Abstract
Introduction
In this study, 27 genetic polymorphisms that were previously reported to be associated with clinical outcomes in colorectal cancer patients were investigated in relation to overall survival (OS) and disease free survival (DFS) in colorectal cancer patients from Newfoundland.
Methods
The discovery and validation cohorts comprised of 532 and 252 patients, respectively. Genotypes of 27 polymorphisms were first obtained in the discovery cohort and survival analyses were performed assuming the co-dominant genetic model. Polymorphisms associated with disease outcomes in the discovery cohort were then investigated in the validation cohort.
Results
When adjusted for sex, age, tumor stage and microsatellite instability (MSI) status, four polymorphisms were independent predictors of OS in the discovery cohort MTHFR Glu429Ala (HR: 1.72, 95%CI: 1.04–2.84, p = 0.036), ERCC5 His46His (HR: 1.78, 95%CI: 1.15–2.76, p = 0.01), SERPINE1 −675indelG (HR: 0.52, 95%CI: 0.32–0.84, p = 0.008), and the homozygous deletion of GSTM1 gene (HR: 1.4, 95%CI: 1.03–1.92, p = 0.033). In the validation cohort, the MTHFR Glu429Ala polymorphism was associated with shorter OS (HR: 1.71, 95%CI: 1.18–2.49, p = 0.005), although with a different genotype than the discovery cohort (CC genotype in the discovery cohort and AC genotype in the validation cohort). When stratified based on treatment with 5-Fluorouracil (5-FU)-based regimens, this polymorphism was associated with reduced OS only in patients not treated with 5-FU. In the DFS analysis, when adjusted for other variables, the TT genotype of the ERCC5 His46His polymorphism was associated with shorter DFS in both cohorts (discovery cohort: HR: 1.54, 95%CI: 1.04–2.29, p = 0.032 and replication cohort: HR: 1.81, 95%CI: 1.11–2.94, p = 0.018).
Conclusions
In this study, associations of the MTHFR Glu429Ala polymorphism with OS and the ERCC5 His46His polymorphism with DFS were identified in two colorectal cancer patient cohorts. Our results also suggest that the MTHFR Glu429Ala polymorphism may be an adverse prognostic marker in patients not treated with 5-FU.
Introduction
Colorectal cancer has a high incidence in the developed countries [1]. In 2004 this disease was the 4th leading cause of death due to cancer with over 600,000 deaths worldwide [2]. In Canada, it is a major health concern with an estimated 22,200 new cases and 8,900 deaths expected in 2011 [3]. There are significant inter-provincial variations in incidence and mortality rates, and the province of Newfoundland and Labrador (NL) has the highest age-standardized incidence and mortality rates for colorectal cancer among the Canadian provinces [3]. Both genetic and environmental factors play a role in susceptibility to colorectal cancer. While the majority of the colorectal cancer patients are sporadic cases, nearly 5% of the colorectal cancers are caused by inherited high-penetrant mutations [4]. Thirty-five per cent of the risk for developing sporadic colorectal cancer is also attributed to the inherited factors [5].
Important colorectal cancer outcomes include recurrence, metastasis and death. Currently, the most valuable prognostic criterion in colorectal cancer patients is the TNM (tumor-node-metastasis) staging defined by the American Joint Committee on Cancer [6]. Generally, patient prognosis worsens with increasing stage.
A number of clinical and molecular parameters have also been investigated for their prognostic utility in colorectal cancer. For instance, Popat et al [7] reported in their meta-analysis that patients with microsatellite instability-high (MSI-H) tumors have a more favorable prognosis when compared to patients with microsatellite instability-low (MSI-L) or microsatellite stable (MSS) tumors. Several other clinicopathological and molecular features have also been reported to be associated with prognosis, such as high tumor grade [8], mucinous histology [9], lymphovascular invasion [10], chromosomal instability [11], and the presence of the BRAF1 Val600Glu somatic mutation in tumors [12], though contradictory reports have also been published [13]–[15]. Inconsistent results on the association between familial risk status and survival of colorectal cancer patients were also reported [16], [17]. Additionally, demographic factors such as gender and ethnicity may be modifiers of prognosis [6]. These factors only partly account for the variations in cancer patient outcomes and it is possible that genetic factors (such as single nucleotide polymorphisms (SNPs), insertion/deletion (indel) polymorphisms, and somatic mutations) may influence prognosis. Their investigation thus may help understanding the reasons for the inter-patient outcome variability and the underlying biological mechanisms.
Several studies have previously reported significant associations between genetic variations and outcomes in colorectal cancer patients. In the present study, we investigated 27 such polymorphisms ( Table 1 ) as potential prognostic factors in a colorectal cancer patient cohort (discovery cohort, n = 532) and subsequently tested the validity of the positive associations in an additional colorectal cancer patient cohort (validation cohort, n = 252).
Table 1. Polymorphisms investigated in the discovery cohort.
| Pathway | Gene | Polymorphism | SNP ID | Minor alleleand MAF inthe discoverycohort(replicationcohort) | ****MAF(Caucasian) | Chromosome,location | Functional impact of the polymorphism |
| Cell cycle | CCND1 | Pro241Pro, A/G | *rs9344 | A, 45.28% | 48–63% | Chr 11, 69462910 | G allele causes an alternative transcript of CCND1 mRNA [55] |
| Cell adhesion & signaling | DCC | Arg201Gly, C/G | *rs2229080 | G, 36.98% | 33–42% | Chr 18, 50432602 | G allele associated with reduced gene expression [56] |
| Cell signaling | EGFR | Arg521Lys, G/A | *rs2227983 | A, 26.89% | 22–30% | Chr 7, 55229255 | A allele associated with reduced ability of EGFR to induce cell growth [57] |
| FGFR4 | Gly388Arg, A/G | *rs351855 | T, 31.26% | 26–31% | Chr 5, 176520243 | G allele associated with greater motility and progression of cancer cells [58] | |
| DNA repair | ERCC1 | Asn118Asn, C/T | *rs11615 | C, 37.57% | 33–45% | Chr 19, 45923653 | T allele associated with reduced gene expression [59] |
| ERCC2 | Lys751Gln, G/T | *rs13181 | G, 35.69% | 27–42% | Chr 19, 45854919 | G allele linked with inefficient DNA repair [60] | |
| ERCC5 | His46His, C/T | *rs1047768 | T, 41.13% (42.15%) | 32–51% | Chr 13, 103504517 | unknown | |
| EXO1 | Pro757Leu, C/T | *rs9350 | T, 14.6% | 15–27% | Chr 1, 242048674 | unknown | |
| OGG1 | Ser326Cys, C/G | *rs1052133 | G, 23.54% | 15–22% | Chr 3, 9798773 | G allele reduces DNA binding and repair activity [61] | |
| MLH1 | Ile219Val, A/G | *rs1799977 | G, 28.63% | 0–35% | Chr 3, 3705356 | unknown | |
| XRCC1 | Arg399Gln, G/A | *rs25487 | A, 34.36% | 37–58% | Chr 19, 44055726 | A allele associated with impaired DNA repair function [62] | |
| XRCC3 | Thr241Met, C/T | *rs861539 | T, 39.74% | 37–65% | Chr 14, 104165753 | T allele associated with defective DNA repair mechanism [63] | |
| Apoptosis | FAS | c.-24+733T>C | *rs1800682 | C, 44.91% | 39–50% | Chr 10, 90749963 | unknown |
| Drug metabolism | GSTM1 | gene deletion | **na | non-deletion allele, 45.10% (44.54%) | *****38–62% | loss of gene function | |
| GSTP1 | Ile105Val, A/G | *rs1695 | G, 36.67% | 29–42% | Chr 11, 67352689 | G allele associated with reduced enzymatic activity [64] | |
| GSTT1 | gene deletion | **na | deletion allele, 17% | *****15–20% | loss of gene function | ||
| Inflammation | IL6 | −174G/C in promoter | *rs1800795 | C, 44.25% | 50–57% | Chr 7, 22766645 | C allele linked with reduced gene expression [65] |
| PTGS2 | c.3618A/G in 3′-UTR | *rs4648298 | G, 1.63% | 1.7–1.8% | Chr 1, 186641682 | unknown | |
| Tissue remodeling | MMP1 | −1607 indel G inpromoter | ***rs1799750 | G, 46.9% | 43.30% | Chr 11, 102670496 | insG allele associated with enhanced gene transcription [66] |
| MMP2 | −1306C/T in promoter | *rs243865 | T, 22.92% | 18–25% | Chr 16, 55511806 | T allele associated with reduced gene expression [67] | |
| DNA synthesis | MTHFR | Ala222Val, C/T | *rs1801133 | T, 31.77% | 21–37% | Chr 1, 11856378 | T allele associated with reduced protein activity [68] and amount [69] |
| MTHFR | Glu429Ala, A/C | *rs1801131 | C, 30.61% (30%) | 33–38% | Chr 1, 11854476 | C allele associated with reduced enzymatic activity [31] | |
| TYMS | 2R/3R in 5′-UTR | **rs34743033 | 2R, 46.6% | 44.60% | Chr 18, 657646∶657730 | 3R allele confers increased translational efficiency [70] | |
| TYMS | indel 6 bp in 3′-UTR | *rs16430 | del, 34.13% | 37.00% | Chr 18, 673444 | deletion of 6 bp associated with lower mRNA stability [71] | |
| Angiogenesis | SERPINE1 | −675 indelG inpromoter | ***rs1799889 | G, 46.71% (46.53%) | 54.30% | Chr 7, 100769710∶100769711 | insG allele linked with lower transcriptional activity [72] |
| VEGFA | −634G/C in 5′-UTR | *rs2010963 | C, 29.1% | 20–43% | Chr 6, 43738350 | G allele associated with low gene expression [73] | |
| VEGFA | +936C/T in 3′-UTR | *rs3025039 | T, 10.73% | 10–22% | Chr 6, 43752536 | T allele associated with lower plasma VEGF levels [74] |
na: not available, MAF: minor allele frequency, VNTR: variable number of tandem repeats, 2R: 2 VNTR repeats, 3R: 3 VNTR repeats. The EGFR rs2227983 polymorphism is also known as rs11543848. The PTGS2 c.3618A/G excluded from analysis due to its low minor allele frequency.
genotyped by MassArray® technology,
genotyped by gel electrophoresis of PCR-amplified fragments,
genotyped by TaqMan® SNP genotyping assays.
MAF information was retrieved from the dbSNP database [75].
Materials and Methods
Ethics Statement
This study includes two patient cohorts. For both cohorts, collection of the patient clinical data and biospecimens was approved for research purposes by the Regional Health Boards and the Human Investigation Committee (HIC) of Memorial University of Newfoundland. In the discovery cohort, written informed consent was obtained from the patients recruited or their proxies. The Human Investigation Committee of Memorial University of Newfoundland waived the need for written informed consent from the participants in the replication cohort. Ethics approval for this particular project was also obtained from the Human Investigation Committee of Memorial University of Newfoundland.
Patient Cohorts
a) The discovery cohort
This cohort consisted of 532 colorectal cancer patients from the Newfoundland Colorectal Cancer Registry (NFCCR). NFCCR was established in 1999 and recruited 736 stage I–IV colorectal cancer patients between 1999 and 2003 [18]. All patients were ≤75 years old and their diagnosis was confirmed by pathological examination. The molecular and genetic characteristics of this cohort and other details have been previously reported by others [19], [20]. For all patients, the clinical data was compiled (although there were also missing values for some variables; Table 2 ). In this study, 532 of the 736 colorectal cancer patients from the NFCCR were investigated for whom the genomic DNA (extracted from blood) was also available. Patient data on clinicopathological features, recurrence and metastasis, and the date of death were retrieved from clinical reports (medical, pathology, radiology, autopsy, and surgical reports, lab investigations, physicians’ assessment and progress notes, inpatient discharge summaries), the Newfoundland Cancer Treatment and Research Foundation database, or patient follow-up questionnaires. In this cohort, 62% of the patients were treated with 5-FU based chemotherapy in either neoadjuvant or adjuvant settings or upon diagnosis of local and distant recurrences, whereas the remaining patients were either not treated with chemotherapy, or were treated with cisplatin/etoposide (n = 1). Patients in this cohort were followed until April 2010. The median follow-up time in this cohort for overall survival and disease free survival was 6.4 and 6 years, respectively ( Table 2 ).
Table 2. Baseline characteristics of the discovery and the validation cohorts.
| Variable | Discovery cohort n (%) | Validation cohort n (%) | p-value |
| Sex | |||
| Male | 327 (61.50%) | 133 (52.78%) | |
| Female | 205 (38.50%) | 119 (47.22%) | p = 0.021 |
| Median age in years (range) | 61.4 (20.7–75) | 68.7 (25.3–91.6) | p<0.001 |
| Histology | |||
| non-mucinous | 471 (88.50%) | 211 (83.73%) | |
| Mucinous | 61 (11.50%) | 41 (16.27%) | p = 0.062 |
| Location | |||
| Colon | 353 (66.40%) | 202 (80.16%) | |
| Rectum | 179 (33.60%) | 50 (19.84%) | p<0.001 |
| Stage | |||
| I | 99 (18.60%) | 48 (19.05%) | |
| II | 206 (38.70%) | 88 (34.92%) | |
| III | 175 (32.90%) | 68 (26.98%) | |
| IV | 52 (9.80%) | 41 (16.27%) | |
| Unknown | – | 7 (2.78%) | p = 0.034 |
| Grade | |||
| well diff./moderately diff. | 489 (91.90%) | 211 (83.73%) | |
| poorly diff./undiff. | 39 (7.30%) | 37 (14.68%) | |
| Unknown | 4 (0.80%) | 4 (1.59%) | p = 0.001 |
| * Invasion | |||
| Absence | 326 (61.30%) | 64 (25.40%) | |
| Presence | 166 (31.20%) | 101 (40.08%) | |
| Unknown | 40 (7.50%) | 87 (34.52%) | p<0.001 |
| OS status | |||
| Dead | 177 (33.30%) | 155 (61.51%) | |
| Alive | 354 (66.60%) | 97 (38.49%) | |
| Unknown | 1 (0.10%) | – | p<0.001 |
| Median OS follow-up time in years (range) | 6.4 (0.4–10.9) | 5.4 (0–12.48) | |
| DFS status | |||
| Event | 208 (39.10%) | 167 (66.27%) | |
| no event | 323 (60.71%) | 85 (33.73%) | |
| Unknown | 1 (0.19%) | – | p<0.001 |
| Median DFS follow-up time in years (range) | 6 (0.2–10.9) | 3.3 (0–12.5) | |
| MSI Status | |||
| MSI-H | 56 (10.50%) | 24 (9.52%) | |
| MSI-L/MSS | 455 (85.50%) | 228 (90.48%) | |
| Unknown | 21 (4%) | p = 0.543 | |
| Familial risk | |||
| Low | 256 (48.10%) | Na | nd |
| Intermediate/high | 276 (59.10%) | ||
| BRAF1 Val600Glu mutation | |||
| Presence | 49 (9.20%) | Na | nd |
| Absence | 435 (81.80%) | ||
| Unknown | 48 (9%) | ||
| 5-FU based treatment | |||
| 5-FU treated | 330 (62.03%) | 88 (34.92%) | |
| other/no chemotherapy | 199 (37.41%) | 148 (58.73%) | |
| Unknown | 3 (0.56%) | 16 (6.35%) | p<0.001 |
Vascular invasion and lymphatic invasion were highly correlated in the discovery cohort (p<0.001). Since data for vascular invasion in the validation cohort was not available, vascular invasion in the discovery cohort and lymphatic invasion in the validation cohort were compared with each other. diff: differentiated, n: number of patients, na: not available, nd: not done.
b) The validation cohort
This is a retrospective cohort comprised of 280 previously collected colorectal cancer patients from the Avalon Peninsula of Newfoundland. For all 280 patients the clinical data was collected, however genomic DNA (extracted from non-tumor tissues) was available for only 252 patients. Hence, 252 of 280 patients were included into the present study. Patients in this cohort were diagnosed with primary colorectal cancer in a two-year period (between 1997–1998). The patient selection criteria are as follows: a) patients with carcinoma in polyp were included only if the tumor invaded into the stalk, b) patients whose colorectal cancer was a recurrence of an earlier colorectal cancer or a metastasis from a distant organ, and those with carcinoid tumors, familial adenomatous polyposis, carcinoma in situ and mucosal carcinoma were excluded, and c) patients were selected regardless of their age of diagnosis. Prognostic data of these patients was collected from the medical and hospital records and the Newfoundland and Labrador Centre for Health Information. In the validation cohort, 34.9% of the patients were treated with 5-FU-based chemotherapy in either neoadjuvant or adjuvant settings or upon diagnosis of local or distant recurrences. The remaining patients were either not treated with chemotherapy or were treated with other agents such as irinotecan, tomudex or oxaliplatin. Patients in this cohort were followed until July 2009. The median follow-up time for this cohort was 5.4 and 3.3 years for overall survival and disease free survival, respectively ( Table 2 ).
Selection of Polymorphisms
The dbCPCO database [21] (http://www.med.mun.ca/cpco/) summarizes literature on genetic markers studied for their prognostic associations in colorectal cancer patients. In August 2010, a search of the entries in this database for survival measures (e.g. overall survival) was performed. As a result of this search, 31 polymorphisms were identified. Out of 31, one polymorphism (EGFR (CA)n repeat) was not included in this study because of the lack of a suitable equipment in our lab required to obtain its genotypes. In addition, three polymorphisms (EGF A61G, TP53 Arg72Pro and PTGS2 −765 G/C) could not be genotyped using the MassArray® technology. As a result, 27 polymorphisms from 24 different genes that were a) reported to be associated with overall survival in at least one study (either univariate or multivariate analyses) (Table S1 in File S1), b) suitable to be genotyped using the genotyping techniques used in this project (e.g. single nucleotide polymorphisms, indels, microsatellite repeats, gene deletions), and c) successfully genotyped (i.e. did not fail to be genotyped using the MassArray® method) were investigated in the current study ( Table 1 ).
Genotyping Methods
The genotypes for 27 polymorphisms were obtained in the discovery cohort and the genotypes for four polymorphisms that were associated with OS in the discovery cohort (MTHFR Glu429Ala, ERCC5 His46His, SERPINE1 −675indelG, and GSTM1 gene deletion) were obtained in the validation cohort. Genotypes were obtained using the Sequenom MassArray® platform, TaqMan® SNP genotyping assays and gel electrophoresis of PCR-amplified fragments. Further details related to genotyping experiments can be found in the Methods S1 and the Table S2 in File S1. Each genotyping reaction included non-template amplifications as negative controls. At least 5.9% of the genotypes were successfully duplicated with a minimum 99.7% concordance rate. Samples with discordant genotypes were either re-genotyped (genotypes obtained by using the TaqMan® SNP genotyping assays and gel electrophoresis of PCR-amplified fragments) or excluded from analysis (genotypes obtained by using the Sequenom MassArray® technique). The minimum successful genotyping rates were 97.4% for the discovery cohort and 94.44% for the validation cohort. In the case of in-house genotyping experiments (i.e. TaqMan® SNP genotyping assays and the gel-electrophoresis of PCR-amplified fragments), genotyping reactions for failed DNA samples were attempted at least two additional times, depending on the availability of DNA.
Statistical Analyses
a) Hardy Weinberg Equilibrium (HWE) test
HWE test was manually performed for polymorphisms using the Pearson’s Chi-square test (Table S3 in File S1). For the GSTM1 and GSTT1 gene deletions HWE was not tested as heterozygote genotypes cannot be detected using the genotyping methodology applied in this study.
b) Survival endpoints
OS time was the time from diagnosis of colorectal cancer until death from any cause. DFS time was the time from diagnosis of colorectal cancer until the occurrence of metastasis, recurrence or death from any cause, whichever was earlier. Patients who did not experience the outcome of interest were censored at the time of last follow up.
c) Variables
Categorical variables analyzed were sex (males vs females), tumor histology (mucinous vs non-mucinous), tumor location (rectal vs colon), stage (stages II, III and IV vs stage I), tumor grade (poorly differentiated/undifferentiated vs well/moderately differentiated), vascular and lymphatic invasions (present vs absent), familial risk (high/intermediate risk vs low risk), microsatellite instability status (MSI-H vs MSI-L/MSS) and BRAF1 Val600Glu mutation status (present vs wildtype). For the discovery set, the familial risk status was determined previously by the NFCCR investigators using the Amsterdam II and revised Bethesda criteria [18]. Tumor MSI status and BRAF1 Val600Glu status analyses were also previously performed by NFCCR [19], [20], [22]. Vascular and lymphatic invasions were highly correlated in the discovery cohort (>95% of tumors with vascular invasion also had lymphatic invasion). Please note that in some models, confidence intervals for the stage IV patients were wide, reflecting the small sample size for this group of patients. These results therefore should be interpreted cautiously.
We categorized the genotypes for each polymorphism assuming the co-dominant genetic model (i.e. minor allele homozygotes and heterozygotes were individually compared to the major allele homozygotes). In the case of the MTHFR Glu429Ala polymorphism, we also performed multivariable analyses under the recessive (CC vs CA+AA genotypes) and the dominant (CC+CA vs AA genotypes) genetic models. The PTGS2 c.3618A/G polymorphism was excluded from the statistical analysis due to its very low minor allele frequency (1.63%). For the VNTR polymorphism in the TYMS gene (rs34743033), 0.93% of the samples in the discovery cohort had a rare 4R allele. These patients were combined with 3R/3R genotypes for analyses. Age was the only continuous variable in our analysis.
d) Univariate analyses
Time-to-event survival plots were constructed using the Kaplan-Meier method and were compared by the log-rank test.
e) Multivariable analysis
The variables used in the construction of the final multivariable models were selected by backward elimination method for OS and DFS separately using the Cox regression method. Selected variables were then re-entered in the final models. The proportionality assumption was verified by examining the log-minus-log (log(-log(S(t)))) plots. We also tested the interaction between the MTHFR Glu429Ala genotypes (co-dominant genetic model) and the 5-FU treatment using the Cox regression method.
f) Stratified analyses
Since the MTHFR enzyme (thus the Glu429Ala polymorphism by modifying the MTHFR enzymatic activity) plays a biological role in the 5-FU metabolism/efficacy (See Discussion), 5-FU stratification was done only for this polymorphism. Since this polymorphism was not associated with DFS, 5-FU stratification analysis for DFS was not performed.
g) Comparisons of cohorts
To test if the differences between the baseline characteristics of the discovery and the validation cohorts were significant, we performed the Chi-square test for the categorical variables. Since age was not normally distributed in both cohorts, the non-parametric Mann Whitney-U test was used to compare differences in distribution of age between these two cohorts. Similar analyses were also performed to compare the entire NFCCR cohort (n = 736) and the discovery cohort (n = 532) and also the entire second cohort (n = 280) and validation cohort included in our analysis (n = 252).
PASW Statistics 18 software release 18.0.2 (IBM, NY, USA) was used to perform the statistical analyses. All tests were double sided and the significance threshold was set at p = 0.05. To avoid false-negative results, correction for multiple testing was not performed in the discovery cohort analysis. While this also increases the potential number of false-positive associations, analysis of the associations detected in the discovery cohort in an additional patient cohort (i.e. the replication cohort) helped eliminate the false-positive findings.
Results
The Discovery Cohort Characteristics
Baseline characteristics of the discovery cohort are listed in Table 2 . The median age at diagnosis was 61.4 years. One-third (33.3%) of the patients had died and 39% of patients had experienced recurrence, metastasis or death by the time of last follow-up. The discovery cohort has a significantly lower proportion of stage IV patients (9.8%) when compared to the entire NFCCR cohort (20.9%) (p<0.001). The discovery cohort also significantly differed from the NFCCR cohort in terms of proportions of tumors with vascular (p = 0.007) and lymphatic invasions (p = 0.021), and deceased patients (p<0.001).
Overall Survival Analysis in the Discovery Cohort
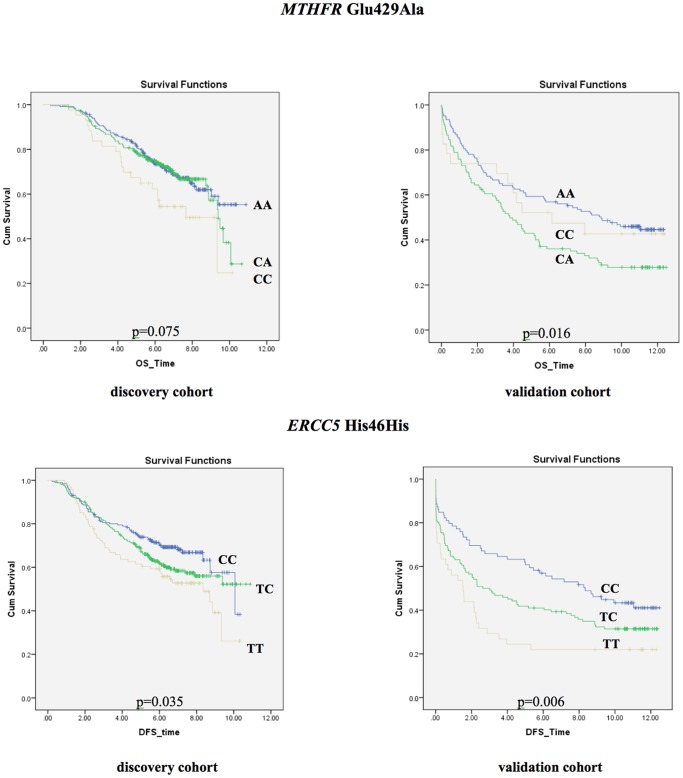
Out of 26 polymorphisms investigated, four polymorphisms were significantly associated with OS when adjusted for sex, age, stage and MSI status ( Table 3 ). Briefly, for the MTHFR Glu429Ala polymorphism, patients homozygous for the C allele had shorter survival (HR: 1.72, 95% CI: [1.04–2.84], p = 0.036) compared to patients homozygous for the A allele. For the ERCC5 His46His polymorphism, patients with the TT genotype had a greater risk of death (HR: 1.78, 95% CI: [1.15–2.76], p = 0.01) compared to those patients with the CC genotype. In the case of the SERPINE1 −675indelG polymorphism, the minor allele homozygotes (insG/insG) had increased survival (HR: 0.52, 95% CI: [0.32–0.84], p = 0.008) compared to the patients with delG/delG genotype. Genotype distributions of these three polymorphisms were in HWE (p>0.05; Table S3 in File S1). In addition, patients with at least one copy of GSTM1 gene had a greater risk of death compared to patients with no copy of the gene (HR: 1.40, 95% CI: [1.03–1.92], p = 0.033) ( Table 3 ).
Table 3. Multivariable Cox regression analysis results for overall survival in the discovery and validation cohorts.
| Discovery cohort (n = 504, deaths = 168) | Validation cohort (n = 224, deaths = 134) | |||||
| *Co-dominant genetic model | ||||||
| #Variable | HR (95% CI) | p-value | n | HR (95% CI) | p-value | n |
| MTHFR Glu429Ala | 0.105 | 0.010 | ||||
| CA vs AA | 1.18 (0.84–1.64) | 0.342 | 230 vs 232 | 1.71 (1.18–2.49) | 0.005 | 92 vs 112 |
| CC vs AA | 1.72 (1.04–2.84) | 0.036 | 42 vs 232 | 0.89 (0.45–1.74) | 0.730 | 20 vs 112 |
| ERCC5 His46His | 0.034 | 0.609 | ||||
| TC vs CC | 1.37 (0.94–1.97) | 0.098 | 240 vs 173 | 1.20 (0.80–1.80) | 0.387 | 112 vs 76 |
| TT vs CC | 1.78 (1.15–2.76) | 0.010 | 91 vs 173 | 1.26 (0.74–2.16) | 0.398 | 36 vs 76 |
| SERPINE1 − 675indelG | 0.029 | 0.716 | ||||
| insG/delG vs delG/delG | 0.81 (0.57–1.15) | 0.238 | 258 vs 141 | 1.19 (0.78–1.80) | 0.420 | 103 vs 69 |
| insG/insG vs delG/delG | 0.52 (0.32–0.84) | 0.008 | 105 vs 141 | 1.08 (0.67–1.73) | 0.766 | 52 vs 69 |
| GSTM1 gene deletion | ||||||
| present vs absent | 1.40 (1.03–1.92) | 0.033 | 228 vs 276 | 1.23 (0.86–1.78) | 0.261 | 99 vs 125 |
| Sex | ||||||
| male vs female | 1.46 (1.04–2.05) | 0.031 | 313 vs 191 | 1.28 (0.90–1.84) | 0.175 | 118 vs 106 |
| Age at diagnosis (per year) | 1.02 (1–1.04) | 0.046 | 1.05 (1.03–1.07) | <0.001 | ||
| Stage | <0.001 | <0.001 | ||||
| II vs I | 1.47 (0.84–2.59) | 0.180 | 194 vs 95 | 1.14 (0.63–2.09) | 0.662 | 80 vs 44 |
| III vs I | 2.08 (1.19–3.64) | 0.010 | 165 vs 95 | 2.61 (1.45–4.71) | 0.001 | 64 vs 44 |
| IV vs I | 11.69 (6.45–21.16) | <0.001 | 50 vs 95 | 11.32 (5.92–21.67) | <0.001 | 36 vs 44 |
| MSI status | ||||||
| MSI-H vs MSI-L/MSS | 0.23 (0.09–0.64) | 0.004 | 56 vs 448 | 0.26 (0.11–0.61) | 0.002 | 21 vs 203 |
| ** Dominant genetic model | ||||||
| MTHFR Glu429Ala | 1.19 (0.87–1.61) | 0.277 | 272 vs 232 | 1.56 (1.12–2.17) | 0.009 | 122 vs 121 |
| (CA+CC vs AA) | ||||||
| *** Recessive genetic model | ||||||
| MTHFR Glu429Ala | 1.80 (1.13–2.86) | 0.014 | 42 vs 462 | 0.69 (0.38–1.25) | 0.219 | 21 vs 222 |
| (CC vs CA+AA) | ||||||
The multivariable Cox regression model assuming the co-dominant genetic model contained the MTHFR Glu429Ala, ERCC5 His46His, SERPINE1 −675indelG, GSTM1 gene deletion genotypes as well as sex, age, stage and MSI status as covariates.
Only the multivariable Cox regression analysis result for the MTHFR Glu429Ala polymorphism when adjusted for sex, age, stage and MSI status is shown (assuming the dominant genetic model). The complete multivariable models for the dominant genetic model can be found in Tables S4 and S6 in File S1 for the discovery and validation cohorts.
The multivariable Cox regression analysis result for the MTHFR Glu429Ala polymorphism when adjusted for sex, age, stage and MSI status is shown (assuming the recessive genetic model). The complete multivariable models can be found in Tables S5 and S7 in File S1 for the discovery and validation cohorts. CI: confidence interval, HR: hazard ratio, n: number of patients, vs: versus.
#The major homozygote genotypes and other referent categories are underlined.
Disease-free Survival Analysis in the Discovery Cohort
Out of 26 polymorphisms, the ERCC5 His46His and OGG1 Ser326Cys polymorphisms were associated with shorter DFS in the discovery cohort when adjusted for other variables ( Table 4 ). Specifically, patients homozygous for the T allele of the ERCC5 His46His (HR: 1.54, 95% CI: [1.04–2.29], p = 0.032) and G allele of the OGG1 Ser326Cys (HR: 1.81, 95% CI: [1.08–3.04], p = 0.025) had worse survival compared to the major allele homozygotes.
Table 4. Multivariable model for disease-free survival in the discovery and validation cohorts.
| Discovery cohort (n = 504, events = 198) | Validation cohort (n = 227, events = 148) | |||||
| #Variable | HR (95% CI) | p-value | n | HR (95% CI) | p-value | n |
| ERCC5 His46His | 0.098 | 0.036 | ||||
| TC vs CC | 1.24 (0.89–1.72) | 0.211 | 240 vs 172 | 1.48 (1.02–2.17) | 0.041 | 114 vs 77 |
| TT vs CC | 1.54 (1.04–2.29) | 0.032 | 92 vs 172 | 1.81 (1.11–2.94) | 0.018 | 36 vs 77 |
| OGG1 Ser326Cys | 0.082 | Nd | Nd | |||
| GC vs CC | 1.09 (0.80–1.48) | 0.590 | 167 vs 304 | |||
| GG vs CC | 1.81 (1.08–3.04) | 0.025 | 33 vs 304 | |||
| ERCC1 Asn118Asn | 0.152 | Nd | Nd | |||
| TC vs TT | 1.19 (0.87–1.64) | 0.281 | 215 vs 206 | |||
| CC vs TT | 1.48 (0.99–2.20) | 0.054 | 83 vs 206 | |||
| TYMS indel 6 bp | 0.171 | Nd | nd | |||
| ins 6 bp/del 6 bp vs ins 6 bp/ins 6 bp | 0.83 (0.61–1.13) | 0.235 | 226 vs 221 | |||
| del 6 bp/del 6 bp vs ins 6 bp/ins 6 bp | 1.25 (0.80–1.96) | 0.325 | 57 vs 221 | |||
| GSTM1 gene deletion | ||||||
| present vs absent | 1.28 (0.96–1.70) | 0.090 | 229 vs 275 | 1.17 (0.84–1.63) | 0.366 | 101 vs 126 |
| Location | ||||||
| rectum vs colon | 1.33 (0.99–1.79) | 0.055 | 166 vs 338 | 1.07 (0.71–1.60) | 0.743 | 44 vs 183 |
| Stage | <0.001 | <0.001 | ||||
| II vs I | 1.51 (0.93–2.47) | 0.099 | 194 vs 95 | 1.82 (1.04–3.19) | 0.036 | 81 vs 45 |
| III vs I | 2.09 (1.28–3.41) | 0.003 | 164 vs 95 | 3.14 (1.79–5.51) | <0.001 | 65 vs 45 |
| IV vs I | 6.24 (3.69–10.53) | <0.001 | 51 vs 95 | 130.16 (52.48–322.83) | <0.001 | 36 vs 45 |
| MSI status | ||||||
| MSI-H vs MSI-L/MSS | 0.35 (0.17–0.71) | 0.004 | 55 vs 449 | 0.37 (0.18–0.76) | 0.007 | 22 vs 205 |
The multivariable model contained location, stage and MSI status in addition to the ERCC5 His46His, OGG1 Ser326Cys, ERCC5 Asn118Asn, TYMS indel 6 bp, and GSTM1 gene deletion genotypes in the discovery cohort and the ERCC5 His46His and GSTM1 gene deletion genotypes in the validation cohort as covariates. The genotypes for the OGG1 Ser326Cys, ERCC5 Asn118Asn, and TYMS indel 6 bp polymorphisms were not available in the validation cohort. CI: confidence interval, HR: hazard ratio, n: number of patients, nd: not done, vs: versus. Event refers to recurrence, metastasis or death in the patient, whichever had occurred earlier.
#The referent categories are underlined. Please note that reflecting the small numbers of patients in the validation cohort, the CIs for the effect estimate in stage IV patients are quite wide and should not be interpreted as an accurate estimation.
The Validation Cohort Characteristics
Baseline characteristics of the validation cohort are listed in Table 2 . The median age of diagnosis was 68.7 years. By the time of last follow-up 61.5% of patients had died and 66.3% of the patients had experienced recurrence, metastasis or death. There were no statistically significant differences between the initial 280 patients in this cohort and the 252 patients included in this study in terms of clinical and molecular features (data not shown).
However, there were significant differences between the discovery and validation cohorts in terms of clinicopathological characteristics. First, there was a higher proportion of stage IV patients in the validation cohort (16.3%) compared to the discovery cohort (9.8%) (p = 0.034). Second, the median age at diagnosis in the validation cohort (68.7 years) was significantly higher (p<0.001) compared to that of the discovery cohort (61.4 years). The proportions of patients in terms of sex, tumor location, grade, OS and DFS status, vascular and lymphatic invasions, and treatment with 5-FU-based regimens were also different between the two cohorts ( Table 2 ).
Overall Survival Analysis in the Validation Cohort
The genotype distribution of four polymorphisms tested in the validation cohort did not deviate from HWE. Out of these four polymorphisms, only the MTHFR Glu429Ala polymorphism was associated with shorter survival times when adjusted for other variables ( Table 3 ). However, in contrast to the discovery set, in the validation cohort, the heterozygotes (Glu/Ala) had shorter survival (HR: 1.71, 95% CI: [1.18–2.49], p = 0.005) when compared to homozygotes for glutamate (Glu/Glu) ( Figure 1 ). Thus the genotype associated with worse survival in the validation cohort (AC) was different than the genotype associated in the discovery cohort (CC). Therefore, we also performed multivariable analyses assuming the recessive and dominant genetic models in these two cohorts ( Table 3 , Tables S4–S7 in File S1). As a result, in the discovery set, the MTHFR Glu429Ala polymorphism was associated with OS in the recessive genetic model (CC vs AC+AA; HR: 1.80, 95% CI: [1.13–2.86], p = 0.014), but not in the dominant genetic model. In contrast, in the validation set, this polymorphism was associated with OS in the dominant genetic model (CC+AC vs AA; HR: 1.56, 95% CI: [1.12–2.17], p = 0.009), but not in the recessive genetic model. Analysis of this polymorphism assuming the additive genetic model did not yield significant association with OS in either cohort (data not shown). Thus, the association of the MTHFR Glu429Ala polymorphism with OS in these two cohorts is detected under different genetic models.
Figure 1. Kaplan-Meier survival curves for the MTHFR Glu429Ala (overall survival) and the ERCC5 His46His polymorphisms (disease-free survival) assuming the co-dominant genetic model. P-values are based on log-rank test.
Explorative Analyses for Overall Survival and the MTHFR Glu429Ala Polymorphism
While this interesting association pattern of the MTHFR Glu429Ala polymorphism with OS in two cohorts may also be explained by the reduced statistical power to detect the effects of each genotype groups, we also performed additional explorative analyses to investigate other possibilities. First, to test whether the association of different genotypes could be due to relatively higher median age in the validation cohort when compared to the discovery cohort (see Discussion), we performed a multivariable analysis in patients from the validation set who were under 75 years of age at the time of diagnosis (n = 149). Yet, we found that the pattern of association observed in this analysis (AC vs AA, HR: 2.02, 95% CI: [1.20–3.41], p = 0.008) was similar to that obtained in the validation cohort, suggesting age at diagnosis was not the reason for this disparity. Second, since the 5-FU efficacy might be modified by the activity of the MTHFR enzyme (see Discussion), we also tested the association of the MTHFR Glu429Ala genotypes with OS in patient groups stratified based on their treatment characteristics (patients treated with 5-FU based regimens versus patients not treated with it). Interestingly, in this analysis, we have found that when adjusted for age, stage, MSI status, this polymorphism was significantly associated with OS in the patients not treated with 5-FU (both CC vs AA and AC vs AA genotypes in the discovery set and AC vs AA genotypes in the validation cohort), but not in the patients treated with 5-FU based regimens ( Tables 5 and 6 ). Analysis of the interaction between the MTHFR Glu429Ala polymorphism and the 5-FU treatment status in both the discovery and the validation cohorts did not reveal a statistically significant interaction between these two variables.
Table 5. The MTHFR Glu429Ala polymorphism and overall survival in the discovery cohort patients (stratified by treatment with 5-Fluorouracil).
| Treated with 5-FU | Not treated with 5-FU | |||||
| Discovery cohort (n = 310) | Discovery cohort (n = 191) | |||||
| #Variable | HR (95% CI) | p-value | n | HR (95% CI) | p-value | n |
| MTHFR Glu429Ala | 0.206 | 0.005 | ||||
| AC vs AA | 0.88 (0.61–1.28) | 0.514 | 143 vs 140 | 2.47 (1.15–5.27) | 0.020 | 85 vs 92 |
| CC vs AA | 1.51 (0.85–2.68) | 0.161 | 27 vs 140 | 6.08 (1.95–18.93) | 0.002 | 14 vs 92 |
| Age | 1.02 (1.00–1.04) | 0.132 | 1.09 (1.04–1.14) | 0.001 | ||
| Stage | <0.001 | <0.001 | ||||
| II vs I | 2.56 (0.35–18.81) | 0.356 | 104 vs 7 | 0.67 (0.31–1.41) | 0.289 | 89 vs 87 |
| III vs I | 2.58 (0.36–18.76) | 0.349 | 154 vs 7 | 7.70 (2.76–21.48) | <0.001 | 10 vs 87 |
| IV vs I | 15.84 (2.16–115.98) | 0.007 | 45 vs 7 | 2.98 (0.83–10.66) | 0.093 | 5 vs 87 |
| MSI-H vs MSI-L/MSS | 0.08 (0.01–0.57) | 0.012 | 23 vs 287 | 0.53 (0.16–1.82) | 0.315 | 32 vs 159 |
5-FU: 5-fluorouracil, CI: confidence interval, HR: hazard ratio, n: number of patients, vs: versus.
#The referent categories are underlined. Please note that reflecting the small numbers of patients, the CIs for the effect estimate in stage IV patients are quite wide and should not be interpreted as an accurate estimation.
Table 6. The MTHFR Glu429Ala polymorphism and overall survival in the validation cohort patients (stratified by treatment with 5-Fluorouracil).
| Treated with 5-FU | Not treated with 5-FU | |||||
| Validation cohort (n = 87) | Validation cohort (n = 141) | |||||
| #Variable | HR (95% CI) | p-value | n | HR (95% CI) | p-value | n |
| MTHFR Glu429Ala | 0.676 | 0.032 | ||||
| AC vs AA | 1.34 (0.70–2.56) | 0.382 | 41 vs 39 | 1.70 (1.10–2.63) | 0.017 | 57 vs 71 |
| CC vs AA | 1.07 (0.35–3.29) | 0.903 | 7 vs 39 | 0.82 (0.36–1.88) | 0.637 | 13 vs 71 |
| Age | 1.02 (0.99–1.05) | 0.189 | 1.06 (1.03–1.08) | <0.001 | ||
| Stage | <0.001 | <0.001 | ||||
| II vs I | 0.54 (0.14–2.10) | 0.372 | 26 vs 6 | 1.36 (0.73–2.54) | 0.331 | 56 vs 36 |
| III vs I | 1.42 (0.42–4.78) | 0.570 | 42 vs 6 | 3.61 (1.83–7.12) | <0.001 | 23 vs 36 |
| IV vs I | 6.32 (1.72–23.16) | 0.005 | 13 vs 6 | 11.32 (5.52–23.20) | <0.001 | 26 vs 36 |
| MSI-H vs MSI-L/MSS | 0.34 (0.08–1.46) | 0.146 | 8 vs 79 | 0.37 (0.15–0.94) | 0.037 | 13 vs 128 |
5-FU: 5-fluorouracil, CI: confidence interval, HR: hazard ratio, n: number of patients, vs: versus.
#The referent categories are underlined. Please note that reflecting the small numbers of patients, the CIs for the effect estimate in stage IV patients are quite wide and should not be interpreted as an accurate estimation.
Disease Free Survival Analysis in the Validation Cohort
In the multivariable analysis of DFS in the validation cohort, the association of the ERCC5 His46His polymorphism with DFS was also detected as follows: compared to those patients with the CC genotype, patients with TT and the CT genotypes had shorter DFS (HR: 1.81, 95% CI: [1.11–2.94], p = 0.018 and HR: 1.48, 95%CI: [1.02–2.17], p = 0.041), respectively ( Table 4 ).
Discussion
The main result of our study is that the associations of the MTHFR Glu429Ala polymorphism with the overall survival and the ERCC5 His46His polymorphism with the disease-free survival were detected in two separate cohorts of colorectal cancer patients.
Two other studies have also reported the MTHFR Glu429Ala polymorphism to be associated with OS in colorectal cancer patients. In one study conducted in a Spanish cohort [23], patients carrying the C allele (AC+CC genotype) had worse survival than those with AA genotype, when adjusted for clinicopathological variables. Similarly, in another study [24] performed in metastatic colon cancer patients, female patients with AC+CC genotypes for the MTHFR Glu429Ala had worse OS than female patients with AA genotype in univariate analysis. However, in six other studies, no association was observed between the MTHFR Glu429Ala polymorphism and OS in colorectal cancer in univariate or multivariable analyses [25]–[30].
The role of the MTHFR enzyme and its Glu429Ala polymorphism in colorectal cancer prognosis is not well-known. However, MTHFR has been biologically investigated in great detail (i.e. its role in folate metabolism and 5-FU mechanism of action). In addition, the Glu429Ala polymorphism has been previously shown to cause moderate reduction in the activity of MTHFR enzyme; the Ala/Ala homozygotes have close to 60% of the normal MTHFR activity and the heterozygotes have ∼80% of the normal MTHFR activity [31], [32].
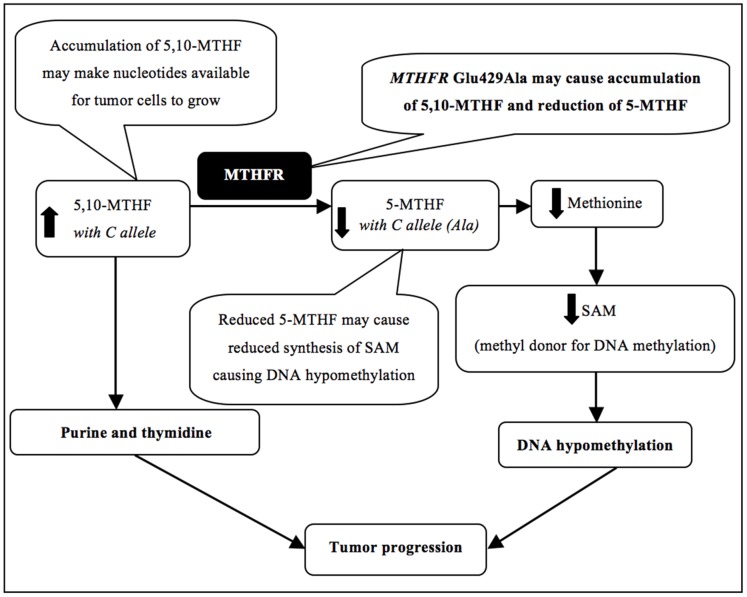
In folate metabolism, one of the biological activities of the MTHFR enzyme is to convert the 5,10-methylenetetrahydrofolate (5,10-MTHF) to 5-methyltetrahydrofolate (5-MTHF) [33]. 5,10-MTHF is predominantly used in the synthesis of purines and thymidine, the nucleotides used by the dividing cells in DNA synthesis. In addition, 5-MTHF is used in the synthesis of S-adenosyl-methionine (SAM), a key mediator in a number of methylation reactions including DNA methylation [33]. Thus reduction in MTHFR enzymatic activity due to Glu429Ala polymorphism may result in accumulation of 5,10-MTHF and concurrent reduction of 5-MTHF to a certain extent ( Figure 2 ). Accumulation of 5,10-MTHF form of folate may provide increased amounts of nucleotides for DNA synthesis to the rapidly proliferating tumor cells to grow. This theory is supported by recent reports which suggest that once a colorectal adenoma has developed, folate supplementation can aid its growth and progression [34]–[37], presumably by facilitating large amounts of nucleotide precursors for tumor growth [33], [35], [36]. In another study, folate supplementation was found to be associated with progression of already developed colorectal cancer in rats [36]. Also in the Aspirin/Folate Polyp Prevention Study [34], [36], folate supplementation was associated with higher risk of advanced adenomas as well as increased number of adenomas in patients with previously developed colorectal adenomas. These findings suggest a negative effect of high folate levels in colorectal cancer prognosis. Therefore, in our cohorts, the association of the MTHFR Glu429Ala polymorphism with worse prognosis of colorectal cancer patients may be due to reduced activity of MTHFR and accumulation of folate which can facilitate tumor growth.
Figure 2. 5,10-MTHF: 5,10-methylene tetrahydrofolate, 5-MTHF: 5-methyl tetrahydrofolate, MTHFR: methylene tetrahydrofolate reductase, SAM: S-adenosyl methionine.
Arrows indicate the potential consequences of the polymorphism on biological processes depicted.
In addition, due to the inefficient MTHFR enzyme function (for example, due to the Glu429Ala polymorphism), the optimal conversion of 5,10-MTHF to 5-MTHF may also be reduced, causing reduction in the levels of 5-MTHF. This may ultimately lead to a decrease in synthesis of SAM ( Figure 2 ). SAM is an important methyl donor for a large number of reactions and its deficiency can induce DNA hypomethylation. In MTHFR gene knockout mice, the levels of SAM as well as the extent of DNA methylation were found to be significantly reduced [38]. Thus by reducing the enzymatic activity, the MTHFR Glu429Ala polymorphism may lead to a similar, although a less severe consequence. In a study conducted in tumor cells from colon cancer patients, for example, DNA hypomethylation was associated with unfavorable cancer-specific survival and OS [39]. Accordingly, DNA hypomethylation due to the MTHFR Glu429Ala polymorphism mediated reduction in SAM levels may have also contributed to unfavorable prognosis in our cohorts.
Another finding in our study was the association of different genotypes of the MTHFR Glu429Ala polymorphism with OS in two separate colorectal cancer patient cohorts. To explore the potential causes of this disparity, we have focused on the differences between the discovery and validation cohorts that were known to play a biological role in MTHFR activity or the folate metabolism; namely age and treatment with 5-FU. Older individuals are known to have an impaired ability to absorb dietary folate [40] and the validation cohort in our study had a significantly higher median age compared to the discovery cohort (p<0.001). We therefore hypothesized that along with the low folate absorption in older patients, the mildly reduced MTHFR activity due to heterozygosity of MTHFR Glu429Ala polymorphism might have been sufficient to contribute to the unfavorable prognosis observed in the validation cohort (in this case, the Ala/Ala homozygotes would also be expected to have shorter OS; however, it was possible that this association might have been missed due to insufficient study power for comparison of CC vs AA genotypes). Therefore, we conducted a multivariable analysis in the validation cohort patients, who were <75 years of age at the time of diagnosis (similar to the patients in the discovery set). These results also showed the association of the AC genotype of the MTHFR Glu429Ala polymorphism with OS when compared to the AA genotype, similar to the association detected in the entire validation cohort. Therefore, it is not likely that increased age together with the MTHFR Ala429 variant and their effect on folate mechanism may explain the association of two different genotypes in our discovery and validation cohorts.
We then focused on the 5-FU and its effect on the folate metabolism. 5-FU is routinely used in the colorectal cancer chemotherapy and one of its anti-neoplastic mechanisms is the inhibition of thymidine synthesis. In the process of thymidine synthesis inhibition, 5,10-MTHF stabilizes the chemical complex necessary for inhibition of thymidylate synthase (TYMS) enzyme [41]. In the presence of increased concentration of 5,10-MTHF, the inhibition of thymidine synthesis and thus the efficacy of 5-FU is expected to increase and this has been demonstrated in vitro in human colon cancer cells [42]. However, in multiple prognostic studies, statistical association of the MTHFR Glu429Ala polymorphism with response to treatment with 5-FU based chemotherapy in colorectal cancer patients was not detected [25], [26], [43]–[49] suggesting that MTHFR Glu429Ala polymorphism may not affect the efficacy of 5-FU based treatments. In the present study too, no significant association of MTHFR Glu429Ala was found in patients treated with 5-FU based chemotherapy (although we cannot fully rule out the possibility of insufficient study power to detect an effect). However, in our study, MTHFR Glu429Ala polymorphism was associated with shorter OS in patients, who were not treated with 5-FU in both the discovery and the validation cohorts ( Tables 5 and 6 ). On further analyses, we found that the majority of non-5-FU treated patients were stage I and II (92%) and had colon tumors (81.4%), who generally receive surgical treatment without 5-FU-based chemotherapy. Therefore, these results suggest that the reduced MTHFR activity due to Glu429Ala polymorphism may be associated with shorter OS, and thus may be a promising adverse prognostic marker, in early stage colon cancer patients or those patients not treated with 5-FU. Alternatively, other polymorphisms highly linked with MTHFR Glu429Ala polymorphism may be the reason for this association (Methods S2 and Figure S1 in File S1). While these results are needed to be confirmed with further studies, to our knowledge, this is the first study that identified a potential prognostic significance of the MTHFR Glu429Ala polymorphism in colorectal cancer patients not treated with 5-FU.
In the present study, we also show that the TT genotype of the ERCC5 His46His polymorphism is associated with shorter DFS in the two colorectal cancer patient cohorts investigated ( Table 4 ). To our knowledge, this is the first study that reports the association of the ERCC5 His46His polymorphism with DFS in colorectal cancer patients. ERCC5 is one of the endonucleases functioning in the nucleotide excision repair. ERCC5 His46His is a synonymous and non-splice site polymorphism and its impact on function of ERCC5 protein is uncertain. Previously, the TT genotype of this polymorphism was reported to be associated with short progression free survival (PFS) in advanced colorectal cancer patients receiving oxaliplatin [49] and short PFS and OS times in stage I and II head and neck cancer patients receiving radiotherapy [50]. Radiotherapy-resistant lung cancer cells have also shown an up-regulation of ERCC5 [51]. Additionally, in a study of ovarian cancer patients treated with platinum-based chemotherapeutic drugs, loss of heterozygosity (LOH) of ERCC5 and down regulation of this gene were associated with a favorable PFS, presumably due to increased efficacy of these drugs [52]. Whether His46His polymorphism causes up-regulation or down-regulation of ERCC5 is presently unknown and functional characterization of the polymorphism is required to understand its potential prognostic role in colorectal cancer. Alternatively, since this synonymous polymorphism does not alter the amino acid sequence in the protein, it is also likely that rather than the polymorphism itself, another highly correlated polymorphism in the same LD block may have a biological impact on disease progression and survival (Methods S2 and Figure S2 in File S1).
Twenty-four of the 26 selected polymorphisms (excluding one polymorphism which was not included into the statistical analysis in this study due to its low minor allele frequency) did not show an association with survival in our discovery cohort and previously reported associations were thus not replicated in colorectal cancer patients from Newfoundland. Such a lack of concordance in results of genetic prognostic studies is a common observance due to significant heterogeneity in the cohort characteristics and study design amongst different studies. For example, variations in patient ethnicities, treatment characteristics, follow-up times and clinical characteristics are regarded as critical reasons for the discordance in results of genetic prognostic studies in different study cohorts [53], [54]. In addition, the discovery and validation cohorts used in this study are predominantly composed of Caucasian patients, with follow-up times of up to 10 years and other clinicopathological features described in Table 2 . These features may not be shared by the other published cohorts (Table S1 in File S1), which may have contributed to differences in the results. Finally, there are differences between the discovery and validation cohorts in terms of several demographic and clinicopathological features (sex, age, stage, grade and invasion status, 5-FU treatment status) and OS and DFS follow up times ( Table 2 ). These differences as well as the small sample size of the validation cohort may also explain why no associations of ERCC5 His46His, SERPINE1 −675indelG and GSTM1 gene deletion polymorphisms with OS were detected in the validation cohort. Therefore, it is possible that associations of ERCC5 His46His, SERPINE1 −675indelG and GSTM1 gene deletion polymorphisms with OS may be detected in other cohorts with similar characteristics to the discovery cohort. Alternatively, the associations detected in the discovery cohort could be false-positive associations.
The limitations of this study are the dissimilarities between the discovery and validation cohorts, the fact that the discovery cohort was biased towards early stage patients, small size of the validation cohort, the short follow-up time in the validation cohort, especially for DFS, the limited number of genes and polymorphisms investigated, and the limited gene coverage (i.e. other polymorphisms in these genes were not studied). The main strength of this study is the relatively large sample size of the discovery cohort. This is also one of the few studies in colorectal cancer that attempted to replicate results in an additional patient cohort.
Supporting Information
Supporting information. Figure S1 The circled SNP is rs1801131 (MTHFR Glu429Ala), which lies in a 12kb LD block. The black squares indicate other highly correlated SNPs (r2>0.80). Figure S2 The circled SNP is rs1046678 (ERCC5 His46His), which lies in a 23kb LD block. The black squares indicate other highly correlated SNPs (r2>0.80). Table S1 n: number. Table S2 *Assay ID by Applied Biosystems (CA, USA). Underlined are the sequences on probes that are complementary to alleles they recognize. Assays for rs1799750 in MMP1 gene and rs1799889 in SERPINE1 gene were custom designed. Assays for rs1801131 in MTHFR gene and rs1047768 in ERCC5 gene were predesigned by Applied Biosystems. Primer and probe sequences for these assays are not available since they are proprietary of Applied Biosystems. Seq: sequence. Table S3 n/a: not applicable. Polymorphisms with χ2 value greater than 3.84 were considered to be deviating from HWE (p<0.05). *For these gene deletions, since heterozygote genotype cannot be determined by the genotyping method applied, HWE was not calculated. All polymorphisms were investigated in this study regardless of their deviations from the HWE as these deviations may also be attributed to the fact that the Newfoundland population is considered a genetically isolated population [49]. Nevertheless, it is worth noting that while the OGG1 Ser326Cys polymorphism that deviated from the HWE was included in the DFS multivariable model of the discovery cohort, its genotype data was not available for the validation cohort patients. Thus, the main conclusion on the disease-free survival analysis that the ERCC5 His46His polymorphism was associated with DFS in both the discovery and the validation patient cohorts is not affected by including this polymorphism in the DFS analysis of the discovery cohort. Table S4 CI: confidence interval, HR: hazard ratio, MSI: microsatellite instability, n: number of patients, vs: versus. Table S5 CI: confidence interval, HR: hazard ratio, MSI: microsatellite instability, n: number of patients, vs: versus. Table S6 CI: confidence interval, HR: hazard ratio, MSI: microsatellite instability, n: number of patients, vs: versus. Table S7 CI: confidence interval, HR: hazard ratio, MSI: microsatellite instability, n: number of patients, vs: versus. Methods S1 Genotyping reactions. Methods S2 Construction of linkage disequilibrium (LD) maps.
(DOC)
Acknowledgments
We thank the study participants in the discovery and the validation cohorts, the NFCCR personnel and Dr. Jane Green for their valuable contribution to this study. Technical assistance by Michelle Simms and Payal Sipahimalani and statistical guidance by Dr. XiaoQing Liu are gratefully acknowledged.
Funding Statement
This study is funded by grants from the Memorial University; the 2010 Cox Award by the Faculty of Medicine, Memorial University; the Canadian Institute of Health Research (CIHR) fund for the Colorectal Cancer Interdisciplinary Health Research Team at the University of Toronto and Memorial University, the National Cancer Institute of Canada (grants 18223 and 18226) and the Atlantic Innovation Fund for the Interdisciplinary Research Team in Human Genetics. AAN was supported by the CIHR Interdisciplinary Health Research Team Fellowship. AH was supported by the Walter and Jessie Boyd & Charles Scriver MD/PhD Studentship Award (Canadian Institute of Health Research Institute of Genetics and the Canadian Gene Cure Foundation). The funding sources had no involvement in the study design; in the collection, analysis or interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
References
- 1. Center MM, Jemal A, Smith RA,Ward E (2009) Worldwide variations in colorectal cancer. CA Cancer J Clin 59(6): 366–378. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO) (2008) The global burden of disease: 2004 update. World Health Organization website. Available: http://www.who.int/healthinfo/global_burden_disease/2004_report_update/en/index.html. Accessed 2013 March 19.
- 3.Canadian Cancer Society’s Steering Committee on Cancer Statistics (2011) Canadian Cancer Statistics 2011. May 2011. Canadian Cancer Society website. Available: http://www.cancer.ca/~/media/CCS/Canada%20wide/Files%20List/English%20files%20heading/PDF%20-%20Policy%20-%20Canadian%20Cancer%20Statistics%20-%20English/Canadian%20Cancer%20Statistics%202011%20-%20English.ashx. Accessed 2013 March 19.
- 4. Kemp Z, Thirlwell C, Sieber O, Silver A, Tomlinson I (2004) An update on the genetics of colorectal cancer. Hum Mol Genet 13 Spec No 2: R177–185. [DOI] [PubMed] [Google Scholar]
- 5. Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, et al. (2000) Environmental and heritable factors in the causation of cancer–analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343(2): 78–85. [DOI] [PubMed] [Google Scholar]
- 6.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, et al.. (2010) AJCC (American Joint Committee on Cancer) Cancer Staging Handbook, 7th Edition. In: ed. New York: Springer; p.192–206.
- 7. Popat S, Hubner R, Houlston RS (2005) Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 23(3): 609–618. [DOI] [PubMed] [Google Scholar]
- 8. Compton CC (2002) Pathologic prognostic factors in recurrence of rectal cancer. Clin Colorectal Cancer 2(3): 149–160. [DOI] [PubMed] [Google Scholar]
- 9. Song W, Wu SJ, He YL, Cai SR, Zhang CH, et al. (2009) Clinicopathologic features and survival of patients with colorectal mucinous, signet-ring cell or non-mucinous adenocarcinoma: experience at an institution in Southern China. Chin Med J (Eng) 122(13): 1486–1491. [PubMed] [Google Scholar]
- 10. Lim SB, Yu CS, Jang SJ, Kim TW, Kim JH, et al. (2010) Prognostic significance of lymphovascular invasion in sporadic colorectal cancer. Dis Colon Rectum 53(4): 377–384. [DOI] [PubMed] [Google Scholar]
- 11. Walther A, Houlston R, Tomlinson I (2008) Association between chromosomal instability and prognosis in colorectal cancer: A meta-analysis. Gut 57(7): 941–950. [DOI] [PubMed] [Google Scholar]
- 12. Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, et al. (2005) Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res 65(14): 6063–6069. [DOI] [PubMed] [Google Scholar]
- 13. Xie L, Villeneuve PJ, Shaw A (2009) Survival of patients diagnosed with either colorectal mucinous or non-mucinous adenocarcinoma: a population-based study in Canada. Int J Oncol 34(4): 1109–1115. [DOI] [PubMed] [Google Scholar]
- 14. Maestro ML, Vidaurreta M, Sanz-Casla MT, Rafael S, Veganzones S, et al. (2007) Role of the BRAF mutations in the microsatellite instability genetic pathway in sporadic colorectal cancer. Ann Surg Oncol 14(3): 1229–1236. [DOI] [PubMed] [Google Scholar]
- 15. French AJ, Sargent DJ, Burgart LJ, Foster NR, Kabat BF, et al. (2008) Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin Cancer Res 14(11): 3408–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zell JA, Honda J, Ziogas A, Anton-Culver H (2008) Survival after colorectal cancer diagnosis is associated with colorectal cancer family history. Cancer Epidemiology Biomarkers Prev 17(11): 3134–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kune GA, Kune S, Watson LF (1992) The effect of family history of cancer, religion, parity and migrant status on survival in colorectal cancer. The Melbourne Colorectal Cancer Study. Eur J Cancer 28A((8–9)): 1484–1487. [DOI] [PubMed] [Google Scholar]
- 18. Green RC, Green JS, Buehler SK, Robb JD, Daftary D, et al. (2007) Very high incidence of familial colorectal cancer in Newfoundland: a comparison with Ontario and 13 other population-based studies. Fam Cancer 6(1): 53–62. [DOI] [PubMed] [Google Scholar]
- 19. Woods MO, Younghusband HB, Parfrey PS, Gallinger S, McLaughlin J, et al. (2010) The genetic basis of colorectal cancer in a population-based incident cohort with a high rate of familial disease. Gut 59(10): 1369–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wish TA, Hyde AJ, Parfrey PS, Green JS, Younghusband HB, et al. (2010) Increased Cancer Predisposition in Family Members of Colorectal Cancer Patients Harboring the p.V600E BRAF Mutation: a Population-Based Study. Cancer Epidemiology Biomarkers Prev 19(7): 1831–1839. [DOI] [PubMed] [Google Scholar]
- 21. Savas S, Younghusband HB (2010) dbCPCO: A database of genetic markers tested for their predictive and prognostic value in colorectal cancer. Hum Mutat 31(8): 901–907. [DOI] [PubMed] [Google Scholar]
- 22. Hyde A, Fontaine D, Stuckless S, Green R, Pollett A, et al. (2010) A histology-based model for predicting microsatellite instability in colorectal cancers. Am J Surg Pathol 34(12): 1820–1829. [DOI] [PubMed] [Google Scholar]
- 23. Fernández-Peralta AM, Daimiel L, Nejda N, Iglesias D, Medina Arana V, et al. (2010) Association of polymorphisms MTHFR C677T and A1298C with risk of colorectal cancer, genetic and epigenetic characteristic of tumors, and response to chemotherapy. Int J Colorectal Dis 25(2): 141–151. [DOI] [PubMed] [Google Scholar]
- 24. Zhang W, Press OA, Haiman CA, Yang DY, Gordon MA, et al. (2007) Association of methylenetetrahydrofolate reductase gene polymorphisms and sex-specific survival in patients with metastatic colon cancer. J Clin Oncol 25(24): 3726–3731. [DOI] [PubMed] [Google Scholar]
- 25. Marcuello E, Altés A, Menoyo A, Rio ED, Baiget M (2006) Methylenetetrahydrofolate reductase gene polymorphisms: genomic predictors of clinical response to fluoropyrimidine-based chemotherapy? Cancer Chemother Pharmacol 57(6): 835–840. [DOI] [PubMed] [Google Scholar]
- 26. Sharma R, Hoskins JM, Rivory LP, Zucknick M, London R, et al. (2008) Thymidylate synthase and methylenetetrahydrofolate reductase gene polymorphisms and toxicity to capecitabine in advanced colorectal cancer patients. Clin Cancer Res 14(3): 817–825. [DOI] [PubMed] [Google Scholar]
- 27. Afzal S, Jensen SA, Vainer B, Vogel U, Matsen JP, et al. (2009) MTHFR polymorphisms and 5-FU-based adjuvant chemotherapy in colorectal cancer. Ann Oncol 20(10): 1660–1666. [DOI] [PubMed] [Google Scholar]
- 28. Gusella M, Frigo AC, Bolzonella C, Marinelli R, Barile C, et al. (2009) Predictors of survival and toxicity in patients on adjuvant therapy with 5-fluorouracil for colorectal cancer. Br J Cancer 100(10): 1549–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Etienne-Grimaldi MC, Milano G, Maindrault-Goebel F, Chibaudel B, Formento JL, et al. (2010) Methylenetetrahydrofolate reductase (MTHFR) gene polymorphisms and FOLFOX response in colorectal cancer patients. Br J Clin Pharmacol 69(1): 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boige V, Mendiboure J, Pignon J, Loriot M, Castaing M, et al. (2010) Pharmacogenetic assessment of toxicity and outcome in patients with metastatic colorectal cancer treated with LV5FU2, FOLFOX, and FOLFIRI: FFCD 2000–05. J Clin Oncol 28(15): 2556–2564. [DOI] [PubMed] [Google Scholar]
- 31. van der Put NMJ, Gabreëls F, Stevens EMB, Smeitink JAM, Trijbels FJM, et al. (1998) A second common mutation in the methylenetetrahydrofolate reductase gene: An additional risk factor for neural-tube defects? Am J Hum Genet 62(5): 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weisberg I, Tran P, Christensen B, Sibani S, Rozen R (1998) A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol Genet Metab 64(3): 169–172. [DOI] [PubMed] [Google Scholar]
- 33. Kim YI (2007) Folate and colorectal cancer: an evidence based critical review. Mol Nutr Food Res 51(3): 267–292. [DOI] [PubMed] [Google Scholar]
- 34. Ulrich CM, Potter JD (2007) Folate and Cancer–Timing Is Everything. JAMA 297(21): 2408–2409. [DOI] [PubMed] [Google Scholar]
- 35. Holmes RS, Zheng Y, Baron JA, Li L, McKeown-Eyssen G, et al. (2010) Use of folic acid–containing supplements after a diagnosis of colorectal cancer in the colon cancer family registry. Cancer Epidemiology Biomarkers Prev 19(8): 2023–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim Y (2006) Folate: a magic bullet or a double edged sword for colorectal cancer prevention? Gut 55(10): 1387–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duthie SJ (2011) Folate and cancer: how DNA damage, repair and methylation impact on colon carcinogenesis. J Inherit Metab Dis 34(1): 101–109. [DOI] [PubMed] [Google Scholar]
- 38. Chen Z, Karaplis AC, Ackerman SL, Pogribny IP, Melnyk S, et al. (2001) Mice deficient in methylenetetrahydrofolate reductase exhibit hyperhomocysteinemia and decreased methylation capacity, with neuropathology and aortic lipid deposition. Hum Mol Gen 10(5): 433–443. [DOI] [PubMed] [Google Scholar]
- 39. Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Chan AT, et al. (2008) A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst 100(23): 1734–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Crott JW, Choi S, Ordovas JM, Ditelberg JS, Mason JB (2004) Effects of dietary folate and aging on gene expression in the colonic mucosa of rats: implications for carcinogenesis. Carcinogenesis 25(1): 69–76. [DOI] [PubMed] [Google Scholar]
- 41. Longley DB, Harkin DP, Johnston PG (2003) 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 3(5): 330–338. [DOI] [PubMed] [Google Scholar]
- 42. Sohn K, Croxford R, Yates Z, Lucock M, Kim Y (2004) Effect of the Methylenetetrahydrofolate Reductase C677T Polymorphism on Chemosensitivity of Colon and Breast Cancer Cells to 5-Fluorouracil and Methotrexate. J Natl Cancer Inst 96(2): 134–144. [DOI] [PubMed] [Google Scholar]
- 43. Jakobsen A, Nielsen JN, Gyldenkerne N, Lindeberg J (2005) Thymidylate synthase and methylenetetrahydrofolate reductase gene polymorphism in normal tissue as predictors of fluorouracil sensitivity. J Clin Oncol 23(7): 1365–1369. [DOI] [PubMed] [Google Scholar]
- 44. Ruzzo A, Graziano F, Loupakis F, Rulli E, Canestrari E, et al. (2007) Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFOX-4 chemotherapy. J Clin Oncol 25(10): 1247–1254. [DOI] [PubMed] [Google Scholar]
- 45. Zarate R, Rodríguez J, Bandres E, Patiño-Garcia A, Ponz-Sarvise M, et al. (2010) Oxaliplatin, irinotecan and capecitabine as first-line therapy in metastatic colorectal cancer (mCRC): a dose-finding study and pharmacogenomic analysis. Br J Cancer 102(6): 987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Balboa E, Duran G, Lamas MJ, Gomez-Caamaño A, Celeiro-Muñoz C, et al. (2010) Pharmacogenetic analysis in neoadjuvant chemoradiation for rectal cancer: high incidence of somatic mutations and their relation with response. Pharmacogenomics 11(6): 747–761. [DOI] [PubMed] [Google Scholar]
- 47. Lamas MJ, Duran G, Balboa E, Bernardez B, Touris M, et al. (2011) Use of a comprehensive panel of biomarkers to predict response to a fluorouracil-oxaliplatin regimen in patients with metastatic colorectal cancer. Pharmacogenomics 12(3): 433–442. [DOI] [PubMed] [Google Scholar]
- 48. Cecchin E, Agostini M, Pucciarelli S, De Paoli A, Canzonieri V, et al. (2011) Tumor response is predicted by patient genetic profile in rectal cancer patients treated with neo-adjuvant chemo-radiotherapy. The Pharmacogenomics Journal 11(3): 214–226. [DOI] [PubMed] [Google Scholar]
- 49. Kweekel DM, Antonini NF, Nortier JW, Punt CJ, Gelderblom H, et al. (2009) Explorative study to identify novel candidate genes related to oxaliplatin efficacy and toxicity using a DNA repair array. Br J Cancer 101(2): 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carles J, Monzo M, Amat M, Jansa S, Artells R, et al. (2006) Single-nucleotide polymorphisms in base excision repair, nucleotide excision repair, and double strand break genes as markers for response to radiotherapy in patients with Stage I to II head-and-neck cancer. Int J Radiat Oncol Biol Phys 66(4): 1022–1030. [DOI] [PubMed] [Google Scholar]
- 51. Guo WF, Lin RX, Huang J, Zhou Z, Yang J, et al. (2005) Identification of differentially expressed genes contributing to radioresistance in lung cancer cells using microarray analysis. Radiat Res 164(1): 27–35. [DOI] [PubMed] [Google Scholar]
- 52. Walsh CS, Ogawa S, Karahashi H, Scoles DR, Pavelka JC, et al. (2008) ERCC5 is a novel biomarker of ovarian cancer prognosis. J Clin Oncol 26(18): 2952–2958. [DOI] [PubMed] [Google Scholar]
- 53. McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, et al. (2005) REporting recommendations for tumor MARKer prognostic studies (REMARK). Nat Clin Pract Urol 2(8): 416–422. [PubMed] [Google Scholar]
- 54. Hopkins J, Cescon DW, Tse D, Bradbury P, Xu W, et al. (2008) Genetic polymorphisms and head and neck cancer outcomes: a review. Cancer Epidemiol Biomarkers Prev 17(3): 490–499. [DOI] [PubMed] [Google Scholar]
- 55. Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WD, et al. (1995) Alternate splicing produces a novel cyclin D1 transcript. Oncogene 11(5): 1005–1011. [PubMed] [Google Scholar]
- 56. Schmitt CA, Thaler KR, Wittig BM, Kaulen H, Meyer zum Büschenfelde KH, et al. (1998) Detection of the DCC gene product in normal and malignant colorectal tissues and its relation to a codon 201 mutation. Br J Cancer 77(4): 588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moriai T, Kobrin MS, Hope C, Speck L, Korc M (1994) A variant epidermal growth factor receptor exhibits altered type alpha transforming growth factor binding and transmembrane signaling. Proc Natl Acad Sci USA 91(21): 10217–10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bange J, Prechtl D, Cheburkin Y, Specht K, Harbeck N, et al. (2002) Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res 62(3): 840–847. [PubMed] [Google Scholar]
- 59. Yu JJ, Mu C, Lee KB, Okamoto A, Reed EL, et al. (1997) A nucleotide polymorphism in ERCC1 in human ovarian cancer cell lines and tumor tissues. Mutat Res 82(1–2): 13–20. [DOI] [PubMed] [Google Scholar]
- 60. Lunn RM, Helzlsouer KJ, Parshad R, Umbach DM, Harris EL, et al. (2000) XPD polymorphisms: effects on DNA repair proficiency. Carcinogenesis 21(4): 551–555. [DOI] [PubMed] [Google Scholar]
- 61. Hill JW, Evans MK (2006) Dimerization and opposite base-dependent catalytic impairment of polymorphic S326C OGG1 glycosylase. Nucleic Acids Res 34(5): 1620–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang Y, Spitz MR, Zhu Y, Dong Q, Shete S, et al. (2003) From genotype to phenotype: correlating XRCC1 polymorphisms with mutagen sensitivity. DNA Repair 2(8): 901–908. [DOI] [PubMed] [Google Scholar]
- 63. Yoshihara T, Ishida M, Kinomura A, Katsura M, Tsuruga T, et al. (2004) XRCC3 deficiency results in a defect in recombination and increased endoreduplication in human cells. EMBO J 23(3): 670–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Watson MA, Stewart RK, Smith GB, Massey TE, Bell DA (1998) Human glutathione S-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis 1998 19(2): 275–280. [DOI] [PubMed] [Google Scholar]
- 65. Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, et al. (1998) The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest 102(7): 1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rutter JL, Mitchell TI, Butticè G, Meyers J, Gusella JF, et al. (1998) A single nucleotide polymorphism in the matrix metalloproteinase-1 promoter creates an Ets binding site and augments transcription. Cancer Res 58(23): 5321–5325. [PubMed] [Google Scholar]
- 67. Price SJ, Greaves DR, Watkins H (2001) Identification of novel, functional genetic variants in the human matrix metalloproteinase-2 gene: role of Sp1 in allele-specific transcriptional regulation. J Biol Chem 276(10): 7549–7558. [DOI] [PubMed] [Google Scholar]
- 68. van der Put NM, van den Heuvel LP, Steegers-Theunissen RP, Trijbels FJ, Eskes TK, et al. (1996) Decreased methylene tetrahydrofolate reductase activity due to the 677C–>T mutation in families with spina bifida offspring. J Mol Med (Ber) 74(11): 691–694. [DOI] [PubMed] [Google Scholar]
- 69. Odin E, Wettergren Y, Carlsson G, Danenberg PV, Termini A, et al. (2006) Expression and clinical significance of methylenetetrahydrofolate reductase in patients with colorectal cancer. Clin Colorectal Cancer 5(5): 344–349. [DOI] [PubMed] [Google Scholar]
- 70. Kaneda S, Takeishi K, Ayusawa D, Shimizu K, Seno T, et al. (1987) Role in translation of a triple tandemly repeated sequence in the 5′-untranslated region of human thymidylate synthase mRNA. Nucleic Acids Res 15(3): 1259–7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mandola MV, Stoehlmacher J, Zhang W, Groshen S, Yu MC, et al. (2004) A 6 bp polymorphism in the thymidylate synthase gene causes message instability and is associated with decreased intratumoral TS mRNA levels. Pharmacogenetics 14(5): 319–327. [DOI] [PubMed] [Google Scholar]
- 72. Eriksson P, Kallin B, van’t Hooft FM, Båvenholm P, Hamsten A (1995) Allele-specific increase in basal transcription of the plasminogen-activator inhibitor 1 gene is associated with myocardial infarction. Proc Natl Acad Sci USA 92(6): 1851–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Koukourakis MI, Papazoglou D, Giatromanolaki A, Bougioukas G, Maltezos E, et al. (2004) VEGF gene sequence variation defines VEGF gene expression status and angiogenic activity in non-small cell lung cancer. Lung Cancer 46(3): 293–298. [DOI] [PubMed] [Google Scholar]
- 74. Krippl P, Langsenlehner U, Renner W, Yazdani-Biuki B, Wolf G, et al. (2003) A common 936 C/T gene polymorphism of vascular endothelial growth factor is associated with decreased breast cancer risk. Int J Cancer 103(4): 468–471. [DOI] [PubMed] [Google Scholar]
- 75. Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, et al. (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29(1): 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lin HJ, Han C, Bernstein DA, Hsiao W, Lin BK, et al. (1994) Ethnic distribution of the glutathione transferase Mu 1–1 (GSTM1) null genotype in 1473 individuals and application to bladder cancer susceptibility. Carcinogenesis 15(5): 1077–1081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information. Figure S1 The circled SNP is rs1801131 (MTHFR Glu429Ala), which lies in a 12kb LD block. The black squares indicate other highly correlated SNPs (r2>0.80). Figure S2 The circled SNP is rs1046678 (ERCC5 His46His), which lies in a 23kb LD block. The black squares indicate other highly correlated SNPs (r2>0.80). Table S1 n: number. Table S2 *Assay ID by Applied Biosystems (CA, USA). Underlined are the sequences on probes that are complementary to alleles they recognize. Assays for rs1799750 in MMP1 gene and rs1799889 in SERPINE1 gene were custom designed. Assays for rs1801131 in MTHFR gene and rs1047768 in ERCC5 gene were predesigned by Applied Biosystems. Primer and probe sequences for these assays are not available since they are proprietary of Applied Biosystems. Seq: sequence. Table S3 n/a: not applicable. Polymorphisms with χ2 value greater than 3.84 were considered to be deviating from HWE (p<0.05). *For these gene deletions, since heterozygote genotype cannot be determined by the genotyping method applied, HWE was not calculated. All polymorphisms were investigated in this study regardless of their deviations from the HWE as these deviations may also be attributed to the fact that the Newfoundland population is considered a genetically isolated population [49]. Nevertheless, it is worth noting that while the OGG1 Ser326Cys polymorphism that deviated from the HWE was included in the DFS multivariable model of the discovery cohort, its genotype data was not available for the validation cohort patients. Thus, the main conclusion on the disease-free survival analysis that the ERCC5 His46His polymorphism was associated with DFS in both the discovery and the validation patient cohorts is not affected by including this polymorphism in the DFS analysis of the discovery cohort. Table S4 CI: confidence interval, HR: hazard ratio, MSI: microsatellite instability, n: number of patients, vs: versus. Table S5 CI: confidence interval, HR: hazard ratio, MSI: microsatellite instability, n: number of patients, vs: versus. Table S6 CI: confidence interval, HR: hazard ratio, MSI: microsatellite instability, n: number of patients, vs: versus. Table S7 CI: confidence interval, HR: hazard ratio, MSI: microsatellite instability, n: number of patients, vs: versus. Methods S1 Genotyping reactions. Methods S2 Construction of linkage disequilibrium (LD) maps.
(DOC)