Abstract
Background
The epigenome refers to marks on the genome, including DNA methylation and histone modifications, that regulate the expression of underlying genes. A consistent profile of gene expression changes in end-stage cardiomyopathy led us to hypothesize that distinct global patterns of the epigenome may also exist.
Methods and Results
We constructed genome-wide maps of DNA methylation and histone-3 lysine-36 trimethylation (H3K36me3) enrichment for cardiomyopathic and normal human hearts. More than 506 Mb sequences per library were generated by high-throughput sequencing, allowing us to assign methylation scores to ≈28 million CG dinucleotides in the human genome. DNA methylation was significantly different in promoter CpG islands, intragenic CpG islands, gene bodies, and H3K36me3-enriched regions of the genome. DNA methylation differences were present in promoters of upregulated genes but not downregulated genes. H3K36me3 enrichment itself was also significantly different in coding regions of the genome. Specifically, abundance of RNA transcripts encoded by the DUX4 locus correlated to differential DNA methylation and H3K36me3 enrichment. In vitro, Dux gene expression was responsive to a specific inhibitor of DNA methyltransferase, and Dux siRNA knockdown led to reduced cell viability.
Conclusions
Distinct epigenomic patterns exist in important DNA elements of the cardiac genome in human end-stage cardiomyopathy. The epigenome may control the expression of local or distal genes with critical functions in myocardial stress response. If epigenomic patterns track with disease progression, assays for the epigenome may be useful for assessing prognosis in heart failure. Further studies are needed to determine whether and how the epigenome contributes to the development of cardiomyopathy.
Keywords: genes, genome-wide analysis, genomics, heart failure
Altered gene expression is a characteristic hallmark of heart failure.1–4 Moreover, analysis of the transcriptome in human cardiomyopathy shows that altered gene expression may be used to predict disease severity.5 Although gene expression changes can be explained by pathways that link to the assembly of cardiac transcription factors at gene promoters during myocardial stress,6 our better understanding of epigenomics7 now allows us to determine whether epigenomic changes also occur in heart failure.
The epigenome refers to marks on the genome, including DNA methylation and histone modification. These marks do not change the underlying genetic sequence but regulate how underlying genes are expressed.7 Unlike the genome, which is largely stable, the epigenome is dynamic and allows organisms to respond and adapt to environmental cues. The environment therefore promotes epigenomic changes with consequent plasticity in gene expression and phenotype.8
Strong evidence indicates that drifts in DNA methylation occur with increasing age.9,10 In addition, the recent discovery of active DNA demethylation in postmitotic cells11 and rapid and dynamic DNA methylation/demethylation in vivo12,13 further challenges the conventional view that DNA methylation is a stable epigenetic mark. Dynamic changes in DNA methylation may therefore link the environment to disease pathogenesis.8 The role of DNA methylation has been proven in cancer14 and implicated in other complex diseases, including psychosis,15 type II diabetes mellitus,16 and atherosclerosis,17,18 but its role in heart failure is unknown.
DNA methylation occurs on cytosine nucleotides in the mammalian epigenome. Methylated cytosines usually precede guanines as CpG dinucleotides, and clusters of CpG-rich regions in the genome are called CpG islands (CGIs). We have previously reported that a large population of CGIs and gene promoters are more significantly hypomethylated in end-stage cardiomyopathic (EsCM) hearts.19 Here, we sought to extend our study to examine DNA methylation profiles in EsCM across the whole cardiac genome. Although myocardial gene expression may differ according to the type of heart failure and other confounding variables, etiology-linked gene expression changes are relevant in early heart failure when gene expression patterns segregate, for example between ischemic and idiopathic cardiomyopathy. In contrast, gene expression in end-stage heart failure follows a generic and convergent “final common pathway” that disregards the initial cause.4,20 The final common pathway pattern of gene expression concurs with generic final pathway processes such as apoptosis, fibrosis, altered contractility, and altered energetics that are found in all forms of end-stage heart failure, regardless of the initial cause. To determine whether generic final pathway changes also occur in the epigenomes of heart failure, we compared the composite DNA methylation and histone-3 lysine-36 trimethylation (H3K36me3) enrichment profiles of both ischemic and idiopathic EsCM hearts with that of age- and sex-matched normal controls.
Methods
Ethics Statement
Human left ventricular (LV) tissue was collected with a protocol approved by the Papworth (Cambridge) Hospital Tissue Bank Review Board and the Cambridgeshire Research Ethics Committee (UK). Written consent was obtained from every individual according to the Papworth Tissue Bank protocol.
Human LV
LV tissues were from patients undergoing cardiac transplantation for EsCM, including both ischemic and idiopathic cardiomyopathy. Normal LV tissues were from healthy male individuals (UK Human Tissue Bank, de Montfort University, UK). At the time of transplantation or donor harvest, whole hearts were removed after preservation and transported as previously described.19,21 After analysis by a cardiovascular pathologist (M.G.), the LV segments were cut and immediately stored in RNAlater (Ambion, Applied Biosystems, Warrington, UK). Individual patient details are listed in Table I in the online-only Data Supplement.
Methylated DNA Immunoprecipitation Sequencing
Genomic DNA from LV tissues was isolated with the Qiagen Genomic Tip-500/G anionic columns (catalog No. 10262) and the Genomic DNA Buffer Set (catalog No. 19060) as described by the manufacturer with minor adjustments. Briefly, ≈200 mg tissue was homogenized in G2 lysis buffer containing 80 μg/mL of RNase A with a handheld homogenizer (Polytron), and proteinase K was added to a final concentration of 1 mg/mL and incubated at 50°C for at least 2 hours while rotating until all the tissue was fully digested. Samples were subsequently centrifuged at 5000g for 10 minutes, and genomic DNA was isolated from the supernatant with the Genomic Tip-500/G anionic columns. Subsequently, at least 6 μg of gDNAs from the control and EsCM samples was sheared 3 times for 10 minutes each time with a Bioruptor probe (Diagenode, Belgium) on ice at high setting (30 seconds on, 30 seconds off) and then passed through Qiagen QIAPrep Spin columns (catalog No. 27104) to clean up the samples (and to dispose of fragments <100 bp in size). The extent of shearing was confirmed by running 300 ng of samples on 1.5% agarose gel. All samples were sheared to a similar extent, ranging from 100 to 500 bp, with the majority of fragments at ≈150 to 200 bp.
With the use of the Illumina DNA Sample Prep kit (FC-102–1001-1), 5 μg of each of the sheared gDNAs was end repaired, adenosines bases were added to the blunt 3′ ends, and respective adaptors were added to the DNA fragments, exactly as recommended by the manufacturer. After each step, samples were cleaned up with QIAquick Spin columns (catalog No. 28104). Subsequently, samples were heated at 95°C for 10 minutes and immediately cooled on ice for 10 minutes. Then, 2.2 μg of single-stranded gDNA was used for methylated DNA immunoprecipitation (MeDIP), and the rest was stored at −20°C as input.
MeDIP was performed with 7.5 μg of the 5′-methyl cytosine antibody (catalog No. MAb-5MECYT-500, Diagenode) in 500 μL of immunoprecipitation buffer (10 mmol/L sodium phosphate, pH 7.0, 140 mmol/L NaCl, 0.05% Triton X-100) and incubated for 2.5 hours at 4°C while rotating. Next, 40 μL of 50% protein A agarose slurry (Santa Cruz, sc-2001) in immunoprecipitation buffer was added and incubated for another 2.5 hours while rotating at 4°C. Protein A agarose beads were subsequently spun down and washed 3 times, 10 minutes each time, with immunoprecipitation buffer before eluting with 250 μL of digestion buffer and rotating at 55°C for another 2.5 hours. Finally, the enriched methylated gDNA was purified with ×2 phenol:chloroform isolation and chloroform wash and precipitated with sodium chloride. After another wash with 70% ethanol, samples were quantified on a Nanodrop, and enrichment was performed using a polymerase chain reaction (PCR) step with Illumina primers 1.1 and 1.2. After 14 cycles of nonsaturating PCR (conditions as recommended by Illumina), samples were cleaned up with the QIAquick Spin columns and quantified on a Nanodrop. We used 20 ng of each sample to confirm enrichment of methylated loci (OXT) and a concomitant depletion of unmethylated loci (UBE2B) versus the input by quantitative PCR.19 Finally, MeDIP samples were loaded on a 2% agarose gel, 150- to 250-bp bands were cut, and DNA was eluted with the Qiagen gel extraction kit and quantified with the Bioanalyser. Considering that we used Solexa Library Single End Primers 1 (92 bp long), this short library is expected to contain insert sizes ranging from 50 to 150 bp long. We sequenced each of the short libraries on 2 lanes of the Illumina GAII machine. More than 14 million reads of 36-bp length were generated for each MeDIP-seq library.
H3K36me3 Chromatin Immunoprecipitation Sequencing
About 200 mg LV tissue was cut into small pieces and homogenized in 4 mL of ice-cold PBS on ice. Cell dispersion was verified under the microscope with Trypan blue. The homogenate was centrifuged at 5100g for 5 minutes at 4°C, and the first cross-linking step was performed by resuspending cell pellets in 4 mL of 2 mmol/L diglutarate (Sigma, 80–424)/PBS fixative plus protease inhibitor (PI; Calbiochem, 539131) and rotating for 45 minutes at room temperature. The second cross-linking step was performed after centrifugation at 5100g for 5 minutes at 4°C and resuspending the pellet in 10 mL of 1% formaldehyde/PBS+PI and rotating at room temperature for 10 minutes only. Then, 1.1 mL of 1.375 mol/L (10×) glycine was added directly to each tube to stop-fix. Cell pellets were centrifuged at 5100g for 5 minutes at 4°C again, and each pellet was washed in 10 mL ice-cold PBS+PI. Intact nuclei were released from the cell pellets by resuspension in 600 μL lysis buffer (50 mmol/L Tris-HCl, pH 8.1, 10 mmol/L EDTA, 1% SDS)+PI and sonication in the Bioruptor (5 cycles of 30 minutes on, 30 minutes off). This sonication setting disrupted nuclei and sheared gDNA to a range of 200 to 1000 bp. After centrifugation at 13 000g for 5 minutes at 4°C, a 50-μL aliquot of supernatant containing chromatin was kept aside and reverse cross-linked to quantify the amount of DNA and to confirm the extent of shearing by running on an agarose gel. The rest of the sample was frozen at −80°C for up to 1 month for chromatin immunoprecipitation.
Reverse cross-link was performed by addition of a final concentration of 200 mmol/L NaCl and 20 μg proteinase K (Sigma, P2308) and incubating at 65°C for at least 4 hours while gently rocking. Subsequently, 10 μg RNase A was added to each sample and incubated at 37°C for 15 minutes, and gDNA was immediately extracted using an equal volume of phenol:chloroform. After another chloroform wash, gDNA was precipitated by incubating overnight at −20°C with 300 nmol/L sodium acetate and 20 μg glycogen (Roche; catalog No. 10 901 393 001) in 2.5 vol% ethanol. After centrifugation at 13 000g for 30 minutes at 4°C, the pellet was washed with 1 mL of ice-cold 70% ethanol, dissolved in 20 μL of elution buffer (10 mmol/L Tris-HCl, pH 7.6), and quantified with a Nanodrop. A volume of chromatin corresponding to at least 5 μg gDNA is typically used per chromatin immunoprecipitation (ChIP).
For each ChIP, 1.5 mg protein-G Dyna magnetic beads (Invitrogen catalog No. 100.04D) was prepared by washing at least 5 times with 5 mg/mL BSA/PBS while rotating. Next, beads were conjugated to 12 μg H3K36me3 monoclonal antibody (ab9050, Abcam) in 1.5 mL of 5 mg/mL BSA/PBS+PI by rotating at 4°C for at least 6 hours. Antibody-conjugated beads were made to a final volume of 1.5 mL of ChIP dilution buffer (20 mmol/L Tris-HCl, pH 8.1, 2 mmol/L EDTA, 1% Triton, 150 mmol/L NaCl)+PI containing the volume of chromatin equivalent to 5 μg gDNA (as predetermined by reverse cross-linking the input). ChIP was carried out by rotating overnight at 4°C. Dyna magnetic beads were captured with a magnetic rack, and beads were washed at least 6 times with 1.5 mL of radioimmunoprecipitation assay buffer (50 mmol/L HEPES, pH 7.6, 1 mmol/L EDTA, 1% NP40, 0.7% deoxycholate, 500 mmol/L LiCl)+PI for at least 10 minutes each time and once with 1.5 mL of Tris-EDTA buffer (10 mmol/L Tris, 1 mmol/L EDTA, pH 8.0)+PI. gDNA was eluted twice by adding 200 μL ChIP elution buffer (1% SDS, 100 mmol/L NaHCO3) to the beads while shaking vigorously for 15 minutes at room temperature. ChIP gDNA isolated as described above.
Two Illumina single-end read libraries were constructed by pooling ChIP gDNA from 4 control hearts and 3 cardiomyopathic hearts (Table I in the online-only Data Supplement) for control and EsCM, respectively. The sizes of these libraries were confirmed with the Bioanalyser to range between 200 and 400 bp. Each library was sequenced in 2 lanes of the Illumina GAII yielding >50 million reads of 36-bp reads (>506 Mb) per sequencing library. These data will be deposited on GEO.
Bioinformatics
Reads from high-throughput sequencing for MeDIP were aligned to the human reference genome with MAQ (http://maq.sourceforge.net/maq-man.shtml) with up to 90% alignment success, generating at least 14 million aligned 36-bp reads (504 Mb) per sequencing library. Aligned MeDIP-seq data were analyzed with a bayesian deconvolution strategy, BATMAN.22 Methylation/BATMAN scores from 4 normal hearts and 4 EsCM hearts were averaged, and methylation densities were generated from previously described algorithms.23,24 Error bars in the density plots represent bayesian credible intervals. This method of MeDIP-seq provided wide coverage across the genome, covering ≈24 million of the 28 million CG dinucleotides in the entire genome, except for regions with high repeat density where analysis was technically not reliable. Annotation for protein-coding genes in the human genome (hg18) was obtained from the UCSC Genome Browser. Annotation for CGIs and subclasses of CGIs were as described in Maunakea et al.25 Annotation for CGIs also corresponded to the UCSC Genome Browser.
Reads from high-throughput sequencing for H3K36me3 ChIP were aligned by the use of BWA (http://bio-bwa.sourceforge.net/bwa.shtml) with up to 90% alignment success. Control and EsCM libraries were each constructed by pooling H3K36me3 ChIP from 4 normal and 3 EsCM hearts, respectively. More than 50 million aligned 36-bp reads (1800 Mb) were generated per sequencing library. Enrichment scores were determined by sequence tag counts at 500-bp intervals using BEDTools (http://sourceforge.net/projects/bedtools/webcite). Intersections between all data sets were computed with the Table Browser in UCSC Genome Browser or BEDTools. The data set for both MeDIP-seq and H3K36me3 Chromatin immunoprecipitation sequencing will be deposited on GEO.
HL1 Mouse Cardiac Cell Culture and siRNA Knockdown
The HL1 mouse atrial cardiomyocyte cell line was cultured as described elsewhere.26,27 HL1 cells were cultured in Claycomb culture media with or without the rationally designed specific inhibitor of DNA methyltransferase (RG108, 100 μmol/L)28 for 48 hours before RNA extraction. Transient transfection (48 hours) of siRNA duplexes (Dharmacon, Denver, CO), siGENOME nontargeting siRNA (D-001210–01) and siGENOME SMART pool siDux (M-167163–00), was performed by electroporation with Amaxa Nucleofection according to manufacturer’s instructions (Lonza Ltd, Basel, Switzerland). Methylthiazolyldiphenyl–tetrazolium bromide (MTT) cell viability assay was performed according to manufacturer’s instructions (RND Systems, Abingdon, Oxon, UK).
RNA Preparation
RNA was extracted from LV tissue by homogenizing at least 30 mg of frozen tissue in 0.5 mL of TRIreagent (Sigma-Aldrich, St. Louis, MO) with a handheld homogenizer (Polytron). RNA was extracted from the HL1 mouse cardiac muscle cell line by scrapping adherent cells in TRIreagent. Homogenates were centrifuged at 3000 rpm for 3 minutes; supernatant was transferred to a clean Eppendorf; and RNA extraction was performed according to the manufacturer’s protocol with the following modification. After chloroform extraction, ethanol was added to samples to a final concentration of 35%, and samples were loaded onto PureLink RNA columns (Invitrogen, 12183–018A). On-column DNase treatment was carried out with elution of the RNA. Integrity of all RNA samples was checked with the 2100 Bioanalyser (Agilent Technologies).
Whole-Genome cDNA-Mediated Annealing, Section, Extension, and Ligation Expression Array
The whole-genome cDNA-mediated annealing, section, extension, and ligation (DASL, Illumina) assay analyzes >29 000 human genes by using unique PCR and labeling steps of the GoldenGate Assay. It is particularly suitable even for partially degraded RNA because probe groups in the assay span only about 50 bases (http://www.illumina.com/technology/dasl_assay.ilmn). The DASL assay was performed by Cambridge Genomics Services (http://www.cgs.path.cam.ac.uk/) according to Illumina protocols. We used DNase-treated RNA samples isolated from 8 normal and 16 EsCM hearts. The quality control report generated with the arrayQualityMetrics R package showed that none of the samples failed or was an outlier to their replicates. NPPA and NPPB were the 2 most differentially expressed genes (>9-fold upregulated in EsCM; Figure IA in the online-only Data Supplement); 2266 and 1919 genes were found to be >1.2-fold upregulated and downregulated, respectively (adjusted P<0.05; data to be deposited in GEO). Four genes were selected for validation by quantitative PCR (Figure IB in the online-only Data Supplement).
Quantitative Real-Time PCR
cDNA (20 μL) was synthesized from 1 mg total RNA using a mixture of both oligo-dT and random hexamers and the Superscript-III first-strand cDNA synthesis kit (Invitrogen, Paisley, UK). Quantitative real-time PCR for housekeeping genes (RPLPO and TBP) was performed with 4 μL of 1:20 prediluted cDNA in a 20 μL reaction and Taqman Gene Expression Assays (see Table II in the online-only Data Supplement for primer sequences). Quantitative PCR was performed for target genes using validated Taqman Gene Expression Assay primers (Applied Biosystems, Foster City, CA) and normalized by a normalization factor obtained for each sample with geNorm (http://medgen.ugent.be/≈jvdesomp/genorm/) and the expression of both housekeeping genes. All quantitative PCRs were performed at least in triplicate and on the same diluted cDNA samples.
Statistics
We adapted the statistical analysis of Stegle et al29 to test whether there is a significant difference in the methylation pattern of genomic regions in cardiomyopathy. We used a bayesian model selection to compare a model in which both methylation patterns are draws from the same Gaussian process30 with a model in which each pattern is a draw from its own Gaussian process. A positive log Bayes factor for this analysis represents a significant difference. Comparisons that gave a positive log Bayes factor (ie, presence of a significant difference between the methylation patterns of control and EsCM) were further analyzed to determine specifically where differential methylation was in each comparison. We calculated how different the signal was at each position using the symmetrical Kullback-Lieber divergence (a measure of how different 2 distributions are). Symmetrical Kulback-Lieber distances are charted above each methylation density plot with a positive log Bayes factor. Student t test was performed for the analysis of quantitative PCR.
Results
Genome-Wide DNA Methylation Profiles
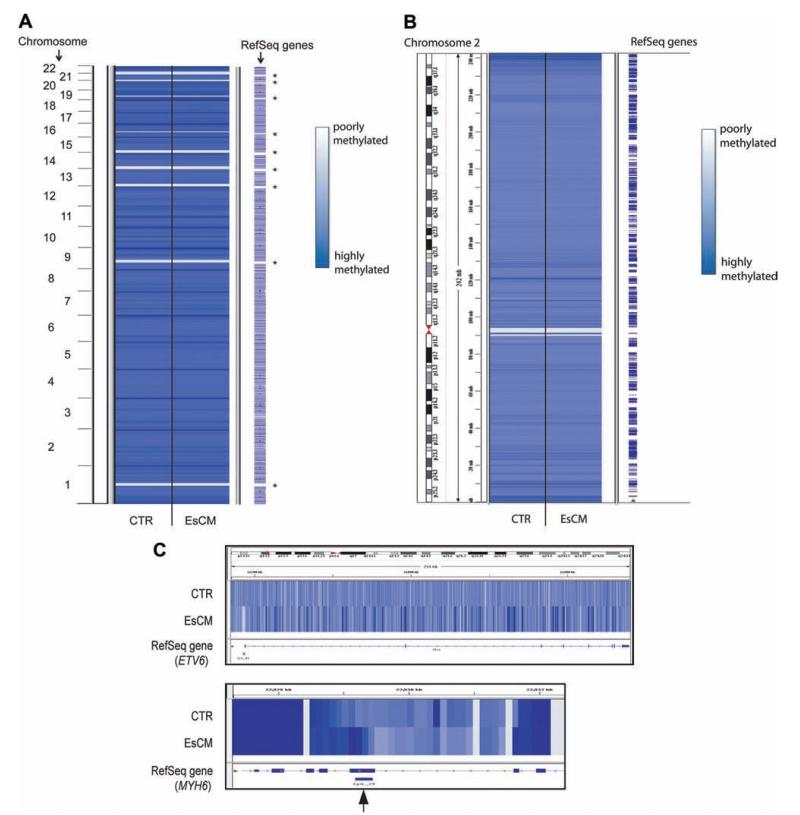
We generated genome-wide profiles of DNA methylation in human hearts by using genomic DNA from LVs and immunoprecipitation with a validated antimethylated cytosine antibody (MeDIP).22,31 Successful immunoprecipitation was confirmed by quantitative PCR for genomic regions, including a positive control methylated region (OXT) and negative control unmethylated region (UBE2B; data not shown).19 High-throughput sequencing was performed and generated >504 Mb of aligned sequences for each of 8 MeDIP libraries constructed from 4 control and 4 cardiomyopathic LVs. Details on patient ages, cause of death, and medication at the time of death are given in Table I in the online-only Data Supplement. Sequences were aligned to the human reference genome (hg18), and DNA methylation/BATMAN scores were assigned at 100-bp windows across the genome for each MeDIP library.22 Average BATMAN scores were obtained for cardiomyopathic (EsCM) and control hearts, covering ≈28 million of the CG dinucleotides in the human genome. DNA methylation profiles across all chromosomes were remarkably similar between EsCM and control (Figure 1A and B). In contrast, marked differences were observed at the level of individual gene loci (example shown in Figure 1C).
Figure 1.
DNA methylation profiles between control (CTR) and end-stage cardiomyopathic (EsCM) hearts are similar at low resolution (A and B), but distinct differences are found at the level of individual genes (C) and at specific CpG islands (An example is shown with the arrow in C). Averaged DNA methylation profiles were generated for control and EsCM from 8 sequencing libraries comprising 4 normal and 4 EsCM hearts and scored at 100-bp resolution genome wide. Analysis was technically not possible in some gene-poor regions where a high density of repetitive elements was present (asterisk in A).
DNA Methylation Profiles in CGIs and Shores
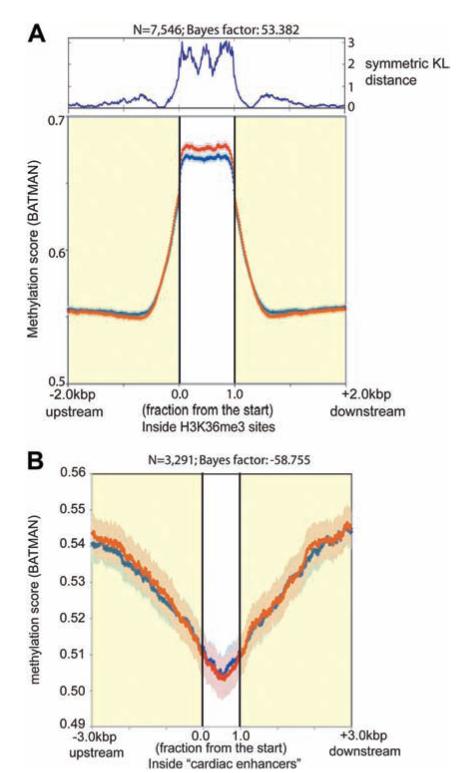
In cancer, differential DNA methylation has been recognized in CGIs32 and “CGI shores.”33 We therefore examined these genomic regions in our DNA methylation maps. An aggregate pattern of hypomethylation was found centered on all CGIs (n=27 639; Figure 2A) in both EsCM and control. Methylation in the center of CGIs was significantly reduced compared with higher methylation scores at CGI shores. To determine whether DNA methylation profiles for all 27 639 CGIs were significantly different between EsCM and control, we compared the 2 averaged curves by using a Gaussian process 2-sample test.29 The log Bayes factor for these curves was 15.9, a positive value indicating a significant difference. To show positions of differential methylation across the x axis, the symmetrical Kulback-Lieber divergence (a measure of how different 2 distributions are) was calculated for each position, and symmetrical Kulback-Lieber distances were charted above the methylation density plot. Positions of higher values in symmetrical Kulback-Lieber distance indicate that the methylation profiles differed both at the center of CGIs and at CGI shores, similar to the finding in some cancers.33
Figure 2.
DNA methylation density plots for all CpG islands (CGIs; A) and subclasses of CGIs (B through E). A, DNA methylation scores (BATMAN) for 27 639 CGIs and CGI shores (3 kb upstream and downstream of CGIs) were plotted for control (CTR; blue) and end-stage cardiomyopathic (EsCM; red; light blue and light red error bars represent bayesian credible intervals for CTR and EsCM, respectively). A Gaussian process 2-sample test returned a log Bayes factor of 15.9 (A positive log Bayes factor implies that the difference between control and EsCM was statistically significant). The symmetrical Kulback-Lieber (KL) divergence was calculated for each position across the x axis and charted above the methylation density plots as symmetrical KL distance and indicates that the methylation profiles differed both at the center of CGIs and at CGI shores. Significant differences in methylation profiles were also found in (B) promoter CGIs and (C) intragenic CGIs but not in (D) intergenic CGIs and (E) 3′ untranslated region (UTR) CGIs. However in the first 2 subclasses, methylation profiles were significantly different at CGIs but not CGI shores.
Methylation profiles in subclasses of CGIs were previously analyzed according to their location in relation to annotated genes.25,34 We found that the methylation profile of EsCM differed significantly from control in CGIs at gene promoters (promoter CGIs; Figure 2B; n=11 967; Bayes factor=533.0) and CGIs within bodies of genes (intragenic CGIs; Figure 2C; n=8092; Bayes factor=59.8) but not intergenic CGIs (Figure 2D; n=7068; Bayes factor=−71.7) and CGIs in 3′ untranslated region of genes (Figure 2E; n=512; Bayes factor=−63.8), as summarized in the Table. In EsCM, promoter CGIs were more hypomethylated and intragenic CGIs were more hypermethylated. Symmetrical Kulback-Lieber distance scores for each set of plots showed that methylation in these subgroups differed in CGIs and but not in CGI shores. Moreover, the methylation score for EsCM was decreased in promoter CGIs to the same extent as methylation was increased in intragenic CGIs. Methylation scores for control and EsCM at promoter CGIs and intragenic CGIs are listed in the online-only Data Supplement.
Table. Summary of Regions in the Cardiac Genome in Which Epigenomic Profiles Were Significantly Different Between Control and End-Stage Cardiomyopathy.
| Sample Size, n |
Bayes Factor |
Significance* | |
|---|---|---|---|
| All CGIs | 27 639 | 15.9 | + |
| Promoter CGIs | 11 967 | 533.0 | + |
| Intragenic CGIs | 8092 | 59.8 | + |
| 3′ UTR CGIs | 7068 | −71.7 | x |
| Intergenic CGIs | 512 | −68.8 | x |
| Promoters of upregulated genes | 2266 | 12.6 | + |
| Promoters of downregulated genes | 1919 | −48.3 | x |
| Active transcribed regions | 7546 | 53.4 | + |
| Cardiac enhancers | 3291 | −58.8 | x |
CGI indicates CpG island; UTR untranslated region; +, statistical significance; x, statistical nonsignificance.
Significant difference between the methylation profiles for normal and cardiomyopathy.
DNA Methylation Profiles in Promoters of Upregulated and Downregulated Genes
Next, we determined whether DNA methylation changes in gene promoters correlated with genes that were upregulated or downregulated in cardiomyopathy. Previous studies have shown that there is a consistent profile of gene expression changes in EsCM hearts.3,35,36 Hence, we performed gene expression profiling using the whole-genome DASL Illumina assay for our series of hearts consisting of 16 EsCM (6 ischemic, 10 idiopathic) and 8 control hearts and generated a list of statistically significant >1.2-fold upregulated (n=2266) and >1.2-fold downregulated (n=1919) genes (adjusted P<0.05; Figure IA in the online-only Data Supplement). A selection of genes from this was validated by standard quantitative PCR (Figure IB in the online-only Data Supplement). Methylation in the promoters of upregulated genes was significantly reduced in EsCM versus control (Figure 3A; n=2266; Bayes factor=12.6) but unchanged in the promoters of downregulated genes (Figure 3B) (n=1919; Bayes factor=−48.3). The former is consistent with the predicted correlation between reduced DNA methylation in the promoters of genes that are more highly expressed. Conversely, downregulated genes in cardiomyopathy were independent of changes in gene promoter methylation.
Figure 3.
DNA methylation was significantly different in promoters of (A) upregulated genes but not (B) downregulated genes.
DNA Methylation Profiles in Actively Transcribed Regions of the Genome
DNA methylation differences in intragenic CGIs (Figure 2C) led us to consider whether altered methylation also exists in actively transcribed regions of the cardiac genome. In general, evidence suggests that highly expressed genes have more densely methylated gene bodies.37 In addition, the Encyclopedia of DNA Elements (ENCODE) Consortium and others have shown that the human genome undergoes pervasive transcription.34,38 Thus, although protein-coding genes account for only a small proportion of the genome, the rest of the genome also undergoes active transcription to form non–protein-coding RNA. The functional role for each of these newly discovered noncoding RNA species remains to be elucidated, but H3K36me3 marks actively transcribed regions of the genome.39 We therefore performed high-throughput sequencing for H3K36me3 ChIP and generated maps of actively transcribed regions of the cardiac genome for both control and EsCM (Figure II in the online-only Data Supplement). Consistent with the finding of more DNA hypermethylation in intragenic CGIs in EsCM, we also found increased DNA methylation in regions of active transcription in EsCM (Figure 4A; n=7546; Bayes factor=53.4). In contrast, DNA methylation profiles did not differ between EsCM and control in cardiac enhancers, as mapped by p300 binding sites in the heart40 (Figure 4B; n=3291; Bayes factor=−58.8).
Figure 4.
DNA methylation was significantly different in (A) actively transcribed regions of the cardiac genome but not (B) cardiac enhancers. Regions of active transcription in the cardiac genome were determined by H3K36me3 chromatin immunoprecipitation sequencing (ChIP-seq), and regions of cardiac enhancers were obtained from a previous p300 ChIP-seq.40
Genome-Wide H3K36me3 Landscape in Human Hearts
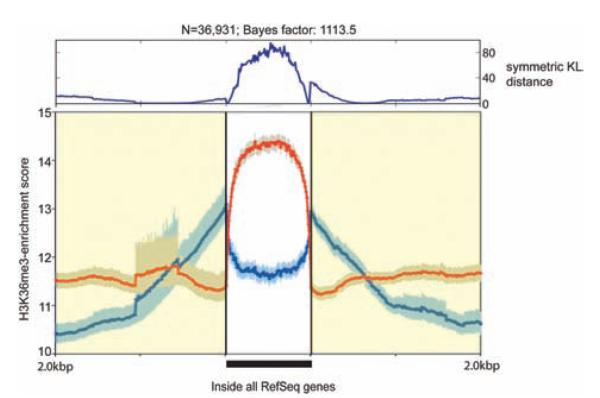
The H3K36me3 enrichment maps were generated for control and EsCM by scoring sequence tag counts per 500-bp window, normalized to 1 million reads per sequencing library (Figure II in the online-only Data Supplement). We assessed regions of all Reference Sequence (RefSeq) genes (n=36 931) in these maps and found a distinct pattern of enrichment in EsCM (Figure 5). This pattern of difference was consistent for both upregulated and downregulated genes (Figure III in the online-only Data Supplement). Enrichment scores for control and EsCM at each RefSeq locus are listed in the online-only Data Supplement.
Figure 5.
H3K36me3 enrichment profiles within regions of annotated Reference Sequence (RefSeq) genes were significantly different between control (CTR) and end-stage cardiomyopathy (EsCM). Composite H3K36me3 enrichment profiles were generated for control (blue) and EsCM (red) from 2 pooled sequencing libraries comprising 4 normal and 3 EsCM hearts, respectively (see Table I in the online-only Data Supplement for details of samples). Enrichment scores were determined as tag counts per 500 bp per 1 million reads for each sequencing library and analyzed for all 36 109 RefSeq genes (as annotated on the UCSC Genome Browser) and 3 kb upstream and downstream of these regions.
Next, we examined the top 4 regions of H3K36me3 enrichment that did not fall within annotated protein-coding genes (Figure IVA and IVB in the online-only Data Supplement). Because H3K36me3 marks active transcription, these genomic regions are predicted to form noncoding RNA in the heart. We therefore performed quantitative PCR with DNase-treated RNA from our series of LV samples (16 cardiomyopathic and 8 control hearts) and discovered first that RNA transcripts were present from these regions even though they have not previously been annotated to represent any known genes. The absence of PCR products from no reverse transcriptase controls in these reactions excluded the likelihood of genomic DNA amplification. Moreover, we found that the abundance of RNA transcripts from these regions correlated to the level of H3K36me3 enrichment. For region of interest 1, higher H3K36me3 enrichment in control correlated with a higher abundance of noncoding RNA in control hearts (Figure IV in the online-only Data Supplement). Conversely, for region of interest 2, region of interest 3, and region of interest 4, higher H3K36me3 enrichment in cardiomyopathy correlated with higher abundance of noncoding RNA in cardiomyopathic hearts. Similarly, mRNA abundance of protein-coding genes (from the DASL expression array) also correlated to the level of H3K36me3 enrichment (Figure VA in the online-only Data Supplement). Finally, H3K36me3 enrichment within all protein-coding genes also correlated to the level of DNA methylation within the bodies of the same genes (Figure VB in the online-only Data Supplement).
Differential Methylation and Expression of the DUX4 Locus
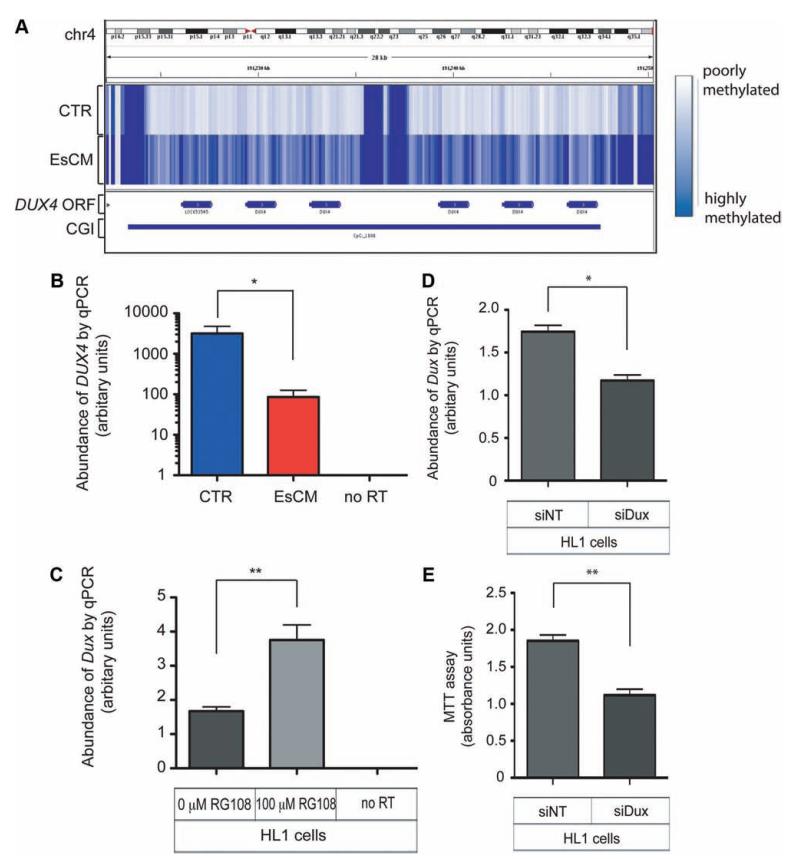
To investigate the physiological relevance of altered DNA methylation in the heart, we chose to examine the CGIs (chr4:191223376–191247590, 4q35.2) with the highest level of MeDIP enrichment associated with an annotated gene locus (DUX4). Methylation of this CGI was higher in EsCM hearts compared with control (see the online-only Data Supplement). Numerous properties of this locus have in fact been reported before because of its association to the hereditary myopathic disorder facioscapulohumeral muscular dystrophy.41 The DUX4 open reading frame is intronless but unusual because it lies within the subtelomeric array of D4Z4 macrosatellite repeat units (4q35.2; Figure 6A). Hence, although it was classified as a retrotransposed gene, DUX4 was eventually shown to code for a double homeobox transcription factor (DUX4),42 and more recent evidence shows that this locus also gives rise to spliced isoforms43 and bidirectional (sense and antisense) non–protein-coding RNA transcripts.44 Moreover, differential DNA methylation and histone modification at this locus have also been reported.45,46 Patients with facioscapulohumeral muscular dystrophy have a contracted number of D4Z4 repeats but can also present with upregulation of DUX4 protein, altered DNA methylation of DUX4, or altered expression of the noncoding RNA transcripts arising from this locus.41
Figure 6.
Differential DNA methylation and expression of DUX4 in human hearts and Dux in the mouse HL1 cardiac cell line. A, The intronless DUX4 open reading frame is embedded within the subtelomeric array of D4Z4 repeat units. The CpG island (CGI) in this locus is hypermethylated in end-stage cardiomyopathy (EsCM) hearts but not in control (CTR). B, The abundance of DUX4 RNA transcripts was quantified with DNase-treated RNA from a panel of left ventricular tissue (8 control and 16 EsCM; Table I in the online-only Data Supplement) and normalized by geNorm that was generated with 2 housekeeping genes, RPLPO and TBP. Absent polymerase chain reaction (PCR) products in “no reverse transcriptase (RT) ” controls excluded the likelihood of genomic DNA amplification. *P<0.05. C, The mouse HL1 cardiac cell line was cultured with or without the specific inhibitor of DNA methyltransferase RG108 for 48 hours, and Dux RNA abundance was quantified with DNase-treated RNA and normalized to the housekeeping gene Gapdh (n=3; **P<0.01). D, HL1 cardiac cells were transfected for 48 hours with either control nontargeting siRNA (siNT) or siRNA targeting mouse Dux (siDux). Dux abundance was quantified as in C (n=3; *P<0.05). E, Methylthiazolyldiphenyl–tetrazolium bromide (MTT) assay was performed as described in Methods and represents survival of HL1 cells 48 hours after transfection with either siNT or siDux (n=3; **P<0.01). Statistical analyses for quantitative PCR (qPCR) and the MTT assay were by the Student t test, and all error bars represent the SEM.
Using our panel of human LV samples, we tested the abundance of RNA transcripts from the DUX4 locus and found that, corresponding to hypermethylation in EsCM, expression of DUX4 was significantly reduced in EsCM hearts compared with control (Figure 6B). Notably, this also corresponded to reduced H3K36me3 enrichment in EsCM hearts at this locus (see the online-only Data Supplement).
Analyzing sequence data from primates, rodents, afrotherian, and other species, Clapp et al47 concluded that the D4Z4-like repeat family is evolutionally conserved and the mouse representative Dux is embedded within repeat units on chromosome 10. We therefore used the mouse HL1 cardiac cell line and assayed for the abundance of Dux RNA transcripts. Treatment of HL1 cells with the rationally designed specific inhibitor of DNA methyltransferase (RG108)28 for 48 hours resulted in upregulation of Dux compared with unchanged levels of the housekeeping gene Gapdh (Figure 6C), indicating that Dux expression, but not Gapdh, was responsive to inhibition of DNA methylation. Bosnakovski et al48 previously demonstrated the biphasic myopathic phenotype of mouse Dux in which high Dux expression led to myoblast death and low Dux levels blocked myogenic differentiation. Similarly, different isoforms of noncoding RNA from the Dux locus affected myogenic differentiation and myosin heavy chain synthesis.44 In HL1 cells, we found that Dux knockdown by siRNA resulted in a corresponding decrease in cell viability as measured by the MTT assay (Figure 6D and 6E).
Discussion
This study is the first to demonstrate global epigenomic profiles in human cardiomyopathy. We have generated DNA methylation and H3K36me3 maps from human hearts, characterized the profiles of them, and uncovered distinct differences in important elements of the cardiac genome.
We found that DNA methylation differs between EsCM and control human hearts in CGIs globally within gene promoters and gene bodies but not in intergenic CGIs and in 3′ untranslated region CGIs. A significant decrease in global gene promoter methylation also correlated with genes that were upregulated in cardiomyopathy but not with genes that were downregulated. The former is consistent with the paradigm that promoter demethylation leads to increased expression of the corresponding gene, whereas the latter suggests that downregulation of genes in cardiomyopathy occurs independently of promoter methylation. This study is also consistent with our previous report in which we used a different series of hearts and a different assay and found that large numbers of CGIs and gene promoters were significantly more hypomethylated in human cardiomyopathy.19 Several animal models have demonstrated the link between altered DNA methylation, gene expression changes, and consequent changes in phenotype.49–53 Although the cause or consequence effect of our observations remains to be proven in similar animal models, the finding of DNA methylation differences in specific elements of the cardiac genome opens up important new horizons for research. For example, our finding that Dux RNA transcript abundance was upregulated by inhibition of DNA methylation in vitro corresponds to the correlation between DUX4 hypermethylation and downregulated DUX4 expression in EsCM hearts. This implicates a mechanism by which DNA methylation changes may be functionally relevant to gene expression changes found in heart failure.
Although the role of DUX4 in facioscapulohumeral muscular dystrophy is increasingly understood,41 its function in myocardial disease has never been examined. The presence of RNA transcripts from these DUX4 repeat units and the other non–protein-coding loci that we have examined and the observation that these RNA transcripts are regulated in EsCM hearts certainly highlight the likelihood that many functionally relevant noncoding RNA in the heart remain to be identified.
We have listed sites of specific differential methylation and tabulated them with their adjacent annotated genes. However, sites of differential DNA methylation are also known to control the expression of remote genes up to 20 kb away.54 This is especially so when considering intrachromosomal and interchromosomal interactions55 that may be brought together by transcription factors such as CTCF,56 the binding of which to its cognate motif is dependent on DNA methylation of the locus.57 However, it is not yet possible to link specific sites of differential DNA methylation to distal or remote genes.
As a practical tool, however, we have identified epigenomic differences in DNA methylation in promoter CGIs, DNA methylation in intragenic CGIs, and DNA methylation and H3K36me3 enrichment in gene bodies or actively transcribed regions of the genome. Subsequent studies are needed to determine whether these features, like profiles of gene expression, predict disease severity or normalize with recovery of LV function.
The genome-wide map of H3K36me3 has also allowed us to identify loci of active transcription in the cardiac genome. We validated that these corresponded to protein-coding genes in our DASL microarray, 4 novel noncoding RNA, and DUX4. This demonstrates the means by which the H3K36me3 map may be used to identify other noncoding RNA transcripts in the heart.38
The prohibitive cost of high-throughput sequencing currently limits our ability to screen large numbers of hearts to identify specific loci of differential methylation or genes that may be specifically controlled by DNA methylation or histone modification in cardiomyopathy. The use of human cardiac samples also limits our ability to develop these descriptive findings beyond an association between epigenomic differences and disease. However, our approach of a meta-analysis using large numbers of global DNA elements and composite epigenomic maps revealed the presence of distinct differences in important elements of the cardiac genome. If DNA methylation or H3K36me3 is a driver of disease development or progression, these may be new means of screening or predicting heart failure. Although quantification of mRNA transcripts is useful for assessing prognosis in heart failure,5 assays for epigenomic profiles will be less complex because of the stability of DNA compared with RNA. Further studies are necessary to determine whether these profiles track with disease progression and indeed whether pathways that lead to these epigenomic changes may also be novel targets for heart failure therapy.
Supplementary Material
CLINICAL PERSPECTIVE.
Molecular research in heart failure points to the hypothesis that a generic cardiac genomic response is activated in the failing myocardium regardless of the original inciting cause. This cardiac genomic stress response is typified by the re-expression of fetal genes, upregulation of fibrosis-related genes, and others. It may also reflect unifying features of a failed myocardium such as fibrosis, altered energy use, vascular rarefaction, cell death, and altered contractility, all of which are features found in the failing myocardium again regardless of the inciting cause. The epigenome refers to “marks” on the genome, including DNA methylation and histone modification. The epigenome regulates the expression of underlying genes, and recent evidence suggests that part of the epigenome may be altered by diet and the environment. In our research, we have examined the genome-wide landscape of the epigenome of healthy and end-stage cardiomyopathic human hearts. We have determined that distinct epigenomic patterns exist in important DNA elements of the human cardiac genome in the failing myocardium. Individually, sites of differential epigenomic patterns may control the expression of specific genes with critical functions in the progression of heart failure. However whether the altered epigenome is simply a consequence in end-stage disease or actually contributes to disease progression remains to be determined. This work now opens an important new avenue of research because the epigenome may represent a drug discovery target for novel heart failure therapy.
Acknowledgments
We acknowledge the contribution of Dr Julien Bauer (Cambridge Genomic Services), who performed the statistical analysis for the gene expression array data set in Figure IA of the online-only Data Supplement.
Sources of Funding This work was supported by grants from the British Heart Foundation (PG/06/101/21461 and FS/07/035), the Cambridge Biomedical Research Centre, and The Addenbrooke’s Charitable Trust.
Footnotes
Disclosures None.
The online-only Data Supplement is available with this article at http://circ.ahajournals.org/lookup/suppl/doi:10.1161/CIRCULATIONAHA.111.040071/-/DC1.
References
- 1.Tan FL, Moravec CS, Li J, Apperson-Hansen C, McCarthy PM, Young JB, Bond M. The gene expression fingerprint of human heart failure. Proc Natl Acad Sci U S A. 2002;99:11387–11392. doi: 10.1073/pnas.162370099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mudd JO, Kass DA. Tackling heart failure in the twenty-first century. Nature. 2008;451:919–928. doi: 10.1038/nature06798. [DOI] [PubMed] [Google Scholar]
- 3.Dorn GW, 2nd, Matkovich SJ. Put your chips on transcriptomics. Circulation. 2008;118:216–218. doi: 10.1161/CIRCULATIONAHA.108.789933. [DOI] [PubMed] [Google Scholar]
- 4.Creemers EE, Wilde AA, Pinto YM. Heart failure: advances through genomics. Nat Rev Genet. 2011;12:357–362. doi: 10.1038/nrg2983. [DOI] [PubMed] [Google Scholar]
- 5.Heidecker B, Kasper EK, Wittstein IS, Champion HC, Breton E, Russell SD, Kittleson MM, Baughman KL, Hare JM. Transcriptomic biomarkers for individual risk assessment in new-onset heart failure. Circulation. 2008;118:238–246. doi: 10.1161/CIRCULATIONAHA.107.756544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 7.Maunakea AK, Chepelev I, Zhao K, Bruneau B. Epigenome mapping in normal and disease States. Circ Res. 2010;107:327–339. doi: 10.1161/CIRCRESAHA.110.222463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 9.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rakyan VK, Down TA, Maslau S, Andrew T, Yang TP, Beyan H, Whittaker P, McCann OT, Finer S, Valdes AM, Leslie RD, Deloukas P, Spector TD. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2011;20:434–439. doi: 10.1101/gr.103101.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klug M, Heinz S, Gebhard C, Schwarzfischer L, Krause SW, Andreesen R, Rehli M. Active DNA demethylation in human postmitotic cells correlates with activating histone modifications, but not transcription levels. Genome Biol. 2010;11:R63. doi: 10.1186/gb-2010-11-6-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 13.Kim MS, Kondo T, Takada I, Youn MY, Yamamoto Y, Takahashi S, Matsumoto T, Fujiyama S, Shirode Y, Yamaoka I, Kitagawa H, Takeyama K, Shibuya H, Ohtake F, Kato S. DNA demethylation in hormone-induced transcriptional derepression. Nature. 2009;461:1007–1012. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
- 14.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 15.Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Jia P, Assadzadeh A, Flanagan J, Schumacher A, Wang SC, Petronis A. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barres R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, Krook A, Zierath JR. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10:189–198. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Post WS, Goldschmidt-Clermont PJ, Wilhide CC, Heldman AW, Sussman MS, Ouyang P, Milliken EE, Issa JP. Methylation of the estrogen receptor gene is associated with aging and atherosclerosis in the cardiovascular system. Cardiovasc Res. 1999;43:985–991. doi: 10.1016/s0008-6363(99)00153-4. [DOI] [PubMed] [Google Scholar]
- 18.Hiltunen MO, Yla-Herttuala S. DNA methylation, smooth muscle cells, and atherogenesis. Arterioscler Thromb Vasc Biol. 2003;23:1750–1753. doi: 10.1161/01.ATV.0000092871.30563.41. [DOI] [PubMed] [Google Scholar]
- 19.Movassagh M, Choy MK, Goddard M, Bennett MR, Down TA, Foo RS. Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PLoS One. 2010;5:e8564. doi: 10.1371/journal.pone.0008564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, Dorn LE, Watson MA, Margulies KB, Dorn GW., 2nd Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. 2009;119:1263–1271. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choy MK, Movassagh M, Siggens L, Vujic A, Goddard M, Sanchez A, Perkins N, Figg N, Bennett M, Carroll J, Foo R. High-throughput sequencing identifies STAT3 as the DNA-associated factor for p53-NF-kappaB-complex-dependent gene expression in human heart failure. Genome Med. 2010;2:37. doi: 10.1186/gm158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Graf S, Johnson N, Herrero J, Tomazou EM, Thorne NP, Backdahl L, Herberth M, Howe KL, Jackson DK, Miretti MM, Marioni JC, Birney E, Hubbard TJ, Durbin R, Tavare S, Beck S. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choy MK, Movassagh M, Goh HG, Bennett MR, Down TA, Foo RS. Genome-wide conserved consensus transcription factor binding motifs are hyper-methylated. BMC Genomics. 2010;11:519. doi: 10.1186/1471-2164-11-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.George CH, Rogers SA, Bertrand BM, Tunwell RE, Thomas NL, Steele DS, Cox EV, Pepper C, Hazeel CJ, Claycomb WC, Lai FA. Alternative splicing of ryanodine receptors modulates cardiomyocyte Ca2+ signaling and susceptibility to apoptosis. Circ Res. 2007;100:874–883. doi: 10.1161/01.RES.0000260804.77807.cf. [DOI] [PubMed] [Google Scholar]
- 27.Claycomb WC, Lanson NA, Jr, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, Izzo NJ., Jr HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci U S A. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brueckner B, Garcia Boy R, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M, Lyko F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305–6311. doi: 10.1158/0008-5472.CAN-04-2957. [DOI] [PubMed] [Google Scholar]
- 29.Stegle OKD, Denby K, Wild DL, Ghahramani Z, Borgwardt KM. [Accessed October 17, 2011];A robust Bayesian two-sample test for detecting intervals of differential gene expression in microarray time series. Lecture Notes in Computer Science (Research in Computational Molecular Biology) doi: 10.1089/cmb.2009.0175. DOI: 10.1007/978-3-642– 02008-7_14. http://www.springerlink.com/content/n1vx358044812384. [DOI] [PMC free article] [PubMed]
- 30.Rassmussen CE, Williams CKI. Gaussian Processes for Machine Learning. MIT Press; Cambridge, MA: 2006. [Google Scholar]
- 31.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 32.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 33.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP. The human colon cancer methylome shows similar hypoand hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, Micklem G, Piano F, Snyder M, Stein L, White KP, Waterston RH. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bristow MR. Microarray measurements of gene expression before and after left ventricular assist device placement. J Am Coll Cardiol. 2003;41:1107–1108. doi: 10.1016/s0735-1097(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 36.Hannenhalli S, Putt ME, Gilmore JM, Wang J, Parmacek MS, Epstein JA, Morrisey EE, Margulies KB, Cappola TP. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation. 2006;114:1269–1276. doi: 10.1161/CIRCULATIONAHA.106.632430. [DOI] [PubMed] [Google Scholar]
- 37.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Bristow J, Ren B, Black BL, Rubin EM, Visel A, Pennacchio LA. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearson CE. FSHD: a repeat contraction disease finally ready to expand (our understanding of its pathogenesis) PLoS Genet. 2010;6:e1001180. doi: 10.1371/journal.pgen.1001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dixit M, Ansseau E, Tassin A, Winokur S, Shi R, Qian H, Sauvage S, Matteotti C, van Acker AM, Leo O, Figlewicz D, Barro M, Laoudj-Chenivesse D, Belayew A, Coppee F, Chen YW. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci U S A. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snider L, Geng LN, Lemmers RJ, Kyba M, Ware CB, Nelson AM, Tawil R, Filippova GN, van der Maarel SM, Tapscott SJ, Miller DG. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snider L, Asawachaicharn A, Tyler AE, Geng LN, Petek LM, Maves L, Miller DG, Lemmers RJ, Winokur ST, Tawil R, van der Maarel SM, Filippova GN, Tapscott SJ. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum Mol Genet. 2009;18:2414–2430. doi: 10.1093/hmg/ddp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Overveld PG, Lemmers RJ, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- 46.Zeng W, de Greef JC, Chen YY, Chien R, Kong X, Gregson HC, Winokur ST, Pyle A, Robertson KD, Schmiesing JA, Kimonis VE, Balog J, Frants RR, Ball AR, Jr, Lock LF, Donovan PJ, van der Maarel SM, Yokomori K. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clapp J, Mitchell LM, Bolland DJ, Fantes J, Corcoran AE, Scotting PJ, Armour JA, Hewitt JE. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2007;81:264–279. doi: 10.1086/519311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bosnakovski D, Daughters RS, Xu Z, Slack JM, Kyba M. Biphasic myopathic phenotype of mouse DUX, an ORF within conserved FSHD-related repeats. PLoS One. 2009;4:e7003. doi: 10.1371/journal.pone.0007003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 50.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- 51.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 53.LaPlant Q, Vialou V, Covington HE, 3rd, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iniguez SD, Koo JW, Mouzon E, Renthal W, Hollis F, Wang H, Noonan MA, Ren Y, Eisch AJ, Bolanos CA, Kabbaj M, Xiao G, Neve RL, Hurd YL, Oosting RS, Fan G, Morrison JH, Nestler EJ. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Botta M, Haider S, Leung IX, Lio P, Mozziconacci J. Intra- and interchromosomal interactions correlate with CTCF binding genome wide. Mol Syst Biol. 2010;6:426. doi: 10.1038/msb.2010.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mukhopadhyay R, Yu W, Whitehead J, Xu J, Lezcano M, Pack S, Kanduri C, Kanduri M, Ginjala V, Vostrov A, Quitschke W, Chernukhin I, Klenova E, Lobanenkov V, Ohlsson R. The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Genome Res. 2004;14:1594–1602. doi: 10.1101/gr.2408304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.