal.pone.0021650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mochmann LH, Bock J, Ortiz-Tanchez J, Schlee C, Bohne A, Neumann K, Hofmann WK, Thiel E, Baldus CD. Genome-wide screen reveals WNT11, a non-canonical WNT gene, as a direct target of ETS transcription factor ERG. Oncogene. 2011;30:2044–2056. doi: 10.1038/onc.2010.582. [DOI] [PubMed] [Google Scholar]
- 58.Uysal-Onganer P, Kawano Y, Caro M, Walker MM, Diez S, Darrington RS, Waxman J, Kypta RM. Wnt-11 promotes neuroendocrine-like differentiation, survival and migration of prostate cancer cells. Mol Cancer. 2010;9:55. doi: 10.1186/1476-4598-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kunderfranco P, Mello-Grand M, Cangemi R, Pellini S, Mensah A, Albertini V, Malek A, Chiorino G, Catapano CV, Carbone GM. ETS transcription factors control transcription of EZH2 and epigenetic silencing of the tumor suppressor gene Nkx3.1 in prostate cancer. PLoS One. 2010;5:e10547. doi: 10.1371/journal.pone.0010547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abate-Shen C, Shen MM, Gelmann E. Integrating differentiation and cancer: the Nkx3.1 homeobox gene in prostate organogenesis and carcinogenesis. Differentiation. 2008;76:717–727. doi: 10.1111/j.1432-0436.2008.00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mohamed AA, Tan SH, Sun C, Shaheduzzaman S, Hu Y, Petrovics G, Chen Y, Sesterhenn IA, Li H, Sreenath T, McLeod DG, Dobi A, Srivastava S. ERG oncogene modulates prostaglandin signaling in prostate cancer cells. Cancer Biol Ther. 2011;11:410–417. doi: 10.4161/cbt.11.4.14180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, Cai Y, Shao LJ, Siddiqui J, Palanisamy N, Li R, Ren C, Ayala G, Ittmann M. Activation of NF-{kappa}B by TMPRSS2/ERG Fusion Isoforms through Toll-Like Receptor-4. Cancer Res. 2011;71:1325–1333. doi: 10.1158/0008-5472.CAN-10-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS, Gerald WL, Sawyers CL. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, Cordon-Cardo C, Gerald W, Pandolfi PP. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Attard G, Clark J, Ambroisine L, Fisher G, Kovacs G, Flohr P, Berney D, Foster CS, Fletcher A, Gerald WL, Moller H, Reuter V, De Bono JS, Scardino P, Cuzick J, Cooper CS. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nam RK, Sugar L, Yang W, Srivastava S, Klotz LH, Yang LY, Stanimirovic A, Encioiu E, Neill M, Loblaw DA, Trachtenberg J, Narod SA, Seth A. Expression of the TMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. Br J Cancer. 2007;97:1690–1695. doi: 10.1038/sj.bjc.6604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J, Collins C, Bismar TA, Chinnaiyan AM, De Marzo AM, Rubin MA. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 68.Mehra R, Tomlins SA, Shen R, Nadeem O, Wang L, Wei JT, Pienta KJ, Ghosh D, Rubin MA, Chinnaiyan AM, Shah RB. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007;20:538–544. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- 69.Winnes M, Lissbrant E, Damber JE, Stenman G. Molecular genetic analyses of the TMPRSS2-ERG and TMPRSS2-ETV1 gene fusions in 50 cases of prostate cancer. Oncol Rep. 2007;17:1033–1036. [PubMed] [Google Scholar]
- 70.Fine SW, Gopalan A, Leversha MA, Al-Ahmadie HA, Tickoo SK, Zhou Q, Satagopan JM, Scardino PT, Gerald WL, Reuter VE. TMPRSS2-ERG gene fusion is associated with low Gleason scores and not with high-grade morphological features. Mod Pathol. 2010;23:1325–1333. doi: 10.1038/modpathol.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pettersson A, Graff RE, Bauer SR, Pitt MJ, Lis RT, Stack EC, Martin NE, Kunz L, Penney KL, Ligon AH, Suppan C, Flavin R, Sesso HD, Rider JR, Sweeney C, Stampfer MJ, Fiorentino M, Kantoff PW, Sanda MG, Giovannucci EL, Ding EL, Loda M, Mucci LA. The TMPRSS2:ERG rearrangement, ERG expression, and prostate cancer outcomes: a cohort study and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012;21:1497–1509. doi: 10.1158/1055-9965.EPI-12-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thompson IM, Ankerst DP, Chi C, Goodman PJ, Tangen CM, Lucia MS, Feng Z, Parnes HL, Coltman CA Jr. Assessing prostate cancer risk: results from the Prostate Cancer Prevention Trial. J Natl Cancer Inst. 2006;98:529–534. doi: 10.1093/jnci/djj131. [DOI] [PubMed] [Google Scholar]
- 73.Picard JC, Golshayan AR, Marshall DT, Opfermann KJ, Keane TE. The multi-disciplinary management of high-risk prostate cancer. Urol Oncol. 2012;30:3–15. doi: 10.1016/j.urolonc.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 74.Laxman B, Tomlins SA, Mehra R, Morris DS, Wang L, Helgeson BE, Shah RB, Rubin MA, Wei JT, Chinnaiyan AM. Noninvasive detection of TMPRSS2:ERG fusion transcripts in the urine of men with prostate cancer. Neoplasia. 2006;8:885–888. doi: 10.1593/neo.06625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA, Debruyne FM, Ru N, Isaacs WB. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59:5975–5979. [PubMed] [Google Scholar]
- 76.Rice KR, Chen Y, Ali A, Whitman EJ, Blase A, Ibrahim M, Elsamanoudi S, Brassell S, Furusato B, Stingle N, Sesterhenn IA, Petrovics G, Miick S, Rittenhouse H, Groskopf J, McLeod DG, Srivastava S. Evaluation of the ETS-related gene mRNA in urine for the detection of prostate cancer. Clin Cancer Res. 2010;16:1572–1576. doi: 10.1158/1078-0432.CCR-09-2191. [DOI] [PubMed] [Google Scholar]
- 77.van Gils MP, Hessels D, Hulsbergen-van de Kaa CA, Witjes JA, Jansen CF, Mulders PF, Rittenhouse HG, Schalken JA. Detailed analysis of histopathological parameters in radical prostatectomy specimens and PCA3 urine test results. Prostate. 2008;68:1215–1222. doi: 10.1002/pros.20781. [DOI] [PubMed] [Google Scholar]
- 78.Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, Mehra R, Lonigro RJ, Tsodikov A, Wei JT, Tomlins SA, Chinnaiyan AM. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68:645–649. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Young A, Palanisamy N, Siddiqui J, Wood DP, Wei JT, Chinnaiyan AM, Kunju LP, Tomlins SA. Correlation of Urine TMPRSS2:ERG and PCA3 to ERG+ and Total Prostate Cancer Burden. Am J Clin Pathol. 2012;138:685–696. doi: 10.1309/AJCPU7PPWUPYG8OH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Salami SS, Schmidt F, Laxman B, Regan MM, Rickman DS, Scherr D, Bueti G, Siddiqui J, Tomlins SA, Wei JT, Chinnaiyan AM, Rubin MA, Sanda MG. Combining urinary detection of TMPRSS2:ERG and PCA3 with serum PSA to predict diagnosis of prostate cancer. Urol Oncol. 2011 doi: 10.1016/j.urolonc.2011.04.001. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tomlins SA, Bjartell A, Chinnaiyan AM, Jenster G, Nam RK, Rubin MA, Schalken JA. ETS gene fusions in prostate cancer: from discovery to daily clinical practice. Eur Urol. 2009;56:275–286. doi: 10.1016/j.eururo.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 82.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, Yu J, Wang L, Montie JE, Rubin MA, Pienta KJ, Roulston D, Shah RB, Varambally S, Mehra R, Chinnaiyan AM. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 83.Bradford TJ, Tomlins SA, Wang X, Chinnaiyan AM. Molecular markers of prostate cancer. Urol Oncol. 2006;24:538–551. doi: 10.1016/j.urolonc.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 84.Rahim S, Beauchamp EM, Kong Y, Brown ML, Toretsky JA, Uren A. YK-4-279 inhibits ERG and ETV1 mediated prostate cancer cell invasion. PLoS One. 2011;6:e19343. doi: 10.1371/journal.pone.0019343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nhili R, Peixoto P, Depauw S, Flajollet S, Dezitter X, Munde MM, Ismail MA, Kumar A, Farahat AA, Stephens CE, Duterque-Coquillaud M, David Wilson W, Boykin DW, David-Cordonnier MH. Targeting the DNA-binding activity of the human ERG transcription factor using new heterocyclic dithiophene diamidines. Nucleic Acids Res. 2013;41:125–38. doi: 10.1093/nar/gks971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vainio P, Gupta S, Ketola K, Mirtti T, Mpindi JP, Kohonen P, Fey V, Perala M, Smit F, Verhaegh G, Schalken J, Alanen KA, Kallioniemi O, Iljin K. Arachidonic acid pathway members PLA2G7, HPGD, EPHX2, and CYP4F8 identified as putative novel therapeutic targets in prostate cancer. Am J Pathol. 2011;178:525–536. doi: 10.1016/j.ajpath.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vainio P, Lehtinen L, Mirtti T, Hilvo M, Seppanen-Laakso T, Virtanen J, Sankila A, Nordling S, Lundin J, Rannikko A, Oresic M, Kallioniemi O, Iljin K. Phospholipase PLA2G7, associated with aggressive prostate cancer, promotes prostate cancer cell migration and invasion and is inhibited by statins. Oncotarget. 2011;2:1176–1190. doi: 10.18632/oncotarget.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, Patel S, Wang X, Liang H, Yu J, Palanisamy N, Siddiqui J, Yan W, Cao X, Mehra R, Sabolch A, Basrur V, Lonigro RJ, Yang J, Tomlins SA, Maher CA, Elenitoba-Johnson KS, Hussain M, Navone NM, Pienta KJ, Varambally S, Feng FY, Chinnaiyan AM. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shao L, Tekedereli I, Wang J, Yuca E, Tsang S, Sood A, Lopez-Berestein G, Ozpolat B, Ittmann M. Highly Specific Targeting of the TMPRSS2/ERG Fusion Gene Using Liposomal Nanovectors. Clin Cancer Res. 2012;18:6648–57. doi: 10.1158/1078-0432.CCR-12-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]


