Abstract
Background
Germin-like superfamily members are ubiquitously expressed in various plant species and play important roles in plant development and defense. Although several GLPs have been identified in peanut (Arachis hypogaea L.), their roles in development and defense remain unknown. In this research, we study the spatiotemporal expression of AhGLPs in peanut and their functions in plant defense.
Results
We have identified three new AhGLP members (AhGLP3b, AhGLP5b and AhGLP7b) that have distinct but very closely related DNA sequences. The spatial and temporal expression profiles revealed that each peanut GLP gene has its distinct expression pattern in various tissues and developmental stages. This suggests that these genes all have their distinct roles in peanut development. Subcellular location analysis demonstrated that AhGLP2 and 5 undergo a protein transport process after synthesis. The expression of all AhGLPs increased in responding to Aspergillus flavus infection, suggesting AhGLPs' ubiquitous roles in defense to A. flavus. Each AhGLP gene had its unique response to various abiotic stresses (including salt, H2O2 stress and wound), biotic stresses (including leaf spot, mosaic and rust) and plant hormone stimulations (including SA and ABA treatments). These results indicate that AhGLPs have their distinct roles in plant defense. Moreover, in vivo study of AhGLP transgenic Arabidopsis showed that both AhGLP2 and 3 had salt tolerance, which made transgenic Arabidopsis grow well under 100 mM NaCl stress.
Conclusions
For the first time, our study analyzes the AhGLP gene expression profiles in peanut and reveals their roles under various stresses. These results provide an insight into the developmental and defensive roles of GLP gene family in peanut.
Introduction
Peanut (Arachis hypogaea L.) is one of the major worldwide oil crops. Peanut has very high nutritional and commercial value. However, the increase in its production is hampered by pathogens such as fungi, bacteria, viruses, insect pests and physiological stresses caused by chemicals and salt. It is estimated that yield losses of peanut are up to about 30% due to various disease and adverse physiological conditions [1]. So it is an urge task to identify and characterized resistant genes in peanut development and defense. An insight into functions and usage of resistant genes will make a great progress in peanut cultivation.
Germins and germin-like proteins (GLPs) are plant exclusive cupin subfamily water-soluble glycoproteins. Germin was first identified during wheat germination [2] and later was found to be oxalate oxidases (OXOs) [3]. Germins and germin-like protein subfamily are characterized by the presence of germin boxes (PHIHPRATEI) and a conserved cupin superfamily derived-motif [4], [5]. This motif is a conserved beta-barrel protein with a metal ion binding ability [6]. According to their sequence similarities and other characters, Germins and the GLP gene family are divided into two distinct group proteins. The first group named “the true germins” is only identified in “true cereals”, which contain barley, corn, oat, rice, rye and wheat. Members in this group have relatively homogeneous protein sequences [7] and always carry OXO enzyme activity. The second group is designated as germin-like proteins (GLPs), whose members show relatively high sequence divergence. Their amino acid sequence similarity to wheat germin varies from 30% to 70%. The second group contains more numerous members than the first group and only few of the second group members possess OXO activity.
GLPs are a large gene family and have a wide range of distribution among plants. Expressed sequence tags (ESTs) or genomic sequencing have identified more than 100 GLPs. In higher plant Arabidopsis thaliana genome, 27 GLP genes have been discovered [8], [9]. Also, 14 GLP genes in barley and 8 GLP genes in rice have been identified [10], [11]. In lower plants, Nakata et al. have identified 77 EST clones of GLPs from Physcomitrella patens [12]. GLPs also have a wide range of expression in various plant organs and developmental stages. GLPs have been identified to express in a variety of tissues such as roots, leaves and flowers [13]–[15]. The ubiquitous distribution of GLPs implies the GLPs' fundamental and indispensable functions in plants. And their expression in various organs suggests that GLPs may execute roles in the development of various plant organs.
GLPs play critical roles not only in plant development but also in plant defense responses. Several evidences have suggested the functions of GLPs in plant defense [16]. One is the observation of increasing expression of certain GLPs in various plants under stresses like fungal, bacteria, and viruse infections [5], [17]–[20], parasite attacks, insect invasions [21], chemical toxicities, salt pressures [22], [23] and drought stresses [24]. The other evidence of GLPs' roles in plant defense is the enhanced resistance of transgenic GLP plants to various stresses. For example, transformation of a wheat GLP into soybean, sunflower and tobacco provided them the resistance to Sclerotinia sclerotiorum. Transient overexpression of GLPs in Barley resulted in enhanced plant resistance to the powdery mildew fungus [5]. It is proposed that the mechanism by which GLPs function plant defense responses is associated to their enzyme activity of OXO and superoxide dismutase (SOD), which can generate H2O2 to influence plant defense. Additional enzyme activities of GLPs that may function in plant defense include ADP glucose pyrophosphatase/phosphodiesterase (AGPPase) [25] and serine protease inhibitors [26].
Expressed sequence tags (ESTs) have identified 8 Arachis hypogaea L. germin-like proteins (AhGLPs) in peanut [27]. The previous studies in our lab have revealed that the expression level of AhGLP1, originally named oxalate oxidase (OXO), significantly increased in Aspergillus- resistant peanut seeds after both drought and A. flavus treatments [28]. Due to GLPs' key functions in plant development and defense, we determined the expression of AhGLPs during peanut development and characterized the functions of AhGLPs both in biotic and abiotic stresses. Our results in this study suggest that AhGLPs express in various organs during peanut development and play important roles in plant defense. AhGLPs are valuable resistant genes that can be used in improving peanut pathogen resistance, thus increasing the yield in peanut cultivation.
Results
Identification of new AhGLP homologue genes
Through homology matrix, we have identified three new homologous members in the AhGLP family. According to their sequence similarity, we termed them as AhGLP3a, AhGLP5a and AhGLP7a. Sequence alignments revealed that these three new members all share more than 97% nucleotide homologies with other corresponding members in the family (Figure S1). The genetic polymorphism between AhGLP3a and AhGLP3b includes two point mutations and one deletion (Table 1), which lead to 3 amino acid changes. Seven nucleotide variations between AhGLP5a and AhGLP5b, and ten nucleotide variations between AhGLP7a and AhGLP7b were identified, respectively. The genetic variations relative to AhGLP5a and AhGLP7a are all point mutations. Among the seven point mutations between AhGLP5a and AhGLP5b, three were silent mutations, the other four ones result in amino acid variations. Similarly, among the ten point mutations between AhGLP7a and AhGLP7b, three were silent mutations, the other seven ones produce amino acid variations.
Table 1. Difference of closely related multiple AhGLPs.
| Genesa | Nom. | Position | Type | Base Mutation | Protein Mutation |
| AhGLP3a vs AhGLP3b | 1 | 107–108 | Codon | AGC→- | Ser36→- |
| 2 | 124 | Point | G→A | Val43→Met42 | |
| 3 | 562 | Point | T→G | Phe189→Val188 | |
| AhGLP5a vs AhGLP5b | 1 | 31 | Point | G→A | Val11→Ile11 |
| 2 | 90 | Point | T→C | Tyr30 (Silent) | |
| 3 | 370 | Point | A→G | Ile124→Val124 | |
| 4 | 381 | Point | T→C | Asp127 (Silent) | |
| 5 | 429 | Point | C→T | Tyr143 (Silent) | |
| 6 | 553 | Point | C→T | Pro185→Ser185 | |
| 7 | 597 | Point | Nb→G | *199→Glu199 | |
| AhGLP7a vs AhGLP7b | 1 | 29 | Point | T→A | Ile10→Asn10 |
| 2 | 73 | Point | G→A | Ala25→Thr25 | |
| 3 | 150 | Point | T→C | Cys50 (Silent) | |
| 4 | 208 | Point | G→A | Ala70→Thr70 | |
| 5 | 241 | Point | G→A | Ala81→Thr81 | |
| 6 | 295 | Point | A→G | Thr99→Ala99 | |
| 7 | 307 | Point | C→T | Rrg106 (Silent) | |
| 8 | 363 | Point | A→G | Pro121 (Silent) | |
| 9 | 643 | Point | G→A | Glu215→Lys215 | |
| 10 | 651 | Point | Nb→T | *217→Ile217 |
AhGLP3a (GU457419.1), AhGLP5a (GU457421.1) and AhGLP7a (GU457423.1) were the known genes as reported by Chen et al (2011) in NCBI genebank, three newfound genes were designated AhGLP3b, AhGLP5b and AhGLP7b distinguished from AhGLP3a, AhGLP5a and AhGLP7a respectively.
un-know bases in the nucleotide sequence of AhGLP5a and AhGLP7a with indefinable amino acids (*).
Mapping of the developmental expression pattern of AhGLP mRNA in peanut
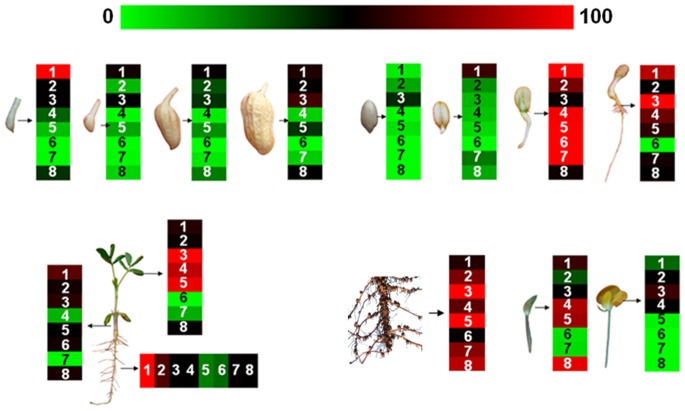
Expression pattern of the AhGLP family genes was first time examined in a variety of peanut tissues and various peanut developmental stages (Figure 1). During pods development (1–35d), AhGLP1 mRNAs level was the highest 1 day after gynophore penetrated into under-soil, and decreased afterward. The mRNA of AhGLP5 increased gradually during pods development. AhGLP2, AhGLP3 and AhGLP8 had similar expression pattern, which have the lowest level in the middle stages. Other three genes (AhGLP4, AhGLP6 and AhGLP7) expressed at a very low level in these stages. During peanut seed germination, the expression level of all the AhGLP genes reached the hightest at day 3, except that of AhGLP3 which was the hightest at day 5.
Figure 1. Expression patterns of peanut AhGLPs in various tissues/organs and developmental stages.
Expression patterns of AhGLPs were examined in (A) developing pods at 1, 4, 15 and 35 days after penetration into soil, (B) seeds at 1 hour, 1, 3 and 5 days after germination, (C) roots, stems and leaves of 14-day-old seedlings, and (D) roots, flower buds and flowers during flowering. Transcript abundance detected using qRT-PCR was normalized to expression of actin gene. Numbers (1–8) inside boxes correspond to AhGLP1, AhGLP2, AhGLP3b, AhGLP4, AhGLP5b, AhGLP6, AhGLP7b and AhGLP8, respectively. Expression levels are color-coded on the bottom bar. Green color indicates the down-regulation of gene expression, red color indicates the up-regulation of gene expression, black indicates the RNA levels unchanged.
The mRNA level of the AhGLP family genes was determined in roots, stems, leaves, flower buds, and flowers. In the stems, AhGLP1 had the highest mRNA level while AhGLP4 and AhGLP7 had the lowest expression level. In the leaves, AhGLP3, AhGLP4 and AhGLP5 were all highly expressed, while both AhGLP6 and AhGLP7 had a very low expression level. In the roots, AhGLP1 also had the highest mRNA level. On the contrary, the mRNA level of AhGLP5 and AhGLP6 was very low. All the AhGLP family genes had a high mRNA level in old roots. The mRNA in flower buds decreased in the sequence of AhGLP8, AhGLP4, AhGLP5, AhGLP1, AhGLP3, AhGLP2 and AhGLP6, AhGLP7. In peanut flowers, only AhGLP2, AhGLP3 and AhGLP4 had a medium level expression. The various expression patterns of different AhGLP family genes suggest their various functions in different tissues and during various development stages.
Determination of the subcellular localization of AhGLP proteins
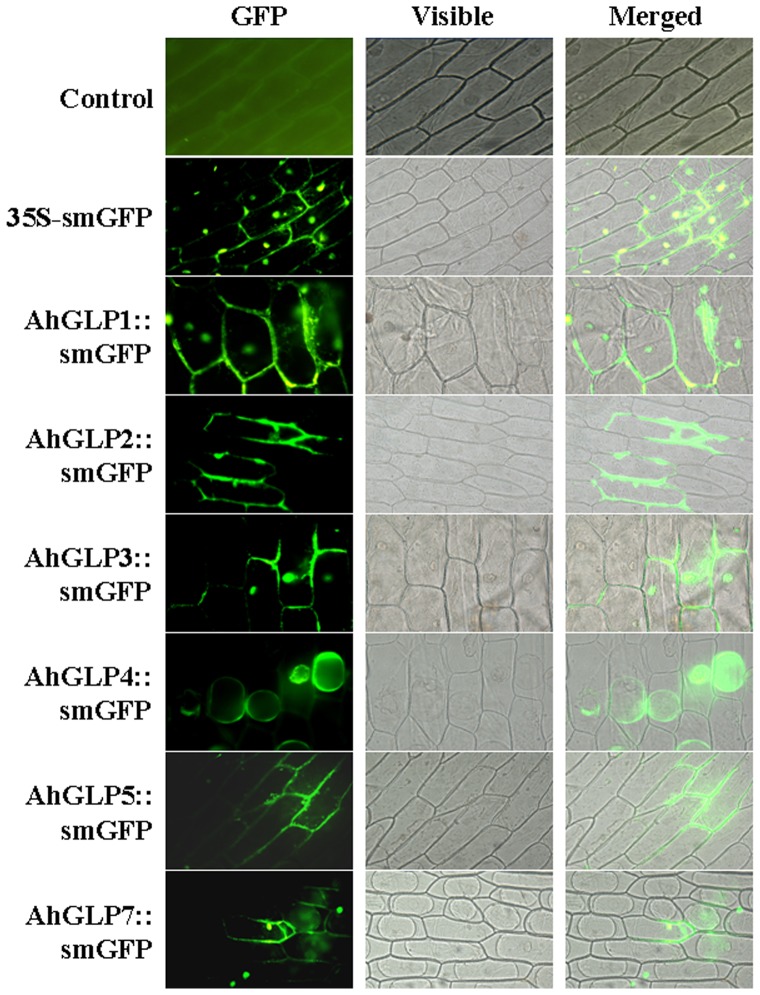
The protein sequence bioinformation analysis showed that most AhGLPs contain putative extracellular localization signal peptides in their N-terminal. The prediction suggests that AhGLP proteins potentially target to the cell membrane or cell wall [27]. To confirm this possibility, the coding region of each AhGLP gene was respectively fused to the N-terminus of soluble modified GFP (smGFP) gene, whose expression was under the control of the constitutive CaMV35S promoter (AhGLPs::smGFP). Onion epidermal cell is a convenient system and widely used for analyzing protein subcellular location [29]. The obtained constructs, together with non-AhGLP control vector were then introduced into onion epidermal cells by Agrobacterium-mediated transient transformation. Subcellular localization of AhGLP proteins in onion (Allium cepa) epidermal cells, indicated by the GFP signal was observed under fluorescence microscopy. The results showed that compared to the control vector (35S-GFP), AhGLP1, 3 and 7 had similar subcellular location pattern being distributed both in cytoplasm and plasma membrane or cell wall whereas AhGLP4 was only localized in cytoplasm (Figure 2). On the contrary, AhGLP2 and 5 were only localized to the plasma membrane or the cell wall. The results suggest that most of AhGLPs can localize to the cell membrane or cell wall in consistence with AhGLPs having putative extracellular localization signal peptides in their N-terminal.
Figure 2. Subcellular localization of AhGLPs-GFP proteins in onion epidermal cells.
Localization of AhGLPs-GFP fusion protein. Control: fluorescence of onion epidermal cells under empty vector. 35S-smGFP: onion epidermal cells expressing the GFP gene only driven by he 35S promoter. GFP fluorescence and differential interference contrast images and Visible/GFP merged images are shown from left to right.
Examination of the expression of AhGLPs in responding to A. flavus infection
Aspergillus flavus is a crop saprophyte that causes severe aflatoxin contamination to peanut seeds. To understand the possible roles of AhGLPs during A. flavus infection, we profiled the expression patterns of AhGLP genes in pre- and post-harvested peanut seeds after A. flavus infection. Compared to untreated control and drought-stresses treated pre-harvested seeds, according to T-test statistical analysis the expression level of AhGLP1, 2, 3, 4 and 5 were significantly up-regulated after A. flavus infections (Figure 3A). Conversely, the expression level of AhGLP6, 7 and 8 did not change much after A. flavus infections. In addition, according to T-test statistical analysis drought treatments significantly decreased the expression of AhGLP6. This suggests that AhGLP6 is the major gene responding to drought stresses.
Figure 3. Expression of AhGLPs in response to A. flavus infection in pre - and post- harvested peanut seeds.
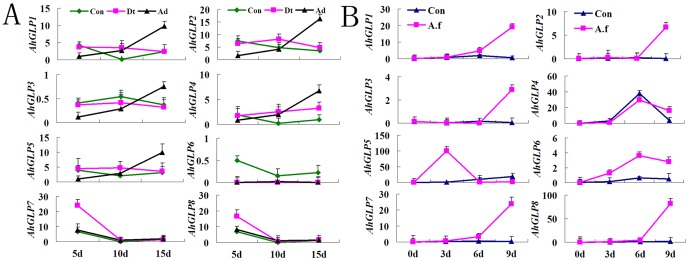
A: Con: control; Dt: drought stress; Ad: A. flavus infection under drought stress condition; B: Changes of the expression of AhGLP family genes in damp-dry peanut seed with 20% RH (relative humidity) under A. flavus infection. Con: control; A. f: A. flavus infection.
For a further confirmation, the expression of AhGLPs in response to A. flavus infection was tested in post-harvested peanut seeds with 20% relative humidity (RH). Compared to untreated control, the expression level of all AhGLPs except AhGLP5 was up-regulated 9 days after A. flavus infections (Figure 3B). And according to T-test statistical analysis, AhGLP5 also significantly increased in day 3 after the infection. This suggests that AhGLP5 is a quick responding gene in A. flavus infections. The data demonstrated that all AhGLPs are A. flavus repsonding genes and may have functions in peanut defense of A. flavus infections.
Profiling of the expression of AhGLPs under various biotic, abiotic and hormone stresses
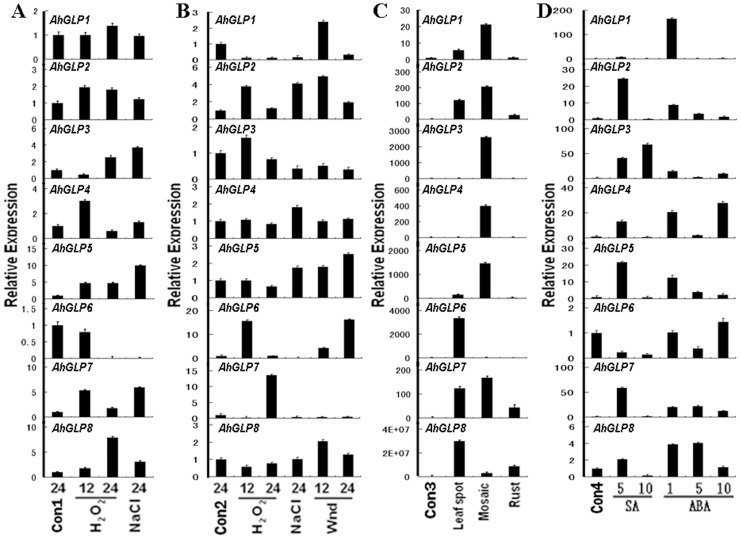
In order to determine the roles of AhGLPs in plant defense, we examined the expression of AhGLPs responding to a variety of biotic, abiotic and hormone stress treatments. 7-day-old peanut seedlings were treated with various abiotic stresses including H2O2, salt and wound. Expression of AhGLPs in leaves and roots was then determined by quantitative RT-PCR analysis. Compared to mock-treated control, according to T-test statistical analysis H2O2 treatments significantly increased the expression of AhGLP2, 3, 4, 5, 7 and 8 and decreased AhGLP6 expression in roots after 12 and/or 24 hours (Figure 4A). According to T-test statistical analysis, NaCl treatments significantly increased the expression of AhGLP3, 5, 7 and 8 and decrease AhGLP6 expression in root in 24 hours (Figure 4A). In the leaves, the expression of AhGLP2, 6 and 7 was significantly up-regulated while AhGLP1 expression significantly down-regulated 12 and/or 24 hours after H2O2 treatments according to T-test statistical analysis (Figure 4B). According to T-test statistical analysis, NaCl treatments significantly increased AhGLP2 level in leaves. The expression of AhGLP4 and 5 in leaves was up-regulated while the expression of AhGLP1, 3, 6 and 7 in leaves was down-regulated after 24 hours under NaCl treatments (Figure 4B). The level of AhGLP1, 2, 5, 6 and 8 was up-regulated, while the level of AhGLP3 and 7 was down-regulated 12 and/or 24 hours after wound treatments on the leaves (Figure 4B).
Figure 4. Differential expression of peanut AhGLP genes in response to various abiotic, biotic and hormone treatment conditions.
AhGLPs transcript abundance was detected by qRT-PCR analysis. Con1 and Con2: control sample of seedling leaf and root to abiotic stresses, respectively; Con3: control of 100-day-old peanut plant leaf to biotic stresses; Con4: control of seedling leaf to hormone treatments; Wnd: wound treatment; Numbers below bars correspond to duration of treatments (h); SA: salicylic acid; ABA: abscisic acid.
To demonstrate the functions of AhGLPs in plant disease defense, 80-day-old peanut plants were treated with a serial of biotic stresses including leaf spot, mosaic and rust. Then, the expression of AhGLPs mRNA in leaves was analyzed by qRT-PCR. According to T-test statistical analysis, leaf spot treatment significantly increased the expression of AhGLP1, 2, 5, 6, 7 and 8 (Figure 4C). According to T-test statistical analysis, mosaic treatment significantly increased the expression of all AhGLPs excpet AhGLP6 (Figure 4C). The level of AhGLP2, 7 and 8 was significantly up-regulated under rust treatments according to T-test statistical analysis (Figure 4C).
In order to examine the responses of AhGLPs to hormone stimulations, their mRNA level was determined in the leaves of 7-day-old peanut seedlings after the stimulations of hormones including salicylic acid (SA) and abscisic acid (ABA). According to T-test statistical analysis, the expression of AhGLP2, 3, 4, 5, 7 and 8 was all significantly up-regulated 5 h after SA stimulation (Figure 4D). Among them, only AhGLP3 expression kept going up, while the level of all others went down 10 hours after SA treatment. The level of AhGLP6 kept going down after SA treatments. The expression of all AhGLPs except AhGLP6 was up-regulated 1 h after ABA stimulation (Figure 4D). Except AhGLP7 and 8, the level of all other AhGLPs went down 5 hours after the treatments. The level of AhGLP7 and 8 also went down in 10 h, while the expression of AhGLP4 and 6 increased again 10 h after ABA treatments.
The results showed that various AhGLP gene expressions were induced in responding to different stresses stimulations. Their expressions induced were different and their expression had unique time windows under the stresses. This suggests that each AhGLP has their unique functions responding to various stresses in the plant defense. To have a comprehensive knowledge of how AhGLPs responding to various stimulations including hormone (including SA and ABA treatments), abiotic stress (including salt, H2O2 stress and wound) and biotic stress (including leaf spot, mosaic and rust), the overlap analysis of AhGLP genes response was performed (Figure 5). Most of the AhGLP genes including AhGLP1, 2, 3, 4, 5, 7 and 8 had increasing expression in responding to all of these three stimulations. Whereas, AhGLP6 expression was down-regulated in responding to most of the stimulations except wound and leaf spot treatments. Additionaly, the expression of AhGLP1, 3 and 7 was down-regulated under abiotic stresses. It is also interesting to note that all peanut GLP family genes primarily had a positive-response to biotic stresses through an increasing mRNA expression.
Figure 5. Venn diagram representing the expression profiles of AhGLP genes commonly or specifically regulated by various environmental stimuli, including plant hormones, abiotic stress and/or biotic stress in peanut leaves.
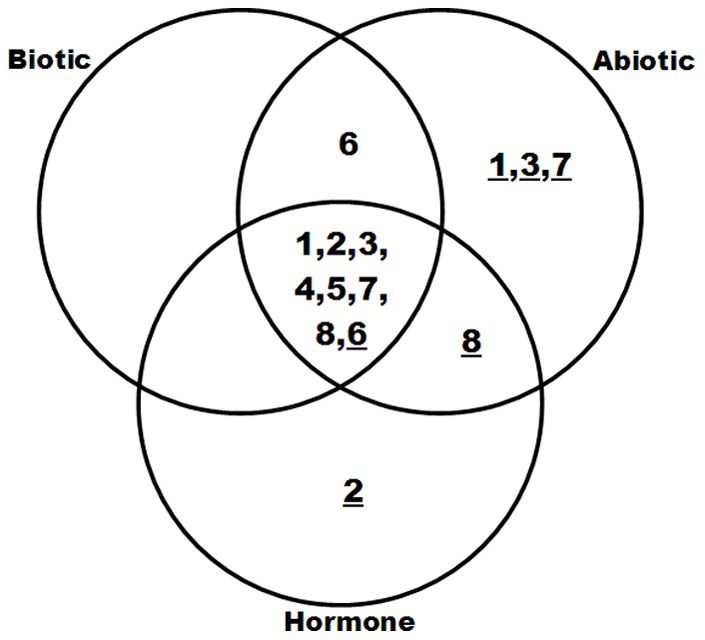
1, 2, 3, 4, 5, 6, 7, 8 were AhGLP1, AhGLP1, AhGLP2, AhGLP3, AhGLP4, AhGLP5, AhGLP6, AhGLP7 and AhGLP8, respectively. The underline numbers indicated down-regulated genes, and the numbers without underlines were up-regulated genes.
Verification of the salt tolerance functions of AhGLPs in transgenic Arabidopsis thaliana
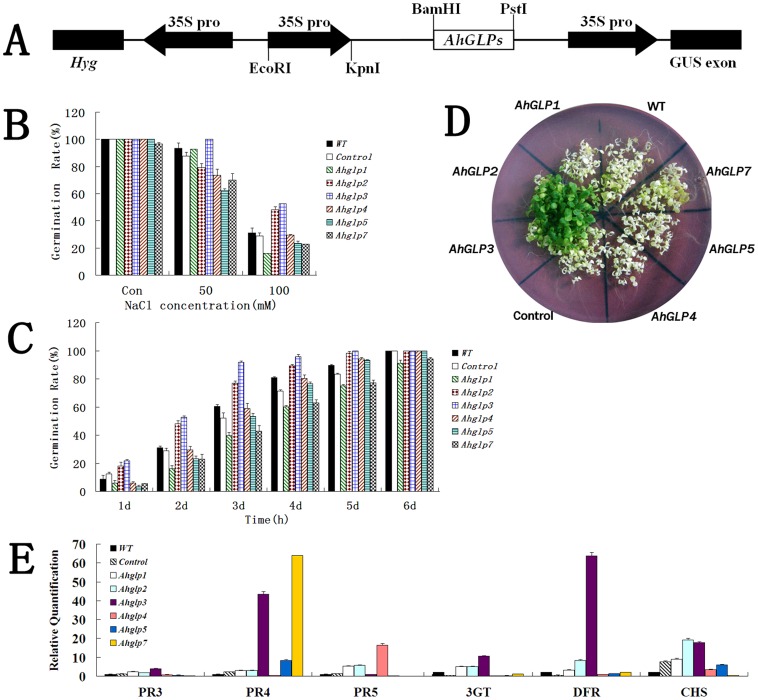
Due to the difficulty of stable transformation of peanut, to study the functions of AhGLPs in vivo we engineered transgenic Arabidopsis plants constitutively expressing AhGLPs through the cauliflower mosaic virus 35S constitutive promoter. After selection with 25 mg/L hygromycin at the T2 generation and confirmation by RT-PCR with the AhGLPs-specific primers, at least three independent transgenic lines carrying a single copy of the each AhGLP insertion were found to constitutively express the AhGLP genes, which could not be detected in wild-type Arabidopsis.
In previous study, NaCl treatments significantly increased the expression of several AhGLPs. To verify the functions of these AhGLPs in vivo, we challenge these transgenic Arabidopsis lines of T3 progeny with salt stresses. The result of the 3 independent lines were similar, here we choose to propose the data of one typical line. Wild type and transgenic Arabidopsis seeds of T3 progeny were grown under untreated and salt treated conditions, and their germination rate was calculated. Under untreated conditions, the germination rates of wild type, empty vector transgenic positive control and AhGLP transgenic Arabidopsis seeds were not significantly different (Figure 6B control). After treatment with various concentrations of NaCl, according to T-test statistical analysis both AhGLP2 and 3 overexpression Arabidopsis seeds displayed significantly higher germination rates than other seeds including wild type and empty vector transgenic control seeds (Figure 6B). The highest germination rates of Arabidopsis seeds overexpressing AhGLP2 and 3 maintained throughout the time points (from day 1 to day 6) (Figure 6C). To further confirm the salt tolerance roles of AhGLP2 and 3, Arabidopsis seedlings were treated with 100 mM NaCl for 15 days. When all other Arabidopsis seedlings grew badly with yellow leaves, the seedlings of AhGLP2- and 3- overexpressing Arabidopsis grew very well with green leaves (Figure 6D). These results suggest that AhGLP2 and 3 have roles in salt tolerance.
Figure 6. Overexpression of AhGLPs and salt tolerance analysis in transgenic Arabidopsis thaliana.
WT: wild type; control: The modified pCAMBIA1301 inserted with 35S only was used as negative control. (A): Diagrams of the constructs (the pCAMBIA1301-35S- AhGLP1, 2, 3, 4, 5 and 7) used for Agrobacterium tumefaciens-mediated transformation of Arabidopsis. (B): Effects of different NaCl content (0, 50 and 100 mM) on the germination of transgenic Arabidopsis seeds for 5 days after germination; (C): Comparison of germination rates and percentages of seedlings with green cotyledons between transgenic lines and wild-type plants under 100 mM NaCl stress. (D): The seeding cultivated on 1/2 MS agar plate containing 50 mM NaCl for 15 days. (E): Transcript analysis of AhGLPs-activated defense related genes (DFR, CHS, 3GT, and AtPR3, 4 and 5) in transgenic Arabidopsis plants.
To define a possible correlation between salt tolerance and defense gene expression, we analyzed the transcript levels of defense genes (PR3 to 5) and antioxidant-related genes (CHS, DFR and 3GT) in transgenic Arabidopsis plants through qRT-PCR. After T-test analysis, the expression of all genes except PR3 was significantly up-regulated in AhGLP2 overexpression Arabidopsis plants. After T-test analysis, the expression of all genes except PR5 was significantly up-regulated in AhGLP3 overexpression Arabidopsis plants (Figure 6E). This result pointed out that the salt tolerance of AhGLP2- and 3 have a positive correlation to defense gene expression level. The expression of PR4 was also up-regulated in AhGLP5- and 7- transgenic Arabidopsis, indicating their correlation. PR5 level was obviously up-regulated in AhGLP4 overexpression Arabidopsis, suggesting their positive correlation. Thus, the increasing level of defense genes in AhGLP2 and 3 transgenic Arabidopsis may partly contribute to their resistance to salt stresses.
Discussion
A great deal of GLP family members have been identified and divided into 5 subfamilies including bryophyte GLP subfamily, gymnosperm GLP subfamily, “ture gemin subfamily”, GLP subfamily 1, GLP subfamily 2 and GLP subfamily 3. Guo et al [27] reported the presence of eight GLP members in peanut based on the analysis of EST database and divided them into three classes including GLP subfamily I (AhGLP3a), GLP subfamily II (AhGLP2, 6 and 8) and GLP subfamily III (AhGLP1, 4, 5a and 7a) according their protein sequence feature. The sequence identities of these subfamily members ranged from 31.3% to 72.0%. In this study, we have identified three new AhGLP homologues named as AhGLP3b, AhGLP5b and AhGLP7b. They all showed highly homology compared with the corresponding AhGLP homologues like AhGLP3a (99.1%), AhGLP5a (98.6%) and AhGLP7a (97.7%) (Additional File 3). It has been found that the closely related GLPs including AtGLP2a/b (97.7% identity) and AtGLP3a/b (99.5% identity) in Arabidopsis had very similar functions [8]. So, we deduced that these closely related AhGLPs could also have the similar functions in peanut.
Although the spatiotemporal expression of germins and GLPs in different plant species has been characterized [30]–[33], their pattern in peanut remained unclear. In this study, we surveyed the transcript accumulation of the eight AhGLP genes across a wide range of tissues/organs and developmental stages of peanut through qRT-PCR analysis. Our results for the basal expression patterns of each of the peanut GLP genes (Figure1) suggest that each AhGLP has its distinct expression pattern in different tissues and stages, indicating that the spatiotemporal regulation of their expression is distinct. Furtheremore each AhGLP had its distinct function during peanut development. For example, AhGLP8 and AhGLP4 had a very high level in flower buds and significantly decreased in flowers, suggesting that these two genes may function in peanut flowering. The low expression level of AhGLP6 and AhGLP7 in both flower buds and flowers indicates that the two genes may not function in flowering. Also, the expression pattern of AhGLP1 suggests its potential roles in roots and pods development and seed germination. Besides, AhGLP3, AhGLP4 and AhGLP5 may function in leaf development and AhGLP3 may play a role during seed germination. Moreover, the specificity of AhGLPs expression paved the way that AhGLPs can be used as markers of various developmental stages in future.
It has been identified that in wheat about 40% germins are associated with cell wall and critical for development [34]. The N-terminal signal peptide in Germin and GLPs has been proved to help their secretion from the cell [35]. Cellular localization studies also confirmed the association of GLPs with the cell surface [20], [32]. In rice, a high abundance of GLPs has been shown to locate in epidermal cells [36]. Sequence analysis identified a signal peptide in AhGLPs N-terminal region, suggesting the possibility of AhGLP apoplastic or plasma membrane localization [27]. Our results showed that all AhGLPs are distributed at plasma membrane or cell wall (Figure 2). This confirmed their N-terminal signal peptide's functions. Among these AhGLPs, only AhGLP2 and 5 were excluded from the nucleus. The results suggest that AhGLP2 and 5 may perform a protein translocation process after synthesis.
The expression of GLPs and germin has been shown to be differentially regulated responding to pathogen attack in several plant species such as barley [5], rice [20], grapevine [31] and Arabidopsis [37]. They are expressed in a diverse range of tissues and have inkling of the broad spectrum of defensive activities in host-pahogen interaction [35]. It is worth mention that the overexpression of barley OXO gene results in enhanced resistance to the Sclerotinia minor in peanut [38]. In contrast, transient silencing of some barley GLP subfamilies increased the susceptibility to powdery mildew fungus [5]. Recently, our previous proteomics study revealed that the level of AhGLP1 protein, originally named as OXO, was positively correlated to A. flavus infection in pre-harvested resistant peanut seed [28]. In our study, the expression of most AhGLP family genes was also induced significantly when attacked by fungal pathogens (leaf spot, A. flavus) and bacteria (Rust disease) in peanut leaves (Figure. 4B). This suggests that AhGLPs play important roles in pathogen defense. To further understand the molecular function of AhGLP family genes in response to A. flavus infection, the expression patterns were analyzed in pre- and post-harvest peanut seeds during A. flavus invasion, respectively (Figure 3A and B). The results showed that all AhGLP mRNAs might response to A. flavus infection in seeds. Like GLPs in other plant species, our results suggested that AhGLPs are also broad spectrum and effective defense proteins against multiple pathogens.
Plants have a variety of strategies to adapt to unfavourable environmental conditions including various abiotic and biotic stresses [39]. Many studies have indicated that the germin and GLPs play important roles in resistance to various abiotic and/or biotic stresses [35]. In some model crops and plants, such as wheat, barley, rice and Arabidopsis, a number of GLP genes have been shown to function in response to stresses mainly thought their SOD [33], [40], [41], OXO [42] and AGPPase enzyme activity [25]. Although some of peanut GLP gene sequences were present in the GenBank database, so far only AhGLP2 has been identified to have SOD enzyme activity [27]. In this study, we analyze the differential expression of AhGLPs between peanut roots treated with different abiotic (NaCl, H2O2, wound) stresses and untreated control (Figure 4A). Same analysis was performed in the leaves. Six barley HvGER family genes [5] appeared to participate in multiple abiotic stress responses in leaf and root. Similarly, this analysis showed that all the peanut GLP genes were significantly differentially expressed under at least one of the abiotic stresses in leaves. Moreover, in roots, only AhGLP2, 3, 4, 5, 7 and 8 changed their expression after NaCl and H2O2 treatments. These results indicated that the expression of peanut GLP genes has tissue-variability and AhGLPs are involved in different regulation pathways in response to abiotic stresses. Phytohormones are important factors that participate in plant gene regulation networks involving abiotic and biotic response and tolerance [43]. The transcript level of some GLPs are enhanced or suppressed after application of phytohormones including salicylic acid (SA) [21], [31], [44] and abscisic acid (ABA) [45]. In this study, all the peanut GLP genes significantly changed their expression level under treatment of exogenous SA and ABA (Figure 4C). Among them, AhGLP6 expression was inhibited, the expression of all other seven members was induced. The result suggests that they might play roles in SA/ABA-dependent signaling transduction pathways during abiotic and biotic stress responses. Germin and GLPs are multi functional proteins, and many expression studies of these genes have shown crosstalk between various stimuli such as biotic, abiotic and hormone [5], [20], [31]. Our overlap analysis also found relation between tissue-/developmental stage-specific expression pattern and stress responses of peanut GLP genes (Figure 1, 2 and 3). It is noteworthy that AhGLP family genes showed broad spectrum stress responses mostly in peanut leaves. However, only AhGLP6 showed response to specific stimuli (Figure 3). GLPs resistance may be broad spectrum and effective against various environmental stimuli [20], [37]. These commonly regulated AhGLPs will provide some promising candidate genes for genetic engineering for improving crop resistance to different stresses.
GLP plays important roles in salt resistance. It has been proved that in responding to salt stress GLP expression increased in barley roots [22]. Moreover, salt stress can prolong the expression of GLP in barley [46]. Also, the GLP genes were proved to response to salt stress in soybean [47]. GLP proteins were gradually up-regulated during the period of salt treatment in wheat leaves [48]. Proteome analysis demonstrated that germin-like protein increased significantly in response to salt stress in the tobacco leaves [49]. In respond to salt stress, not only the expression of GLP but also the expression site changes. In salt-stressed wheat embryos, germin mRNA change their location to coleoptile cells instead of its original site, coleorhiza tissue [50]. In a moss, it is proposed that dissociation of GLP protein from the cell wall into the medium in the cells caused the induction of GLP gene by salt stress during the logarithmic phase [51]. Recently, it has been discovered that several other plant GLP genes can enhance the tolerance to salt stresses in transgenic plants [52]. After salt treatments, the expression of AhGLP3, 5 and 7 significantly increased in root. However, in the leaves only AhGLP2 expression increase obviously in response to salt treatment. The results suggest these 4 AhGLPs may function under salt stresses. To confirm this possibility, we engineered AhGLP transgenic Arabidopsis plants and challenge the plants with salt stresses. Both AhGLP2- and 3- overexpression Arabidopsis seeds displayed significantly higher germination rates than other seeds (Figure 6B). Moreover, the highest germination rates maintained throughout the time points (from day1 to day6) (Figure 5C). After treated with 100 mM NaCl for 15 days, only the seedlings of AhGLP2- and 3-overexpressing Arabidopsis still grew very well with green leaves (Figure 6D). These results suggested that both AhGLP2 and 3 genes were involved in the salt stress response and tolerance in plants.
Salt stress could accelerate production of active oxygen species (AOS) and subsequently cause oxidative damage in plants. Antioxidants, therefore, are elements of the salt stress response in a manner similar to general stress responses in plants [53], and various types of SODs are thought to have important roles in controlling oxidative stress [53]. Thus, one of possible explanation for the tolerance function of AhGLP-2 and 3 in Arabidopsis could be the SOD enzymatic activity, although only AhGLP2 have been identified to have SOD activity [27]. Moreover, it has been reported that the high level of flavonoid may be related to high salt stress tolerance through scavenging of stress-induced AOS [54]. In this study, Arabidopsis plants overexpressing AhGLP2 and 3 enhanced the transcript levels for flavonoid biosynthetic genes including DFR, CHS and 3GT than control plants. Furthermore, the expression of PR proteins (AtPR3, 4 and 5) in transgenic Arabidopsis with AhGLP was enhanced diversely. This result is consistent with the studies by Knecht [37], in which BvGLP1 overexpression in Arabidopsis induced SA- and JA-dependent PR genes transcripts. These transgenic lines could be better protected through activation of the defense genes even in the absence of the pathogens. This strongly suggests that AhGLP2 and 3 may play importantly functional roles in plants by specifically regulating the expression of a set of plant defense-related genes. The function of AhGLPs under pathogen attack will be further studied.
Conclusions
In general, expression patterns of 8 peanut GLP genes were analyzed in different tissues and stages. Their expression in response to various biotic stresses and abiotic stresses, and plant exogenous hormone treatments was also analyzed through qRT-PCR. The results revealed that expression levels of AhGLP family genes are varied greatly in different tissue-/developmental stage and various stresses. Moreover, six AhGLPs have been isolated and were used for expression in Arabidopsis and subcellular localization analysis in onion cells. The results indicated that AhGLP2 and 3 might be salt stress response and tolerance genes and most of the GLP genes located plasma membrane or cell wall.
Materials and Methods
Ethics Statement
No specific permits were required for the described field studies. No specific permissions were required for these locations and activities. The location is not privately-owned or protected in any way and the field studies did not involve endangered or protected species.
Plant material and sampling
A cultivar peanut (Arachis hypogeae. L) YJ-1 with resistance to Aspergillus flavus infection was provided by Crops Research Institute, Guangdong Academy of Agricultural Sciences (GDAAS, China). The seeds were surface sterilized using 70% (v/v) ethanol for 2 min, and then sown in pots of compost soil in a greenhouse under white fluorescent light (16 hr light/8 hr dark) at 30°C and 70% relative humidity. After germination, seedlings and plants were randomly divided into several groups (each containing six samples) and subjected to different stress treatments as explained below. Mature leaves, roots, stems, various stage panicles and seeds were collected. Roots from 7-day-old seedlings were also harvested.
For salt and H2O2 stress treatments, 7-day-old light-grown peanut seedlings were watered with solutions of 100 mM NaCl and 100 mM H2O2, respectively. After 12 and 24 h, the leaves and roots were collected. Likewise, 7-day-old seedlings were incubated with 100 µM ABA-solution and 50 µM salicylic acid (SA)-solution, respectively. And the leaves were collected after 1, 5 and 10 h. For the wound treatment, primary leaves of 7-day-old seedlings were rubbed gently with fine sandpaper and samples were collected after 12 and 24 h. Untreated samples were collected as controls at the same time points.
For peanut leaf disease treatments, 35-day-old plants were sprayed or inoculated with a spore suspension of leaf spot, mosaic and rust [55]. Triplicate samples of control and infected peanut leaves were collected. The post- and pre-harvested peanut seeds of cultivar YJ-1 were challenged with A. flavus according to the method of Liang [56] and Wang [28], respectively. The post-harvest seeds were collected 0, 3, 6 and 9 days after A. flavus infection, and the pre-harvest seeds were sampled 5, 10, 15 and 20 days after treatments with both drought and A. flavus stresses. All plant materials were snap frozen in liquid nitrogen and stored at −80°C.
RNA isolation and purification
Total RNA was isolated from peanut tissues and Arabidopsis leaves (wild-type and AhGLPs-transgenic) using TRIzol reagent (Invitrogen, Carlsbad, CA) according to manufacture's instructions. All samples were collected from three biological replicates [28]. All RNA extracts were treated with RNase-free DNase I (Takara, Dalian, China) then cleaned up with RNeasy Cleanup Kit (Qiagen, Beijing, China). RNA concentration and quality were assessed by Nano Drop ND-100 spectrophotometer (Nano Drop Technologies Inc., Delaware, USA) and electrophoresis on 1% agarose gel. The obtained RNA was stored at −80°C.
Quantitative real time RT-PCR
All qRT-PCRs were performed as described previously [28]. 4 µg of total RNA was reverse transcribed to cDNA using PrimeScript II 1st Strand cDNA Synthesis kit (TaKaRa, Dalian, China) according to the manufacturer's protocols. Quantitative real-time RT-PCR was performed with SYBR® Premix Ex Taq™ II kit (TaKaRa, Dalian, China) in LightCycler 480 instrument (Roche, Germany) equipped with Light-Cycler Software version 1.5 (Roche, Germany) according to the manufacturer's instructions [57].
All the primers specific for peanut AhGLPs and Arabidopsis stress-related genes were designed using the Primer version 5.0 (PREMIER Biosoft International) and listed in Table S1. The 18S rRNA and actin gene were used as internal controls for calculating relative transcript abundance in peanut and Arabidopsis, respectively. All real-time PCR reactions were repeated three times. The relative quantification of RNA expression was calibrated using formula 2−ΔΔCt method [58]. The mean of technical replicates was presented in the results. T-test analysis was performed to determine the statistical significance.
Generation of AhGLPs-transgenic Arabidopsis plants
The full-length coding sequence (ATG to TAA) of AhGLP1, 2, 3, 4, 5 and 7 were amplified by PCR with the gene-specific primers (Table S2). These PCR products were inserted into pCAMBIA 1301 vector, which expression was under the control of Cauliflower mosaic virus (CaMV) 35S promoter. After sequencing confirmation, the recombinant plasmids were introduced into Agrobacterium tumefaciens GV3101 through the freeze-thaw method and then introduced into wild-type Arabidopsis thaliana var. columbia by the floral dip method [59]. Transgenic Arabidopsis seeds were selected on solid Murashige and Skoog (MS) medium supplemented with 25 µg/L hygromycin (hyg). Independent hyg-resistant transgenic plants were further confirmed by PCR amplifications of the insertion cDNA.
Germination and tolerance analysis of AhGLPs in transgenic Arabidopsis
For in vivo salt-tolerance arrays, seeds from wild type and AhGLPs transgenic Arabidopsis were surface-sterilized as described by Clough and Bent [59] and sown on MS plates plus 2% sucrose containing 0, 50 and 100 mM NaCl, respectively. After stratification at 4°C for 3 days, plates were transferred in a growth chamber (100 µE m−2 s−1, 16 hr light/8 hr dark, 22°C). Germination (scored based on radicle emergence) was monitored daily for 6 days. After 100 mM NaCl treatment for 15 days, the seedlings were photographed and the tolerance of different transgenic Arabidopsis lines was observed. All the experiments were performed in duplicates. The mean of technical replicates was presented in the results. T-test analysis was performed to determine the statistical significance.
Subcellular localization of AhGLPs::GFP in onion epidermal cells
The coding sequences of AhGLP1, 2, 3, 4, 5 and 7 were amplified by PCR (Additional File 2) and fused to the 3′ region of the GFP gene, respectively. The AhGLP-GFP fusion genes were subcloned into the pCAMBIA1301 vector, for the expression under the control of CaMV35S promoter. These AhGLP-GFP fusion constructs and empty vector control were introduced into onion epidermal cells through the Agrobacterium-mediated system. The obtained cells were cultured on 1/2 MS medium at 26°C in darkness for 24 h. Subcellular localization of AhGLP proteins in onion (Allium cepa) epidermal cells indicated by the GFP signal was observed under fluorescence microscopy (Axio Observer A1, Zeiss, Germany). All transient expression assays were repeated at least three times.
Supporting Information
Homology matrix of predicted amino acid sequences of AhGLP family.
(DOC)
Primers used to quantify transcripts from the peanut GLP family and 18S genes by qRT-PCR.
(DOC)
Primers used for AhGLP s transgenic and subcellular localization analysis.
(DOC)
Funding Statement
This research was funded by grants from National Natural Science Foundation of China (No. 30971819, 31200211 and 30900907), Natural Science Foundation of Guangdong and Shandong Province (No. 10151064001000002 and ZR2012CQ031), Science and Technology Planning Project of Guangdong Province (2011B010500019), Pearl River Science and Technology Nova of Guangzhou (No. 2011J2200035) and supported by the earmarked fund for Modern Agro-industry Technology Research System (No. nycycx-19, CARS-14). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nelson SC, Simpson CE, Starr JL (1989) Resistance to Meloidogyne arenaria in Arachis spp. Germplasm. J Nematol 21 ((4S)): 654–660. [PMC free article] [PubMed] [Google Scholar]
- 2. Thompson EW Lane BG (1980) Relation of protein synthesis in imbibing wheat embryos to the cell-free translational capacities of bulk mRNA from dry and imbibing embryos. J Biol Chem 255 ((12)): 5965–5970. [PubMed] [Google Scholar]
- 3. Lane BG, Dunwell JM, Ray JA, Schmitt MR, Cuming AC (1993) Germin, a protein marker of early plant development, is an oxalate oxidase. J Biol Chem 268 ((17)): 12239–12242. [PubMed] [Google Scholar]
- 4. Dunwell JM, Khuri S, Gane PJ (2000) Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol Mol Biol Rev 64 ((1)): 153–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zimmermann G, Baumlein H, Mock HP, Himmelbach A, Schweizer P (2006) The multigene family encoding germin-like proteins of barley. Regulation and function in Basal host resistance. Plant Physiol 142 ((1)): 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chakraborty S, Chakraborty N, Jain D, Salunke DM, Datta A (2002) Active site geometry of oxalate decarboxylase from Flammulina velutipes: Role of histidine-coordinated manganese in substrate recognition. Protein Sci 11 ((9)): 2138–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lane BG (2000) Oxalate oxidases and differentiating surface structure in wheat: germins. Biochem J 349 ((Pt 1)): 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rietz S, Bernsdorff FE, Cai D (2012) Members of the germin-like protein family in Brassica napus are candidates for the initiation of an oxidative burst that impedes pathogenesis of Sclerotinia sclerotiorum. Journal of Experimental Botany 63 ((15)) 5507–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. El-Sharkawy I, Mila I, Bouzayen M, Jayasankar S (2010) Regulation of two germin-like protein genes during plum fruit development. Journal of experimental botany 61 ((6)) 1761–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Druka A, Kudrna D, Kannangara CG, von Wettstein D, Kleinhofs A (2002) Physical and genetic mapping of barley (Hordeum vulgare) germin-like cDNAs. Proc Natl Acad Sci U S A 99 ((2)): 850–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Caliskan M, Turet M, Cuming A (2004) Formation of wheat (Triticum aestivum L.) embryogenic callus involves peroxide-generating germin-like oxalate oxidase. Planta 219 ((1)) 132–140. [DOI] [PubMed] [Google Scholar]
- 12. Nakata M, Waanabe Y, Sakurai Y, Hashimoto Y, Matauzaki M, et al. (2004) Germin-like protein gene family of a moss, Physcomitrella patens, phylogenetically falls into two characteristic new clades. Plant Mol Biol 56 ((3)): 381–395. [DOI] [PubMed] [Google Scholar]
- 13. Li HY, Jiang J, Wang S, Liu FF (2010) Expression analysis of ThGLP, a new germin-like protein gene, in Tamarix hispida. Journal of Forestry Research 21 ((3)) 323–330. [Google Scholar]
- 14. Dunwell JM, Gibbings JG, Mahmood T, Naqvi SS (2008) Germin and germin-like proteins: evolution, structure, and function. Critical Reviews in Plant Sciences 27 ((5)) 342–375. [Google Scholar]
- 15. Bhattacharjee S (2005) Reactive oxygen species and oxidative burst: roles in stress, senescence and signal transduction in plants. CURRENT SCIENCE-BANGALORE- 89 ((7)) 1113. [Google Scholar]
- 16. Lane BG (2002) Oxalate, germins, and higher-plant pathogens. IUBMB Life 53 ((2)): 67–75. [DOI] [PubMed] [Google Scholar]
- 17. Schweizer P, Christoffel A, Dudler R (1999) Transient expression of members of the germin-like gene family in epidermal cells of wheat confers disease resistance. Plant J 20 ((5)): 541–552. [DOI] [PubMed] [Google Scholar]
- 18. Liang H, Maynard CA, Allen RD, Powell WA (2001) Increased Septoria musiva resistance in transgenic hybrid poplar leaves expressing a wheat oxalate oxidase gene. Plant Mol Biol 45 ((6)): 619–629. [DOI] [PubMed] [Google Scholar]
- 19. Ficke A, Gadoury DM, Seem RC (2002) Ontogenic resistance and plant disease management: a case study of grape powdery mildew. Phytopathology 92 ((6)): 671–675. [DOI] [PubMed] [Google Scholar]
- 20. Manosalva PM, Davidson RM, Liu B, Zhu X, Hulbert SH, et al. (2009) A germin-like protein gene family functions as a complex quantitative trait locus conferring broad-spectrum disease resistance in rice. Plant Physiol 149 ((1)): 286–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lou Y Baldwin IT (2006) Silencing of a germin-like gene in Nicotiana attenuata improves performance of native herbivores. Plant Physiol 140 ((3)): 1126–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hurkman WJ, Tao HP, Tanaka CK (1991) Germin-Like Polypeptides Increase in Barley Roots during Salt Stress. Plant Physiol 97 ((1)): 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hurkman WJ, Lane BG, Tanaka CK (1994) Nucleotide sequence of a transcript encoding a germin-like protein that is present in salt-stressed barley (Hordeum vulgare L.) roots. Plant Physiol 104 ((2)): 803–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ke Y, Han G, He H, Li J (2009) Differential regulation of proteins and phosphoproteins in rice under drought stress. Biochem Biophys Res Commun 379 ((1)): 133–138. [DOI] [PubMed] [Google Scholar]
- 25. Rodriguez-Lopez M, Baroja-Fernandez E, Zandueta-Criado A, Moreno-Bruna B, Munoz F, et al. (2001) Two isoforms of a nucleotide-sugar pyrophosphatase/phosphodiesterase from barley leaves (Hordeum vulgare L.) are distinct oligomers of HvGLP1, a germin-like protein. FEBS Lett 490 ((1–2)): 44–48. [DOI] [PubMed] [Google Scholar]
- 26. Segarra CI, Casalongue CA, Pinedo ML, Ronchi VP, Conde RD (2003) A germin-like protein of wheat leaf apoplast inhibits serine proteases. J Exp Bot 54 ((386)): 1335–1341. [DOI] [PubMed] [Google Scholar]
- 27. Guo B, Chen X, Hong Y, Liang X, Dang P, et al. (2009) Analysis of Gene Expression Profiles in Leaf Tissues of Cultivated Peanuts and Development of EST-SSR Markers and Gene Discovery. Int J Plant Genomics 2009: 715605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang T, Zhang E, Chen X, Li L, Liang X (2010) Identification of seed proteins associated with resistance to pre-harvested aflatoxin contamination in peanut (Arachis hypogaea L). BMC Plant Biol 10: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lou Y, Ma H, Lin WH, Chu ZQ, Mueller-Roeber B, et al. (2006) The highly charged region of plant beta-type phosphatidylinositol 4-kinase is involved in membrane targeting and phospholipid binding. Plant Mol Biol 60 ((5)): 729–746. [DOI] [PubMed] [Google Scholar]
- 30. Vallelian-Bindschedler L, Mosinger E, Metraux JP, Schweizer P (1998) Structure, expression and localization of a germin-like protein in barley (Hordeum vulgare L.) that is insolubilized in stressed leaves. Plant Mol Biol 37 ((2)): 297–308. [DOI] [PubMed] [Google Scholar]
- 31. Godfrey D, Able AJ, Dry IB (2007) Induction of a grapevine germin-like protein (VvGLP3) gene is closely linked to the site of Erysiphe necator infection: a possible role in defense? Mol Plant Microbe Interact 20 ((9)): 1112–1125. [DOI] [PubMed] [Google Scholar]
- 32. Heintzen C, Fischer R, Melzer S, Kappeler S, Apel K, et al. (1994) Circadian oscillations of a transcript encoding a germin-like protein that is associated with cell walls in young leaves of the long-day plant Sinapis alba L. Plant Physiol 106 ((3)): 905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kukavica B, Vucinic Z, Vuletic M (2005) Superoxide dismutase, peroxidase, and germin-like protein activity in plasma membranes and apoplast of maize roots. Protoplasma 226 ((3–4)): 191–197. [DOI] [PubMed] [Google Scholar]
- 34. Lane BG, Cuming AC, Fregeau J, Carpita NC, Hurkman WJ, et al. (1992) Germin isoforms are discrete temporal markers of wheat development. Pseudogermin is a uniquely thermostable water-soluble oligomeric protein in ungerminated embryos and like germin in germinated embryos, it is incorporated into cell walls. Eur J Biochem 209 ((3)): 961–969. [DOI] [PubMed] [Google Scholar]
- 35. Breen J, Bellgard M (2010) Germin-like proteins (GLPs) in cereal genomes: gene clustering and dynamic roles in plant defence. Funct Integr Genomics 10 ((4)): 463–476. [DOI] [PubMed] [Google Scholar]
- 36. Banerjee J, Maiti MK (2010) Functional role of rice germin-like protein1 in regulation of plant height and disease resistance. Biochem Biophys Res Commun 394 ((1)): 178–183. [DOI] [PubMed] [Google Scholar]
- 37. Knecht K, Seyffarth M, Desel C, Thurau T, Sherameti I, et al. (2010) Expression of BvGLP-1 encoding a germin-like protein from sugar beet in Arabidopsis thaliana leads to resistance against phytopathogenic fungi. Mol Plant Microbe Interact 23 ((4)): 446–457. [DOI] [PubMed] [Google Scholar]
- 38. Livingstone DM, Hampton JL, Phipps PM, Grabau EA (2005) Enhancing resistance to Sclerotinia minor in peanut by expressing a barley oxalate oxidase gene. Plant Physiol 137 ((4)): 1354–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ahuja I, de Vos RC, Bones AM, Hall RD (2010) Plant molecular stress responses face climate change. Trends Plant Sci 15 ((12)): 664–674. [DOI] [PubMed] [Google Scholar]
- 40. Woo EJ, Dunwell JM, Goodenough PW, Marvier AC, Pickersgill RW (2000) Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat Struct Biol 7 ((11)): 1036–1040. [DOI] [PubMed] [Google Scholar]
- 41. Yamahara T, Shiono T, Suzuki T, Tanaka K, Takio S, et al. (1999) Isolation of a germin-like protein with manganese superoxide dismutase activity from cells of a moss, Barbula unguiculata. J Biol Chem 274 ((47)): 33274–33278. [DOI] [PubMed] [Google Scholar]
- 42. Dumas B, Sailland A, Cheviet JP, Freyssinet G, Pallett K (1993) Identification of barley oxalate oxidase as a germin-like protein. C R Acad Sci III 316 ((8)): 793–798. [PubMed] [Google Scholar]
- 43. Moeder W, Ung H, Mosher S, Yoshioka K (2010) SA-ABA antagonism in defense responses. Plant Signal Behav 5 ((10)): 1231–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park CJ, An JM, Shin YC, Kim KJ, Lee BJ, et al. (2004) Molecular characterization of pepper germin-like protein as the novel PR-16 family of pathogenesis-related proteins isolated during the resistance response to viral and bacterial infection. Planta 219 ((5)): 797–806. [DOI] [PubMed] [Google Scholar]
- 45. Tabuchi TKT, Azuma T, Nanmori T, Yasuda T (2003) The expression of a germin-like protein with superoxide dismutase activity in the halophyte Atriplex lentiformis is differentially regulated by wounding and abscisic acid. Physiol Plant 118: 9. [Google Scholar]
- 46. Hurkman WJ, Tanaka CK (1996) Effect of Salt Stress on Germin Gene Expression in Barley Roots. Plant Physiol 110 ((3)): 971–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lu M, Han YP, Gao JG, Wang XJ, Li WB (2011) Identification and analysis of the germin-like gene family in soybean. BMC Genomics 11: 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kamal AH, Cho K, Kim DE, Uozumi N, Chung KY, et al. (2012) Changes in physiology and protein abundance in salt-stressed wheat chloroplasts. Mol Biol Rep 39 ((9)): 9059–9074. [DOI] [PubMed] [Google Scholar]
- 49. Dani V, Simon WJ, Duranti M, Croy RR (2005) Changes in the tobacco leaf apoplast proteome in response to salt stress. Proteomics 5 ((3)): 737–745. [DOI] [PubMed] [Google Scholar]
- 50. Caliskan M (2009) Salt stress causes a shift in the localization pattern of germin gene expression. Genet Mol Res 8 ((4)): 1250–1256. [DOI] [PubMed] [Google Scholar]
- 51. Nakata M, Shiono T, Watanabe Y, Satoh T (2002) Salt stress-induced dissociation from cells of a germin-like protein with Mn-SOD activity and an increase in its mRNA in a moss, Barbula unguiculata. Plant Cell Physiol 43 ((12)): 1568–1574. [DOI] [PubMed] [Google Scholar]
- 52. Patnaik D, Khurana P (2001) Germins and germin like proteins: an overview. Indian J Exp Biol 39 ((3)): 191–200. [PubMed] [Google Scholar]
- 53. Gucciardo S, Wisniewski JP, Brewin NJ, Bornemann S (2007) A germin-like protein with superoxide dismutase activity in pea nodules with high protein sequence identity to a putative rhicadhesin receptor. J Exp Bot 58 ((5)): 1161–1171. [DOI] [PubMed] [Google Scholar]
- 54. Hayashi M, Takahashi H, Tamura K, Huang J, Yu LH, et al. (2005) Enhanced dihydroflavonol-4-reductase activity and NAD homeostasis leading to cell death tolerance in transgenic rice. Proc Natl Acad Sci U S A 102 ((19)): 7020–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. De Jesus WC, Do Vale FXR, Coelho RR, Hau B, Zambolim L, et al. (2001) Effects of Angular Leaf Spot and Rust on Yield Loss of Phaseolus vulgaris. Phytopathology 91 ((11)): 1045–1053. [DOI] [PubMed] [Google Scholar]
- 56. Liang XQ, Holbrook CC, Lynch RE, Guo BZ (2005) beta-1,3-Glucanase Activity in Peanut Seed (Arachis hypogaea) is Induced by Inoculation with Aspergillus flavus and Copurifies with a Conglutin-Like Protein. Phytopathology 95 ((5)): 506–511. [DOI] [PubMed] [Google Scholar]
- 57. Alos E, Roca M, Iglesias DJ, Minguez-Mosquera MI, Damasceno CMB, et al. (2008) An evaluation of the basis and consequences of a stay-green mutation in the navel negra citrus mutant using transcriptomic and proteomic profiling and metabolite analysis. Plant Physiol 147 ((3)): 1300–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 ((4)): 402–408. [DOI] [PubMed] [Google Scholar]
- 59. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 ((6)): 735–743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Homology matrix of predicted amino acid sequences of AhGLP family.
(DOC)
Primers used to quantify transcripts from the peanut GLP family and 18S genes by qRT-PCR.
(DOC)
Primers used for AhGLP s transgenic and subcellular localization analysis.
(DOC)