Abstract
Computational solvent mapping finds binding hot spots, determines their druggability and provides information for drug design. While mapping of a ligand-bound structure yields more accurate results, usually the apo structure serves as the starting point in design. The FTFlex algorithm, implemented as a server, can modify an apo structure to yield mapping results that are similar to those of the respective bound structure. Thus, FTFlex is an extension of our FTMap server, which only considers rigid structures. FTFlex identifies flexible residues within the binding site and determines alternative conformations using a rotamer library. In cases where the mapping results of the apo structure were in poor agreement with those of the bound structure, FTFlex was able to yield a modified apo structure, which lead to improved FTMap results. In cases where the mapping results of the apo and bound structures were in good agreement, no new structure was predicted.
Availability: FTFlex is freely available as a web-based server at http://ftflex.bu.edu/.
Contact: vajda@bu.edu or midas@bu.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Computational solvent mapping emerged as an important tool for the identification and characterization of binding sites in proteins (Brenke et al., 2009), for determining their druggability (Kozakov et al., 2011) and for providing information for fragment-based drug design (Hall et al., 2012). Mimicking the radiographic crystallographic approach of multiple solvent crystal structures (Mattos and Ringe, 1996), this computational method places molecular probes—small organic molecules containing various functional groups—on a dense grid defined around the protein, finds favorable positions using empirical energy functions, further refines the selected poses by energy minimization, clusters the low-energy conformations, calculates the partition function for each cluster and ranks the clusters on the basis of their probabilities. The consensus sites are then defined as those where clusters of different probes overlap; such consensus sites identify the binding hot spots, i.e. regions of interactions that substantially contribute to the binding free energy and hence are of prime importance to drug design.
We have shown that the approximate location of binding hot spots is generally not affected by conformational changes, and can be identified by mapping unbound protein structures even in cases in which the goal is to find small molecule binding sites in protein–protein interfaces (Kozakov et al., 2011). However, the exact shape of hot spots may change on ligand binding, and hence, accounting for protein flexibility provides more accurate mapping results. Mapping snapshots from molecular dynamics simulations is a useful but computationally demanding approach (Ivetac and McCammon, 2012; Landon et al., 2008). Here we describe the FTFlex algorithm, which incorporates side chain flexibility considerations into our previously developed computational mapping method, FTMap (Brenke et al., 2009), in an efficient fashion, while providing similarly accurate results.
2 METHODS
The algorithm, FTFlex, identifies flexible side chains around an initial hot spot, generates their alternative conformers via the use of a previously developed rotamer library (Beglov et al., 2012), and adjusts the side chains one-by-one to generate a modified structure with a maximally opened binding site. Side chains are treated individually, that is, rotamers are generated for a particular side chain, while all other side chains are held fixed in their original conformation. FTFlex was tested on a set of 15 apo structures with varying degrees of side chain movement; the results were compared with the respective bound structures. These targets were selected from the Astex non-native data set (Verdonk et al., 2008) based on either the presence or absence of side chain motion. Details regarding the FTFlex algorithm and selection of the data set can be found in the Supporting Information.
3 RESULTS
To increase efficiency, FTFlex only considers residues that exhibit (i) significant flexibility as evident by the large number of possible rotamers and (ii) significant contributions to binding site size based on RMSD. Thus, FTFlex considers rotamers of the following residues: Arg, His, Lys, Met, Phe, Trp, Tyr, Asp, Asn, Glu and Gln. To test this implementation, the FTFlex algorithm was used to map the apo structures for 15 targets. The apo structures were first mapped using only FTMap—these structures are referred to as ‘apo structures’ throughout.
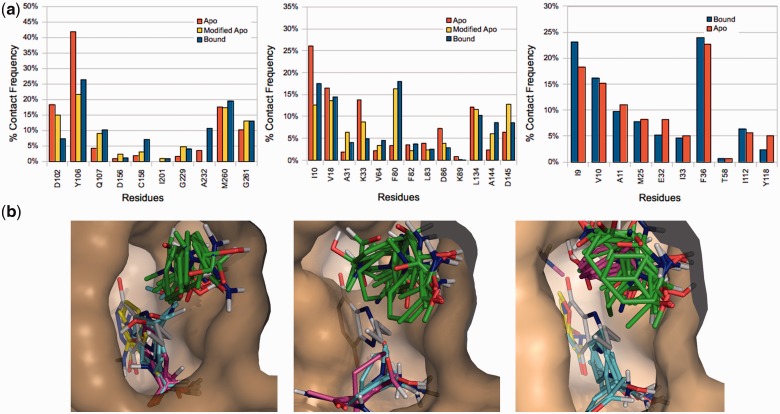
The mapping results for apo, FTFlex-modified apo and bound structures were compared in terms of % contact frequencies, defined as the number of non-bonded contacts between probes and each individual residue, divided by the total number of non-bonded contacts. We also calculated correlation coefficients between the % contact frequencies for apo and bound structures, as well as for modified apo and bound structures. The more similar are the probe–protein interactions, the higher the correlation value. Figure 1a shows % contact frequencies for tRNA-guanine transglycosylase (TGT), left, and cyclin-dependent kinase 2 (CDK2), center. In both cases, there is a deviation in the mapping results obtained using FTMap for the apo and bound structures as indicated by the differences in the probe–protein interactions. For TGT, this deviation can be attributed to Tyr106, which blocks the ligand binding site in the apo structure (Fig. 1b, left and center panels for bound and apo mapping results, respectively). In the FTFlex-modified apo structure, Tyr106 has been rotated out of the site (Fig. 1b, right), which opens the pocket and yields mapping results in better agreement with those of the bound structure. A similar improvement is observed for CDK2 (Fig. 1a, center). For dihydrofolate reductase (DHFR, PDB ID 1ai9), there are minimal structural differences between the apo and bound structures, and FTFlex predicts no new conformations (Fig. 1a).
Fig. 1.
Selected results from FTFlex test set. (a) Distribution of non-bonded contacts between probe clusters and residues from the mapping of apo, FTFlex-modified apo and bound structures. The % contact frequency is defined as the number of non-bonded contacts between probes and each individual residue, divided by the total number of non-bonded contacts. Results are shown for TGT (left), CDK2 (center) and DHFR (right). (b) Probe cluster centers in the bound (left, PDB file 1n2v), apo (center, PDB file 1pud) and modified apo (right) TGT active sites (tan surface), with the target ligand (white sticks) shown
4 DISCUSSION
Using FTFlex, we have shown the following results: (i) Mapping results were improved in cases of poor correlation between bound and apo structures. (ii) When the mapping results for apo and bound structures were similar, no new structure was predicted. (iii) For Tyr, Phe, Trp and His, the new residue conformation predicted by FTFlex for the apo structure was similar to the conformation found in the bound structure.
Funding: This work was supported by grants NIH R01GM064700 and NIH R41GM097907 from NIGMS.
Conflict of Interest: D.K., S.V. and D.B. hold stock in Acpharis Inc.
Supplementary Material
REFERENCES
- Beglov D, et al. Minimal ensembles of side chain conformers for modeling protein-protein interactions. Proteins. 2012;80:591–601. doi: 10.1002/prot.23222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenke R, et al. Fragment-based identification of druggable ‘hot spots' of proteins using Fourier domain correlation techniques. Bioinformatics. 2009;25:621–627. doi: 10.1093/bioinformatics/btp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DR, et al. Hot spot analysis for driving the development of hits into leads in fragment-based drug discovery. J. Chem. Inf. Model. 2012;52:199–209. doi: 10.1021/ci200468p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivetac A, McCammon JA. A molecular dynamics ensemble-based approach for the mapping of druggable binding sites. Methods Mol. Biol. 2012;819:3–12. doi: 10.1007/978-1-61779-465-0_1. [DOI] [PubMed] [Google Scholar]
- Kozakov D, et al. Structural conservation of druggable hot spots in protein-protein interfaces. Proc. Natl Acad. Sci. USA. 2011;108:13528–13533. doi: 10.1073/pnas.1101835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landon MR, et al. Novel druggable hot spots in avian influenza neuraminidase H5N1 revealed by computational solvent mapping of a reduced and representative receptor ensemble. Chem. Biol. Drug Des. 2008;71:106–116. doi: 10.1111/j.1747-0285.2007.00614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattos C, Ringe D. Locating and characterizing binding sites on proteins. Nat. Biotechnol. 1996;14:595–599. doi: 10.1038/nbt0596-595. [DOI] [PubMed] [Google Scholar]
- Verdonk ML, et al. Protein-ligand docking against non-native protein conformers. J. Chem. Inf. Model. 2008;48:2214–2225. doi: 10.1021/ci8002254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.