Abstract
In 2001, we reported linkage of an autosomal dominant form of limb-girdle muscular dystrophy, limb-girdle muscular dystrophy 1F, to chromosome 7q32.1-32.2, but the identity of the mutant gene was elusive. Here, using a whole genome sequencing strategy, we identified the causative mutation of limb-girdle muscular dystrophy 1F, a heterozygous single nucleotide deletion (c.2771del) in the termination codon of transportin 3 (TNPO3). This gene is situated within the chromosomal region linked to the disease and encodes a nuclear membrane protein belonging to the importin beta family. TNPO3 transports serine/arginine-rich proteins into the nucleus, and has been identified as a key factor in the HIV-import process into the nucleus. The mutation is predicted to generate a 15-amino acid extension of the C-terminus of the protein, segregates with the clinical phenotype, and is absent in genomic sequence databases and a set of >200 control alleles. In skeletal muscle of affected individuals, expression of the mutant messenger RNA and histological abnormalities of nuclei and TNPO3 indicate altered TNPO3 function. Our results demonstrate that the TNPO3 mutation is the cause of limb-girdle muscular dystrophy 1F, expand our knowledge of the molecular basis of muscular dystrophies and bolster the importance of defects of nuclear envelope proteins as causes of inherited myopathies.
Keywords: limb-girdle muscular dystrophy 1F, LGMD1F, TNPO3, transportin 3, c.2771del mutation
Introduction
The limb-girdle muscular dystrophies (LGMDs) comprise a group of genetically heterogeneous disorders characterized by progressive and predominantly proximal muscle weakness with histological signs of degeneration and regeneration in muscle (Bushby, 2009). As a result of molecular characterization and improved clinical criteria, the classification and nomenclature of LGMDs have evolved over the last two decades. The canonical categorization of LGMD into autosomal dominant-LGMD (LGMD1) and autosomal recessive (LGMD2) forms is being refined by a classification based on the affected proteins and their correspondent genes (Nigro et al., 2011)
In 2001, we reported the clinical and morphological phenotype of a novel form of autosomal dominant-LGMD affecting 32 individuals in a large Spanish kindred spanning five generations (Gamez et al., 2001). Clinically, the disorder was characterized by muscle weakness primarily affecting the pelvic and shoulder girdles with a wide variability in the age at onset (1–58 years old), disease progression rate and severity. The disease generally ran a benign clinical course, but some individuals with childhood or juvenile onset manifested severe widespread myopathy leading to wheelchair dependency and respiratory insufficiency. Additional clinical features of LGMD1F as well as more detailed descriptions of its time-course and pattern of muscle involvement are presented here.
Initially, the presence of rimmed vacuoles and filamentous inclusions in myofibres of affected subjects prompted us to consider a diagnosis of hereditary inclusion body myopathy (Huizing and Krasnewich, 2009). Hereditary inclusion body myopathy typically presents as an autosomal recessive trait and is due to mutations in GNE (9p13.3; Genebank NM_005476); however, a few cases of autosomal dominant hereditary inclusion body myopathy have also been described and linked to chromosome 17p13.1 (Martinsson et al., 1999) (IBM3 OMIM #605637) or to 7q22.1-31.1 (Lu et al., 2012) (IBM4). Hereditary inclusion body myopathy was ruled out in our family because initial genetic analysis using simple sequence repeat markers indicated that the disorder was not linked to hereditary inclusion body myopathy. Our subsequent studies using genome-wide markers demonstrated a novel locus for this autosomal dominant-LGMD at the chromosomal locus 7q32.1-32.2, between markers D7S1822 and D7S2519, containing 66 genes. These data confirmed that this family has a genetically distinct form of autosomal dominant-LGMD that was classified as LGMD1F (Palenzuela et al., 2003) (OMIM #608423). However, the identity of the mutant gene has been elusive so far, despite attempts to find it following different strategies. Here, using a whole genome sequencing approach, we have identified the causative mutation of the LGMD1F, a single nucleotide deletion in the termination codon of transportin 3 (TNPO3). The histochemical and ultrastructural findings, together with the molecular results at DNA, RNA and protein levels, fully support the pathogenic role of this mutation in LGMD1F.
Materials and methods
Patients
The reported genealogical investigation of the LGMD1F family (Gamez et al., 2001) disclosed a common ancestor born in south-eastern Spain two generations before the oldest living members. The largest branch originated from Subject II-3 (Gamez et al., 2001) and includes 32 patients with LGMD1F, of whom 28 have been closely followed in our centre (University Hospital La Fe, València, Spain). Functional activity was assessed using the Brooke score (from 1: normal; to 6: no function for upper extremity) (Brooke et al., 1981) and the Vignos score (1: able to climb stairs without help; to 10: bedridden for lower limb function) (Vignos et al., 1963). Muscle strength was graded using the Modified Medical Research Council (MMRC) scale. Whole-body muscle imaging was performed on a 1.5 T or 3 T MRI scanner. Abnormal muscle signal intensity was ranked according to Mercuri scale (Mercuri et al., 2002): 1, normal appearance; 2, moth-eaten appearance with scattered small areas of increased signal involving <30% of muscle volume; 3, moderate involvement (a late moth-eaten appearance with numerous discrete areas of increased signal with incipient confluence, involving 30–60% of muscle volume); 4, severe involvement (washed-out or fuzzy appearance due to confluent areas of increased signal, or complete muscle replacement by connective tissue and fat with only a rim of fascia and neurovascular structures).
All pedigree identifiers in this report refer to the updated family tree shown in Supplementary Fig. 1, unless otherwise indicated.
Muscle biopsies
Muscle biopsies from the deltoid or vastus lateralis from 5 of the 32 affected individuals of the family had been performed in the years 1993 and 1994 under informed consent (Gamez et al., 2001) and stored at −80°C at the Neurological Tissue Biobank of Vigo University Hospital. Frozen muscle specimens from Subjects IV-6, IV-11 and IV-21 (Supplementary Fig. 1) (Gamez et al., 2001) were retrieved from the Biobank and further studied by light microscopy. For ultrastructural studies, original electron microscopy micrographs were re-examined. Stored Epon-embedded blocks were used to obtain new ultrathin sections, and were studied under a Philips CM100 transmission electron microscope equipped with a digital camera and the ITEM SYSTEM VELETA (FEI Company) software.
DNA and RNA isolation and complementary DNA synthesis
DNA was extracted from anticoagulated blood from affected and unaffected individuals using a standard phenol–chloroform method. RNA was extracted from skeletal muscle biopsies from Subjects IV-6 and IV-21 and one unrelated healthy control using the RNeasy® Kit (QIAGEN), and treated with the deoxyribonuclease I, amplification grade (Invitrogen) to eliminate any traces of DNA. Then, complementary DNA was synthesized using the High Capacity complementary DNA Reverse Transcription kit, which uses random hexamers (Applied Biosystems).
Whole genome sequencing
Sequencing libraries were constructed according to the TruSeq™ DNA sample preparation protocol (Illumina) with minor modifications, in particular the double size selection. Two micrograms of genomic DNA were fragmented with a Covaris E210 and size selected to 300–700 bp. Resulting fragments were end-repaired, adenylated, ligated to Illumina paired-end adaptors and size selected to very tight sizes using an E-Gel® (Life Technologies). Size selected adapter-insert fragments (two insert sizes: 430 bp and 460 bp) were amplified with 10 PCR cycles and sequenced on an Illumina HiSeq 2000 platform with paired end run of 2 × 100 bp. Base calling and quality control was performed on the Illumina RTA sequence analysis pipeline. Sequence reads were trimmed to the first base with a quality over 30 and mapped to Human genome build hg19 (GRCh37) using GEM mapper (Marco-Sola et al., 2012), allowing up to four mismatches. Reads not mapped by GEM mapper (∼4%) were submitted to a last round of mapping with BFAST (Homer et al., 2009). Results were merged and only uniquely mapping non-duplicate read pairs were used for further analyses. SAM tools suite version 0.1.18 (Li et al., 2009) with default settings was used to call single nucleotide variants and short indels. Variants on regions with low mappability (Derrien et al., 2012), with read depth <10 or with strand bias P-value < 0.001 were filtered out. The population frequency of the variants was assessed by comparing to several databases: the 1000 Genomes Project (http://www.1000genomes.org/), NHLBI Exome Sequencing Project (ESP) release ESP5400 (http://evs.gs.washington.edu/EVS/), and our internal database of sequence variants identified in a set of >100 control samples). The effect prediction was performed with Annovar version 2011 Dec20 (Wang et al., 2010) and snpEff version 2.0.5d (Cingolani et al., 2012).
Dideoxy-DNA sequencing
DNA extracted from blood was used to confirm the segregation of the genotype and the phenotype. A 856 bp fragment encompassing the last coding sequence of the TNPO3 gene was PCR-amplified (forward, 5’-TCCTCAGTCAAGGACCAACCTACCT-3’; reverse, 5’-TCCTGTAAGGGCCAAGCATCCCT-3’), and the product was purified (ExoSAP-IT®, Affimetrix) and sequenced using the dideoxy method (BigDye® Terminator v3.1 Cycle Sequencing kit, Applied Biosystems). In order to analyse the sequences of RNA species, complementary DNA obtained from skeletal muscle of affected and unaffected individuals was PCR-amplified (644 bp fragment, exons 20–24, primers forward 5’-TCTACTACCCTGGACCACCG-3’ and reverse 5’-GCGCTGATTTTCCCTCACAC-3’) and the resulting fragments were sequenced.
Polymerase chain reaction–restriction fragment length polymorphism analysis
A 629 bp fragment was PCR-amplified (Forward primer 5’-TCTACTACCCTGGACCACCG-3’ and Reverse primer 5’-CACACCCCCAAACAGGAACT-3’) from skeletal muscle complementary DNA from Subjects IV-6, IV-21 and one unrelated healthy control subject. The products were digested with the restriction enzyme SfaNI (New England Biolabs). The wild-type sequence of the 629 bp amplicon contains a single SfaNI target generating two fragments: 617 bp + 12 bp. The c.2771del mutation generates an additional target, producing the expected restriction fragment pattern of 400 bp + 216 bp + 12 bp. The fragments were resolved by electrophoresis in a 2% agarose gel, visualized by ethidium bromide staining, and the bands were quantitated by densitometry using ImageJ software.
Western blot
Frozen muscle biopsy samples were homogenized in lysis buffer containing 0.25% NP-40 with protease inhibitor cocktail (cOmplete Mini®, Roche Diagnostic), and after centrifugation, supernatants were collected. Concentrations of protein in the supernatants were measured by bicinchoninic acid assay. Aliquots containing 40 μg protein were separated by SDS-PAGE and transferred to a membrane. After blocking with PBS containing 0.5% skimmed milk, the membrane was incubated at 4°C overnight with primary antibodies: anti-TNPO3 antibody (ab54353, Abcam 1:50) and anti-beta-actin antibody (20536-1-AP, Proteintech, 1:1000). The immunoprobed membrane was washed with PBS containing 0.5% Tween 20 three times, and was incubated for 1 h at room temperature with peroxidase-conjugated anti-mouse IgG antibody or anti-rabbit IgG antibody. After incubation with secondary antibodies, the membrane was washed with PBS containing 0.5% Tween 20 three times, and was developed with ECL Prime Western Blotting Detection Reagents® (GE Healthcare). The membrane was imaged with G:BOX Chemi IR6® (SYNGENE).
Anti-TNPO3 and 4',6-diamidino-2-phenylindole staining
Six-micrometre thick sections of frozen muscle were fixed in ice-cold acetone for 10 min, incubated for 1 h with 1% bovine serum albumin, and stained at 4°C for overnight with murine anti-TNPO3 antibody (ab54353, Abcam) at a concentration of 5 μg/ml. Specimens were then incubated for 1 h with sheep biotinylated anti-mouse IgG antibody (RPN1001, GE Healthcare, 1:100) followed by 1 h with streptavidin-fluorescein (RPN1232, GE Healthcare, 1:250), mounted, and stained with 4’,6’-diamidino-2-phenylindole dihydrochloride (DAPI) using VECTASHIELD® Mounting Medium with DAPI (Vector Laboratories). The stained sections were examined with a confocal microscope (Leica TCS SP5 II®, Leica microsystems), and images were obtained with LAS AF® (Leica microsystems).
Results
Clinical assessments
The cohort reported here comprises 30 individuals spanning Generations III to VI; 13 were included in a previous study (Gamez et al., 2001). They presented with limb-girdle and distal muscle weakness with variable distribution, severity, and rate of progression (Supplementary Table 1). Based on age-at-onset, a predominant group of juvenile-onset (onset before age 15) was delineated from an adult-onset form starting in the third and fourth decades (Gamez et al., 2001). Although a similar distribution was observed in the present cohort, the difference between juvenile- and adult-onset groups was not pronounced.
Six patients in the fifth and sixth generations had infantile onset disease characterized by a mild delay in motor skill (independent walking was never delayed beyond age 22 months), followed by difficulty rising from the floor to a standing position and climbing stairs without the aid of hand rails. Running and jumping were difficult or impossible. Some manifested an abnormal gait with a mixture of waddling and distal leg weakness. When walking or attempting to stand on heels, patients also demonstrated a peculiar posture of the feet with elevation of the big toe, foot drop and weight-bearing on the lateral soles (Supplementary Fig. 2). In addition, all patients had thin legs and thenar muscle atrophy. Two patients presented early contractures at the heels, knees, and elbows with rigid spine and scoliosis reminiscent of Emery-Dreifuss muscular dystrophy, but lacking cardiac involvement.
Common initial symptoms in the late-childhood and adolescent group were difficulty running and playing sports. Overt symptoms of pelvic-femoral weakness, such as difficulty in rising from the floor or climbing steps, were also frequent. One patient had exercise intolerance, myalgia and fatigue reminiscent of a metabolic myopathy.
Symptoms of pelvic-girdle weakness were the most common presentation in late-onset patients. Weakness and atrophy of shoulder girdle muscles appeared in only 70% of cases, always in later or advanced stages of the disease, and usually showing less degree of involvement than muscles of pelvic-femoral and axial territories. While both mild (Brook scale 1–2) and severe (Brook scale 3–4) cases showed minor scapular winging (Supplementary Fig. 2), prominent scapula alata was never observed.
Distal muscle involvement was more frequent (at least 24 of 30 patients) than previously reported. In hands, thenar muscle atrophy was observed in all 24 carefully assessed patients and a high proportion of cases reported difficulty grasping a pencil or opening jars. In lower leg, subclinical muscle weakness was often revealed by asking patients to stand on their heels. Other symptoms associated with the disease were: mild ptosis (five cases), transient dysphagia (nine cases) and episodic vertigo and ataxia (eight cases). Three cases had respiratory muscle involvement; all required non-invasive nocturnal ventilatory support.
The course of the disease was highly variable. The two most severe cases with an Emery-Dreifuss-like phenotype were wheelchair-bound in the third decade. Two other cases reached Vigno stage 8 at the fifth and seventh decade. Three additional patients reached Vigno stages 6 and 7 in their forties through sixties. Two patients died suddenly at age 57 and 78 due to causes unrelated to the myopathy.
Laboratory investigations provided similar results to those previously reported (Gamez et al., 2001); however, electromyography in the current series frequently showed spontaneous activity, and, in 3 out of 8 patients, displayed clear motor neurogenic features. In two patients who presented with intense fatigue, ptosis and transient dysphagia, repetitive stimulation tests and single-fibre electromyography disclosed no abnormalities in neuromuscular transmission. Serum creatine kinase levels were also consistent with the previous report: 40% of the cases showed elevated creatine kinase levels (>500 U/l, maximum 2200). No correlation between clinical severity and creatine kinase levels was identified.
Muscle MRI demonstrated variable involvement of scapular and pelvic-femoral muscles, as well as lower leg muscles (Supplementary Fig. 3). A characteristic relationship between muscle MRI abnormalities and degree of impairment was observed with intensity of the MRI signal changes correlating well with the severity of the clinical involvement. In general, scapular-humeral girdle muscles were much better preserved than pelvic-femoral and leg muscles. The percentage of cases with moderate (3) or severe (4) Mercuri scores by muscle group are: (i) scapular girdle: teres major (80%), pectoral (64%), infraspinatus and serratus anterior (55%), deltoids (46%); (ii) lumbar: paraspinal (90%), abdominal oblique (55%) and rectus abdominus (55%); (iii) thigh: sartorius (100%), vastus lateralis, intermedius and medialis (73%), biceps femoris and semitendinosus (55%); and (iv) lower leg: peroneal (91%), gastrocnemius (91%), soleus (73%) and tibialis anterior (70%). Correlations between the clinical severity and the degree of MRI muscle affectation are presented in Supplementary Fig. 3. Subject V-9 represents a mildly symptomatic subject without overt clinical weakness but with Mercury stage 3 abnormalities in lumbar paraspinal, sartorius and peronei muscles. Subject V-7 represents a moderately affected subject (Vignos scale 5) showing a widespread muscle involvement (Mercuri stage 3) of paravertebral and abdominal lumbar muscles, anterior and posterior thighs and diffuse lower leg muscles. Finally, Subject IV-26 corresponds to a severely affected patient (Vignos rating of 7) manifesting Mercuri 3 and 4 grade abnormalties in scapular, lumbar, thigh and lower leg muscles.
Histological studies
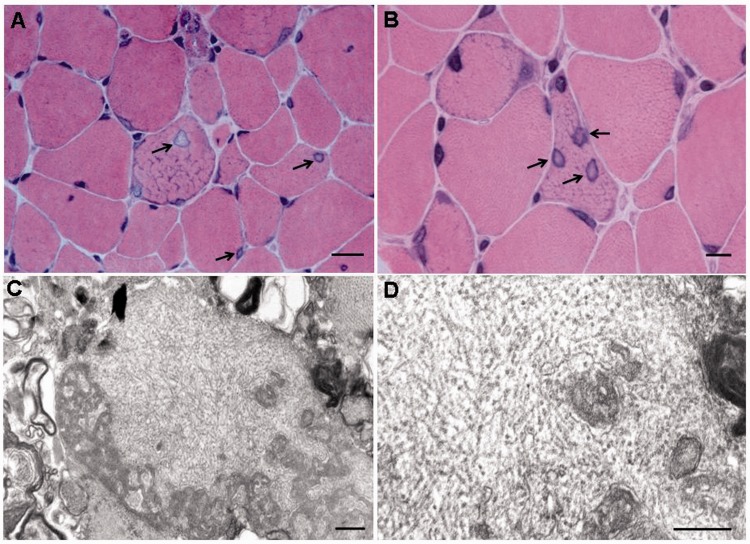
Previous analyses of muscle biopsies from five people affected with LGMD1F had revealed increased variability of fibre size and shape, increased endo- and peri-mysial connective tissue, scattered degenerating fibres, occasional central nuclei, abnormal intermyofibrillar network with abnormal Z bands, rimmed vacuoles and abnormally increased mitochondria with rare paracrystalline inclusions (Gamez et al., 2001). These histological features are similar to those recently reported in the same family (Cenacchi et al., 2012). New analyses of muscle biopsies from seven affected patients (Supplementary Fig. 1) confirmed the described abnormalities in myofibres and connective tissue. In addition, we observed unusually enlarged nuclei with central pallor (Fig. 1). These nuclear abnormalities were identified in all seven biopsies in 11.0–25.8% of muscle fibres (Subject III-14: 18.8%; Subject IV-6: 16.7%; Subject IV-11: 14.8%; Subject IV-18: 16.0%; Subject IV-21: 17.4%; Subject IV-36: 25.8%; Subject V-14: 11.0%). These percentages were derived from counts of the number of affected nuclei and the total number of fibres within each biopsy in haematoxylin–eosin stained slides, using a histometric program (Leica Application Suite v 3.8.0). We have not observed these nuclear abnormalities in other myopathies including Duchenne muscular dystrophy, sarcoglycanopathies, Emery-Dreifuss muscular dystrophy due to emerin or lamin A/C mutations, or FHL1 dystrophy. Ultrastructurally, filamentous inclusions, ∼18 to 20 nm in diameter, were detected within nuclei or in the cytosol of a minority of fibres in two out of five biopsies (Subjects IV-6 and IV-21, Fig. 1). In three biopsies, corresponding to Subjects IV-6, IV-18 and IV-21, light microscopy showed rimmed vacuoles and electron microscopy revealed autophagic vacuoles with prominent pseudomyelin structures, membranous whorls and dense bodies. Immunocytochemical stains for desmin, dystrophin, sarcoglycans, tau, ubiquitin, and amyloid-β proteins, did not show significant alteration or accumulation in muscle fibres.
Figure 1.
Light and electron microscopy findings in muscle biopsies. (A and B) Haematoxylin and eosin staining muscle from affected Subject IV-6. In A, abnormal myonuclei with an ‘empty’ appearance (arrows) (×630). Scale bar = 20 μm. In B, note three abnormal nuclei (arrows) within a myofibre at higher magnification (×1000). Scale bar = 10 μm. (C and D) Electron micrographs showing non-branching tubular filaments 18–20 nm in diameter within a muscle fibre (Subject IV-6). Note myelin and membranous bodies surrounding filaments (C), which are characteristic of rimmed vacuoles. Original magnifications: ×21 000 and ×35 000. Scale bars = 0.5 μm.
Genetic and molecular studies
Initial strategies to identify the genetic cause of the disease included sequencing of candidate genes. Among them, FLNC, encoding filamin c, was extensively investigated because mutations in this gene cause autosomal dominant myofibrillar myopathy (Vorgerd et al., 2005) (OMIM #609524). Studies included dideoxy-DNA sequencing of FLNC exons, flanking introns and promoter regions, Southern and northern blot analyses, and immunohistochemical staining and western blot analyses of muscle biopsies with anti-FLNC antibodies, and revealed normal results compared to controls (data not shown) thereby excluding FLNC as the causative gene.
Dideoxy sequencing of 65 additional genes within the region failed to reveal potentially pathogenic mutations, although the presence of heterozygous changes was difficult to be completely ruled out in some of the electropherograms due to suboptimal quality. Comparative Genomic Hybridization (CGH, NimbleGen Systems Inc.) across the linked region excluded genome copy number variations. Segmentation values across the chromosome 7 regions of interest and other chromosomes showed no differences in DNA from two control subjects and two affected individuals, thereby excluding DNA copy number alterations as the cause of the disease (data not shown).
Because studies at the DNA level were unrevealing, we performed analyses of messenger RNA levels. Expression of 36 genes included in the critical region was analysed in RNA extracts from skeletal muscle of affected members of the family and unaffected unrelated subjects, using a TaqMan® Custom Array 384-well microfluidic card (Supplementary Fig. 4 and Supplementary Table 2). No significant differences could be detected in the expression of the genes analysed, except for a moderate increase of NRF1 transcripts and a pronounced increase of LEP transcripts, in skeletal muscles from affected persons. Because LEP, encoding leptin, is highly expressed in adipose tissue (Meier and Gressner, 2004), we suspected that elevated LEP expression reflected fat replacement of affected muscle, rather than a primary pathogenic alteration. We analysed the expression levels of ADIPOQ, which is expressed exclusively in adipose tissue (Maeda et al., 1996), and also found increased levels of this marker of fat tissue in muscles from affected subjects (Relative quantification (RQ) median; interval: 73.8; 23.9–339.0; n = 5) as compared with levels in unaffected subjects (RQ median; interval: 3.0; undetectable -10.3; n = 6). The levels of ADIPOQ transcripts closely correlated with values for LEP messenger RNA (P > 0.001; Spearman correlation coefficient R = 0.991), thus supporting the notion that increases in LEP transcript reflected fatty replacement of muscle.
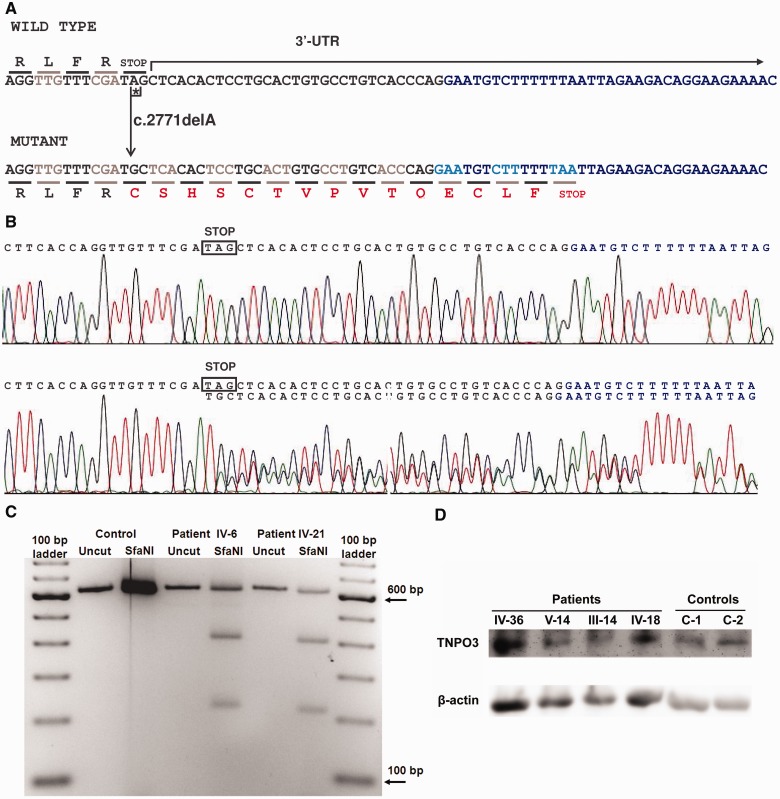
After our initial efforts to find the genetic cause of LGMD1F failed, we applied a more powerful strategy, whole genome sequencing analysis of DNA from one affected individual (Subject III-12, Supplementary Fig. 1). After intersecting the results of whole genome sequencing with the results from previous linkage analysis (Palenzuela et al., 2003) (chromosome 7: 126 287 120–129 963 917), 3888 variants (3125 single nucleotide variants and 763 indels) were identified, from which 718 were novel, not present in the dbSNP database, build 135 (http://www.ncbi.nlm.nih.gov/projects/SNP/). Additional criteria based on the dominant inheritance of the disease, the population frequency of the variants and effect prediction (see ‘Materials and methods’ section), allowed us to rule out all but one of these variants, a heterozygous mutation in the termination codon of the TNPO3 gene, encoding transportin 3, a protein involved in the translocation of proteins from the cytoplasm to the nucleus (Brass et al., 2008; Cribier et al., 2011). The mutation (c.2771del, reference sequence GeneBank NM_012470.3, Fig. 2) is a single adenine nucleotide deletion in the TAG stop codon, common to the two protein isoforms encoded by the gene. The del-A results in conversion of TAG to TGC codon, encoding cysteine, and extension of the reading frame by 15 codons to a downstream of the termination-signal within the transcript. Thus, the frameshift leads to the predicted mutated TNPO3 protein with 15 additional amino acids at the C-terminus [p.(*924Cysext*15) for isoform 1]. The retrospective analysis of the sequences of this gene revealed that this heterozygous mutation had been missed in the past due to the poor quality of the electropherograms. Then, we performed new dideoxy sequence analysis of TNPO3, which demonstrated presence of c.2771del in each of the 29 clinically affected individuals and absence of the mutation in all 20 clinically unaffected relatives tested. Thus, the sequence data indicate that the mutation segregates with the linked chromosome 7q32.1-32.2 region (Palenzuela et al., 2003) and with the phenotype (Supplementary Fig. 1).
Figure 2.
Effects of the c.2771del mutation on TNPO3 messenger RNA and protein. (A) The 3’-terminal coding and untranslated region (UTR) sequences of TNPO3 transcripts, including the 3’-end of the exon 23 (black font) and the 5’-end of the non-coding exon 24 (blue font) of both wild-type and mutant (c.2771del) complementary DNAs. The fragments shown are identical in both transcript variants 1 and 2. The deleted 2771A is labelled with an asterisk. The encoded amino acids are indicated in one-letter code. Changes resulting from the frame-shifted codons in the mutated sequence are indicated in red case, which highlight the disruption of the native TAG stop codon, with a modified C-terminus containing 15 extra amino acids p.(*924Cysext*15) relative to normal isoform 1. (B) Electropherograms showing the complementary DNA sequences from muscle messenger RNA obtained from a healthy control (top, wild-type sequence), and the affected subject IV-6 (bottom) showing the coexistence of both wild-type and c.2771del mutated transcripts at similar amounts. Sequences encompass two different exons (exon 23 in black and exon 24 in blue). A similar result was obtained for the Subject IV-21 (data not shown). (C) PCR-restriction fragment length polymorphism analysis of complementary DNA obtained from skeletal muscle TNPO3 messenger RNA. A 629 bp fragment was PCR-amplified from skeletal muscle complementary DNA from Subjects IV-6, IV-21 and one unrelated healthy control. The products were digested with the restriction enzyme SfaNI. The wild-type sequence of the 629 bp amplicon contains one SfaNI site generating two fragments: 617 bp +12 bp. Because the c.2771del mutation generates an additional SfaNI site, restriction enzyme digestion produces three fragments: 400 bp + 216 bp + 12 bp. Densitometric analysis of the bands showed that the mutated messenger RNA was 64% (Subject IV-6) and 61% (Subject IV-21) of total TNPO3 messenger RNA. (D) Western blot of muscle specimens from affected subjects and controls. Muscle specimens from four subjects with LGMD1F and two control subjects were analysed by western blot. The anti-TNPO3 antibody showed a clear band at approximately 100 kDa in each lane. No differences in the position of bands and no extra bands were observed. There were no significant differences in the amounts of TNPO3 normalized to beta-actin between affected subjects and controls.
To investigate whether the mutated messenger RNA was expressed in skeletal muscle of the affected individuals, complementary DNA was generated using RNA from two skeletal muscle samples (Subjects IV-6 and IV-21, Supplementary Fig. 1), and the 3’-end fragment containing the native stop codon (common to the 3 transcripts described for the gene, TNPO3 gene entry in the NCBI, http://www.ncbi.nlm.nih.gov/gene/23534) was PCR-amplified. Sequence analysis of this amplified complementary DNA revealed the coexistence of both mutated and wild-type transcripts in similar amounts, according to the sizes of the peaks of the two overlapped sequences observed in the electropherograms (Fig. 2). This result was confirmed by PCR-restriction fragment length polymorphism analysis, which indicated that 61–64% of the TNPO3 messenger RNA of the two affected individuals contained the mutant form (Fig. 2). Retrospective review of the real-time PCR results obtained in the microfluidic cards (Supplementary Fig. 4) confirmed that TNPO3 was expressed in skeletal muscle of affected and non-affected persons at similar levels. Taken together, these results demonstrate that the mutated messenger RNA is stable and does not undergo RNA decay.
We performed western blot analysis of biopsied muscle samples from four affected subjects and two unaffected control subjects, to assess changes in amount and molecular weight of TNPO3 protein (Fig. 2). The TNPO3 mutation in the family disrupts the termination codon, and is predicted to extend the C-terminus of TNPO3 by 15 amino acids. Using an anti-TNPO3 antibody that recognizes an N-terminus epitope, present in both normal and mutant TNPO3, western blot analysis showed a single band at the same level in muscle from control subjects and affected individuals, and no extra bands were observed in muscles from affected subjects. However, the 15 amino acid size difference between normal and mutant TNPO3 is likely insufficient to distinguish the two proteins by western blot. Relative to control subject muscles, the amount of muscle TNPO3 normalized to beta-actin was increased in one affected subject (Subject IV-36), and decreased in the other three (Subjects V-14, III-14 and IV-18); therefore, there were no significant difference in TNPO3 quantity in mutant versus normal tissue.
To assess the effects of the TNPO3 mutation on TNPO3 cellular localization, we performed immunohistochemistry of muscle tissue with anti-TNPO3 antibody. Control muscle stained with anti-TNPO3 antibody showed clear nuclear staining and that colocalized with DAPI (Fig. 3I). In muscle of affected individuals, TNPO3 immunostaining was also observed within nuclei, but was unevenly distributed and often limited to the periphery of nuclei, (Fig. 3C and F).
Figure 3.
Anti-TNPO3 and DAPI staining of muscle from affected individuals and controls. Immunofluorescence-stained muscle from two affected individuals (Subject IV-36: A–C, Subject V-14: D–F) and two control subjects (one not shown) (G–I) were observed under a confocal microscope. Each specimen was stained both with anti-TNPO3 antibody (A, D and G) and by DAPI (B, E and H), and merged images were generated (C, F and I). TNPO3 staining colocalized with DAPI in control subjects (I). In affected individuals, signals of TNPO3 were also observed within nuclei, but were unevenly distributed (C and F). Scale bar = 40 μm.
Discussion
LGMD1F is one of the nine autosomal dominant forms of LGMD. Causative genes had been identified for only five forms of autosomal dominant-LGMD: MYOT (LGMD1A, OMIM #159000), LMNA (LGMD1B, OMIM #159001), CAV3 (LGMD1C, OMIM #607801), DES (LGMD1D, OMIM *125660), and DNAJB6 (LGMD1E, OMIM #603511) (Bushby, 2009; Sarparanta et al., 2012) (see GeneReviews™ LGMD Overview). In general, these disorders are characterized by adult-onset and milder clinical phenotypes than LGMD2. Although, most individuals harbouring mutations in these genes fulfil the diagnostic criteria for LGMD, some manifest a wider spectrum of clinical phenotypes. The extreme example is LMNA mutations, which have been associated with a broad spectrum of clinical conditions including Dunnigan lipodystrophy, autosomal dominant Emery-Dreifuss muscular dystrophy, cardiomyopathy, Charcot–Marie–Tooth disease and Hutchinson-Gilford progeria (Worman et al., 2009; Bertrand et al., 2011).
Several lines of evidence strongly support pathogenicity of the TNPO3 mutation in this family with autosomal dominant-LGMD: (i) TNPO3 resides within the chromosome 7q32.1-32.2 locus for LGMD1F; (ii) the mutation segregates with the phenotype; (iii) the microdeletion is absent in publicly available genomic sequence databases (dbSNP build 135, 1000 Genomes Project and 5400 NHLBI exomes) and in our set of >200 Spanish control alleles indicating that all control individuals harbour the canonical TNPO3 TAG termination codon in homozygosity at the position 128 597 311 of the chromosome 7; (iv) the mutation in the termination codon of TNPO3 is predicted to extend the coding sequence at the 3’-end of the messenger RNA and to generate an aberrant protein; (v) the mutated messenger RNA is expressed in the muscle of the affected individuals; and (vi) the detection of histologically abnormal muscle nuclei with atypical nuclear filaments, anomalous TNPO3 immunoreactivity and irregular membranes. These morphological changes of myocyte nuclei indicate that the TNPO3 c.2771del mutation alters nuclear functions, which is consistent with the putative role of the TNPO3 in transport of proteins across the nuclear membrane. We observed similar levels of TNPO3 transcript in skeletal muscle of three healthy control subjects and in five affected subjects (Supplementary Fig. 4). It is likely that the mutant protein, which is predicted to contain 15 additional amino acids at the C-terminus, is expressed in skeletal muscle and exerts a dominant toxic effect.
Although there is evidence that TNPO3 is expressed in skeletal muscle (BioGPS portal for annotation resources (http://biogps.org) (Su et al., 2004), the role of TNPO3 in muscle is currently unknown. TNPO3 was originally identified as TNP-SR2, which encodes a nuclear membrane protein belonging to the importin beta family and transports serine/arginine (SR) rich proteins into the nucleus (Lai et al., 2000, 2001). TNPO3 was subsequently identified by genome-wide RNA interference knockdown as a HIV-dependency factor required for HIV1 infection at a stage between reverse transcription and integration of HIV in human cells (Brass et al., 2008; Konig et al., 2008). The protein mediates nuclear import of the HIV pre-integration complex by binding the viral integrase, both in dividing and non-dividing cells (Christ et al., 2008). The C-terminus domain (CTD) of TNPO3 appears to be required for interactions with HIV1 integrase (Larue et al., 2012); therefore, the abnormal extension of the CTD domain is likely to interfere with its transport function.
Because specific combinations of SR proteins are required for messenger RNA splicing and post-transcriptional processing (Bjork et al., 2009), the TNPO3 mutation may alter muscle transcripts raising the possibility that LGMD1F is RNA-mediated myopathy similar to, but mechanistically distinct from, myotonic dystrophy (Wheeler and Thornton, 2007; Tang et al., 2012). In addition, because mutations in the nuclear envelope proteins emerin and lamin A/C, are known to cause Emery-Dreifuss muscular dystrophy, the TNPO3 mutation causing LGMD1F extends the genetic spectrum of nuclear envelope-related myopathies. In support of this notion is the observation of filamentous inclusions and rimmed vacuoles in all three diseases (Fidzianska et al., 2004). We have noted filaments of ∼18 to 20 nm diameter both in nuclei and within the cytosol of myofibres of affected individuals. Filaments of similar thickness have been observed in several muscle disorders and are ascribed to accumulations of proteins, such as beta-amyloid, tau protein and ubiquitin (Askanas and Engel, 2006; Askanas et al., 2009). Myonuclear breakdown would entail the fragmentation of the nuclear membrane and contribute to the formation of pseudomyelin figures and membranous whorls, which correspond to the rimmed vacuoles seen by light microscopy.
Interestingly, autophagic vacuoles, which we have observed in our affected subjects’ muscle, have been also noted in LGMD1D, which is due to mutations in DNAJB6. In that disease, the presence of autophagy is due to abnormal protein accumulation, which confers a dominant toxic function to the autophagic complex that contains the mutated co-chaperone (Sarparanta et al., 2012). Accordingly, autophagy may be contributing to LGMD1F.
Clinically, LGMD1F was originally described as slowly progressive proximal symmetric weakness with predominantly lower limb onset, normal to mildly raised creatine kinase activity and myopathic electromyography features (Gamez et al., 2001). Great variability in age at onset, distribution of muscle involvement and severity was observed. Two clinical forms were delineated: a benign adult-onset form presenting in the third decade or later, and a juvenile form, beginning before age 15 years and leading to severe functional disability. Relative to the original report, the present study of a cohort of 30 patients with LGMD1F provides a longer and more systematic follow-up, as well as new information about affected individuals from younger generations. While confirming the core phenotype of LGMD1F, which is characterized by pelvic-femoral weakness and less severe and variable shoulder involvement, the new clinical data also demonstrate a broad clinical spectrum and novel clinical features of the disease. The disorder usually begins in childhood or adolescence, and less frequently in adulthood; and typically runs a benign course compatible with a normal working life. We noted mild hand and lower leg weakness producing a stereotyped posture while walking or standing on heels in virtually all patients. In addition, we observed less common but well-characterized presentations, including infantile-onset cases with a congenital myopathy phenotype and a variable course, which can evolve into a severe and rapid progressive phenotype. Interestingly, these severely affected patients also manifested early joint and axial contractures similar to Emery-Dreifuss syndrome, which raises the possibility of pathogenic mechanisms distinct from those involved in the typical cases. In addition, we have also observed very benign patients without complaints of weakness, but rather atypical features such as myalgia, exercise intolerance, and fatigue, mimicking a metabolic myopathy. This broad scenario of clinical nuances highlights the need to deepen the clinical evaluation of this extensive pedigree and other potential families with similar gene defects.
In summary, in this report, we have provided an extensive update of the clinical and morphological features of LGMD1F and have identified a microdeletion mutation in the TNPO3 gene as a cause of this disorder. This finding expands our knowledge on the genetic bases of muscular dystrophies and suggests that other proteins of the nuclear envelope compartment may play a primary role in the pathogeneses of muscular dystrophies and other skeletal muscle-related disorders.
Funding
This work was supported by the Spanish Instituto de Salud Carlos III [PS09/01591 to R.M., PI10/02628 to C.N., PI11/0842 to S.O., PI10/01970 to J.G., RD09/0076/00011 to the activities of Neurological Tissue Biobank, BIOBANCO del CHUVI]; the International Rare Diseases Research Consortium [SpainRDR]; the Conselleria de Economia e Industria, Xunta de Galicia [contract Isidro Parga Pondal to S.O.]; the U.S. National Institutes of Health (NIH) [R01 AR47989 to M.H.]. The CNAG thanks for core funding from the Spanish Ministerio de Economia y Competitividad and the Generalitat de Catalunya - Departament de Salut and Departament d'Economia i Coneixement. M.H. and A.K. also acknowledge support from NIH grants R01 HD057543 and R01 HD056103 from NICHD and the Office of Dietary Supplements (ODS), as well as U54 NS078059 from NINDS and NICHD, and from the Muscular Dystrophy Association USA.
Supplementary material
Supplementary material is available at Brain online.
Acknowledgements
The authors thank the members of the family studied in this work for their collaboration, and Gisela Nogales-Gadea for scientific assistance.
Glossary
Abbreviation
- LGMD
limb-girdle muscular dystrophy
References
- Askanas V, Engel WK. Inclusion-body myositis: a myodegenerative conformational disorder associated with Abeta, protein misfolding, and proteasome inhibition. Neurology. 2006;66:S39–48. doi: 10.1212/01.wnl.0000192128.13875.1e. [DOI] [PubMed] [Google Scholar]
- Askanas V, Engel WK, Nogalska A. Inclusion body myositis: a degenerative muscle disease associated with intra-muscle fiber multi-protein aggregates, proteasome inhibition, endoplasmic reticulum stress and decreased lysosomal degradation. Brain Pathol. 2009;19:493–506. doi: 10.1111/j.1750-3639.2009.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand AT, Chikhaoui K, Yaou RB, Bonne G. Clinical and genetic heterogeneity in laminopathies. Biochem Soc Trans. 2011;39:1687–92. doi: 10.1042/BST20110670. [DOI] [PubMed] [Google Scholar]
- Bjork P, Jin S, Zhao J, Singh OP, Persson JO, Hellman U, et al. Specific combinations of SR proteins associate with single pre-messenger RNAs in vivo and contribute different functions. J Cell Biol. 2009;184:555–68. doi: 10.1083/jcb.200806156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–6. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Brooke MH, Griggs RC, Mendell JR, Fenichel GM, Shumate JB, Pellegrino RJ. Clinical trial in Duchenne dystrophy. I. The design of the protocol. Muscle Nerve. 1981;4:186–97. doi: 10.1002/mus.880040304. [DOI] [PubMed] [Google Scholar]
- Bushby K. Diagnosis and management of the limb girdle muscular dystrophies. Pract Neurol. 2009;9:314–23. doi: 10.1136/jnnp.2009.193938. [DOI] [PubMed] [Google Scholar]
- Cenacchi G, Peterle E, Fanin M, Papa V, Salaroli R, Angelini C. Ultrastructural changes in LGMD1F. Neuropathology. 2012 doi: 10.1111/neup.12003. Advance Access published on December 21, 2012, doi: 10.1111/neup.12003. [DOI] [PubMed] [Google Scholar]
- Christ F, Thys W, De Rijck J, Gijsbers R, Albanese A, Arosio D, et al. Transportin-SR2 imports HIV into the nucleus. Curr Biol. 2008;18:1192–202. doi: 10.1016/j.cub.2008.07.079. [DOI] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribier A, Segeral E, Delelis O, Parissi V, Simon A, Ruff M, et al. Mutations affecting interaction of integrase with TNPO3 do not prevent HIV-1 cDNA nuclear import. Retrovirology. 2011;8:104. doi: 10.1186/1742-4690-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Estelle J, Marco Sola S, Knowles DG, Raineri E, Guigo R, et al. Fast computation and applications of genome mappability. PLoS One. 2012;7:e30377. doi: 10.1371/journal.pone.0030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidzianska A, Rowinska-Marcinska K, Hausmanowa-Petrusewicz I. Coexistence of X-linked recessive Emery-Dreifuss muscular dystrophy with inclusion body myositis-like morphology. Acta Neuropathol. 2004;107:197–203. doi: 10.1007/s00401-003-0794-y. [DOI] [PubMed] [Google Scholar]
- Gamez J, Navarro C, Andreu AL, Fernandez JM, Palenzuela L, Tejeira S, et al. Autosomal dominant limb-girdle muscular dystrophy: a large kindred with evidence for anticipation. Neurology. 2001;56:450–4. doi: 10.1212/wnl.56.4.450. [DOI] [PubMed] [Google Scholar]
- Homer N, Merriman B, Nelson SF. BFAST: an alignment tool for large scale genome resequencing. PLoS One. 2009;4:e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M, Krasnewich DM. Hereditary inclusion body myopathy: a decade of progress. Biochim Biophys Acta. 2009;1792:881–7. doi: 10.1016/j.bbadis.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Lin RI, Huang SY, Tsai CW, Tarn WY. A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J Biol Chem. 2000;275:7950–7. doi: 10.1074/jbc.275.11.7950. [DOI] [PubMed] [Google Scholar]
- Lai MC, Lin RI, Tarn WY. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc Natl Acad Sci USA. 2001;98:10154–9. doi: 10.1073/pnas.181354098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larue R, Gupta K, Wuensch C, Shkriabai N, Kessl JJ, Danhart E, et al. Interaction of the HIV-1 intasome with Transportin 3 (TNPO3 or TRN-SR2) J Biol Chem. 2012;287:34044–58. doi: 10.1074/jbc.M112.384669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Li X, Wang M, Li X, Zhang F, Li Y, et al. A novel autosomal dominant inclusion body myopathy linked to 7q22.1-31.1. PLoS One. 2012;7:e39288. doi: 10.1371/journal.pone.0039288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Okubo K, Shimomura I, Funahashi T, Matsuzawa Y, Matsubara K. cDNA cloning and expression of a novel adipose specific collagen-like factor, apM1 (AdiPose Most abundant Gene transcript 1) Biochem Biophys Res Commun. 1996;221:286–9. doi: 10.1006/bbrc.1996.0587. [DOI] [PubMed] [Google Scholar]
- Marco-Sola S, Sammeth M, Guigo R, Ribeca P. The GEM mapper: fast, accurate and versatile alignment by filtration. Nat Methods. 2012;9:1185–8. doi: 10.1038/nmeth.2221. [DOI] [PubMed] [Google Scholar]
- Martinsson T, Darin N, Kyllerman M, Oldfors A, Hallberg B, Wahlstrom J. Dominant hereditary inclusion-body myopathy gene (IBM3) maps to chromosome region 17p13.1. Am J Hum Genet. 1999;64:1420–6. doi: 10.1086/302375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier U, Gressner AM. Endocrine regulation of energy metabolism: review of pathobiochemical and clinical chemical aspects of leptin, ghrelin, adiponectin, and resistin. Clin Chem. 2004;50:1511–25. doi: 10.1373/clinchem.2004.032482. [DOI] [PubMed] [Google Scholar]
- Mercuri E, Pichiecchio A, Counsell S, Allsop J, Cini C, Jungbluth H, et al. A short protocol for muscle MRI in children with muscular dystrophies. Eur J Paediatr Neurol. 2002;6:305–7. doi: 10.1016/s1090-3798(02)90617-3. [DOI] [PubMed] [Google Scholar]
- Nigro V, Aurino S, Piluso G. Limb girdle muscular dystrophies: update on genetic diagnosis and therapeutic approaches. Curr Opin Neurol. 2011;24:429–36. doi: 10.1097/WCO.0b013e32834aa38d. [DOI] [PubMed] [Google Scholar]
- Palenzuela L, Andreu AL, Gamez J, Vila MR, Kunimatsu T, Meseguer A, et al. A novel autosomal dominant limb-girdle muscular dystrophy (LGMD 1F) maps to 7q32.1-32.2. Neurology. 2003;61:404–6. doi: 10.1212/01.wnl.0000073984.46546.4f. [DOI] [PubMed] [Google Scholar]
- Sarparanta J, Jonson PH, Golzio C, Sandell S, Luque H, Screen M, et al. Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat Genet. 2012;44:450–5, S1–2. doi: 10.1038/ng.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang ZZ, Yarotskyy V, Wei L, Sobczak K, Nakamori M, Eichinger K, et al. Muscle weakness in myotonic dystrophy associated with misregulated splicing and altered gating of Ca(V)1.1 calcium channel. Hum Mol Genet. 2012;21:1312–24. doi: 10.1093/hmg/ddr568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignos PJ, Jr, Spencer GE, Jr, Archibald KC. Management of progressive muscular dystrophy in childhood. JAMA. 1963;184:89–96. doi: 10.1001/jama.1963.03700150043007. [DOI] [PubMed] [Google Scholar]
- Vorgerd M, van der Ven PF, Bruchertseifer V, Lowe T, Kley RA, Schroder R, et al. A mutation in the dimerization domain of filamin c causes a novel type of autosomal dominant myofibrillar myopathy. Am J Hum Genet. 2005;77:297–304. doi: 10.1086/431959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Thornton CA. Myotonic dystrophy: RNA-mediated muscle disease. Curr Opin Neurol. 2007;20:572–6. doi: 10.1097/WCO.0b013e3282ef6064. [DOI] [PubMed] [Google Scholar]
- Worman HJ, Fong LG, Muchir A, Young SG. Laminopathies and the long strange trip from basic cell biology to therapy. J Clin Invest. 2009;119:1825–36. doi: 10.1172/JCI37679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.