Abstract
Context:
Cutaneous leishmaniasis (CL) is a public health problem in several endemic countries. Recent studies on mouse model and also a few clinical experiments showed that the type of immune response generated at the site of infection and especially balance between regulatory and effector T-cells determines the outcome of the disease toward self-limiting or long-lasting lesions.
Aims:
The aim of this study was to evaluate the role of natural regulatory T cells (nTregs) in early and late cutaneous lesions of human Leishmania major (L. major) infection.
Settings and Design:
Skin biopsies were collected from parasitologically proven lesions of 28 CL patients, divided into two groups of early and late lesions. The causative agents were identified to be L. major.
Materials and Methods:
Quantitative real-time reverse transcription polymerase chain reaction (PCR) and immunofluorescent staining of biopsies were used to assess the Foxp3 mRNA expression and frequency of nTregs in two groups. Mann-Whitney U test was used to determine the significance of deference between the two groups.
Results:
Mean relative expressions of Foxp3 mRNA were 0.53 ± 0.23 and 1.26 ± 0.99 in early and late lesions, respectively, which was significantly upper in chronic lesions (P = 0.007). Parallel results were obtained in tissue staining method.
Conclusions:
Increased in gene expression and protein staining of nTreg markers in chronic biopsy samples indicates a role for these cells in chronic L. major induced leishmaniasis and supports the effectiveness of regulatory T cell-based immunotherapy for treatment of chronic CL.
Keywords: Cutaneous, fluorescent antibody technique, Leishmania major, leishmaniasis, real-time PCR, regulatory T cells
INTRODUCTION
Cutaneous leishmaniasis in Southwest Asia and North Africa is a common parasitic disease caused by L. major and L. tropica, the two important primeval species of genus Leishmania. Cutaneous lesions are often self-limiting; appear as papules, nodules, or plaques with or without ulceration and different levels of inflammation.[1] Diverse clinical presentations are due to various parasite species and/or host immune responses to infection.[1,2] Cutaneous lesions caused by L. major often heal in three to four months with more inflammation and ulceration in contrast to L. tropica infections, which have longer durations and less inflammation.[3] However, L. major can occasionally cause chronic lesions lasting more than six months and even years.[4]
Among various immune mechanisms controlling outcome of Leishmania infection, role of different subsets of CD4+ T cells has been vastly investigated. The traditional helper T cells (Th), Th1, and Th2 and related cytokines confer resistance and susceptibility in mouse model, respectively.[5,6] Nevertheless in human disease, the exact role of Th1-Th2 in persistence of cutaneous lesions has not been elucidated and a mixture of Th1-Th2 response seems to deal with both acute and chronic leishmaniasis.[7–10]
To proceed with further mechanisms underlying chronic cutaneous leishmaniasis, CD4+ regulatory T cells (Tregs) which suppress effector T cells have been particularly under investigation. Naturally occurring regulatory T cells (nTregs) are the foremost and best characterized CD4+ Treg subset. Specific nTreg markers especially in human are yet controversial. Nevertheless the surface marker CD25 and the transcription factor Forkhead box P3 (Foxp3) both necessary for nTregs lineage and function are the known common markers of these cell populations.[11] First recognized as suppressors of autoreactive T cells and maintaining immunological self-tolerance, nTregs currently are proven to play a major role in chronic infectious disease. NTregs recognize self-antigens in autoimmunity as well as nonself-antigens in hypersensitivity and infectious disease, limiting inflammation and immune mediated pathology via direct cell contact or secreting different suppressor mediators. However, nTreg down regulation of acute phase reactions may result in failure to efficiently resolve the infections leading to chronic disease.[12,13]
Growing evidence mainly generated in animal models showed a critical role for nTregs in leishmaniasis. In murine models of L. major infection the balance between nTregs and effector T cells appear to control the course of infection.[14]
Currently, few studies have implicated the role of nTregs during human leishmaniasis. In a research, CD4+ CD25+ T cells derived from cutaneous lesions of L. braziliensis patients confirmed functionally to be nTregs but no correlation with pathogenesis was yet detected.[15] In a series of experiments on L. guyanensis infection CD4+ CD25+ Tregs isolated from skin biopsy specimens of acute CL patients had suppressive effect on CD4+ CD25- T cells. Also Foxp3 mRNA expressions were more in skin biopsies than in peripheral blood mononuclear cells (PBMC), confirming recruitment of nTregs to infection site. In L. guyanensis infection, the levels of Foxp3 mRNA in the lesions were correlated with poor prognosis in acute cases and were superior in chronic patients than in acute ones.[16,17]
Current therapies for chronic CL are generally ineffective. Precise understanding of immune regulation in the site of infection can provide new therapeutic approaches, such as immunotherapy, to cure or relieve chronic CL.
Thus far no information is available on nTregs role in old world cutaneous leishmaniasis. In present study, the levels of Foxp3 mRNA in skin biopsy specimens of early and late lesions of CL induced by L. major were measured to investigate the importance of nTregs in persistence of human L. major infection. Furthermore, we detected and quantified the nTregsin patients’ biopsy specimens using specific markers: CD4, CD25, and Foxp3 verifying the last method.
SUBJECTS AND METHODS
Patients
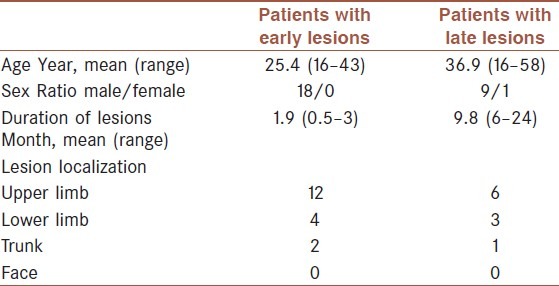
Twenty-eight patients with active CL lesions were selected from the patients referred to Centre for Research in Skin Diseases and Leishmaniasis, Isfahan University of Medical Sciences. Informed consent was obtained from all the patients and the study was approved by ethical committee of Isfahan University of Medical Sciences, Ministry of Health, Iran. Patients were divided into two groups based on duration of disease prior to taking biopsies. Eighteen male patients with disease duration of less than four months and ten patients (M = 9, F = 1) with lesion durations more than six months were included in this study. Subjects with a history of chronic internal or cutaneous disease were excluded from the study. Convenience sampling was chosen as the method of sampling. The reason was mainly the ethical concern of taking biopsies and the low number of patients with late lesions. Clinical characterization of the patients is summarized in Table 1. Parasitological diagnosis was based on direct microscopy, except for two chronic CL patients with negative smears who had positive PCR for Leishmania parasite. All parasitic infections were verified to be L. major by means of high-resolution melting analysis of DNA isolates.[18]
Table 1.
Clinical characterization of the patients
Biopsy specimen collection
After local anesthesia (xylocaine/adrenaline 1: 100,000) punch biopsies of 3.5 mm were obtained from the border of the lesions. Skin defects were closed with one stitch. Two biopsy samples were taken from some chronic patients. For real-time PCR, samples (nine early, eight late) were stored in RNAlater RNA Stabilization Reagent (Qiagen) at -20. For IF staining, biopsies (nine early, seven late) were fixed in 4% paraformaldehyde and dehydrated in 30% phosphate-buffered sucrose solution for 24 h at 4 °C. Then 6 μ m cryostat sections were prepared using a Leica 1800 cryocut (Leica, Germany) and stored at -80 until staining.
RNA extraction and real-time PCR
For tissue disruption and homogenization biopsy samples were loaded into Lysing Matrix D tubes (MP-biomedical, Irvine, CA) containing 1.4 mm ceramic beads in 600 μL buffer RLT (Qiagen) and processed in a Ribolyser reciprocal shaker (Hybaid, UK) as described in Ref.[19]. Total RNA was isolated using the RNeasy Mini kit (Qiagen) and the RNA was reverse transcribed to cDNA by the QuantiTect Reverse Transcription Kit (Qiagen) following the manufacturer's instructions. RNA quality and quantity were verified by means of agarose gel electrophoresis and spectrophotometric analysis of RNA. Real-time reverse transcription PCR was performed on a Rotor-Gene 6000 system (Corbett) using the QuantiFast SYBR Green PCR Kit (Qiagen). PCR reactions were performed in triplicate with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as internal control. Published primer sequences for Foxp3 and GAPDH genes were used in the study.[20] The 2 (-ΔCT) equation was used to quantify mRNA expression of Foxp3 relative to GAPDH gene[21] and the data are expressed as target gene vs. the GAPDH gene mRNA expression.
Immunofluorescent staining
Single staining
The following antibodies were used in this study: mouse monoclonal anti-CD4 (CMG, dilution 1:100), rabbit monoclonal anti-CD25 (Santa Cruz Biotechnology Inc., sc-13946, dilution 1:200), mouse monoclonal anti-Foxp3 (Santa Cruz Biotechnology Inc., sc-53876, dilution 1:30), and secondary antibodies FITC-conjugated goat antirabbit IgG (Sigma, F1262, dilution 1:80) for localization of CD25, and TRITC-conjugated goat antimouse IgG (Sigma, T7782, dilution 1:50) for localization of CD4 and Foxp3 markers. After rehydration with PBS nonspecific sites were blocked with 10 mg/ml BSA, and sections were incubated with CD4 or CD25 antibodies for 1 h at 37 °C, then with appropriate secondary antibody for 45 min at 37 °C. For Foxp3 staining permeabilization was carried out with 0.4% Triton X-100 for 45 min. All sections were counter stained with 4,6-diamidino-2-phenylindole (DAPI) (Sigma, D9542). Omission of primary antibody was used as negative control and tonsillar sections served as positive controls.
Double staining
CD4/CD25 double staining was done by incubating sections with a cocktail of CD4 and CD25 antibodies and then with a mixture of secondary antibodies all with final dilutions the same as single staining procedure. For CD25/Foxp3 double staining after overnight incubation of sections with Foxp3 antibody, incubation with CD25 antibody was done.
Microscopy and quantification
The sections were analyzed with a fluorescent microscope (Olympus, Bx-51, Japan) and images were captured with a digital camera (Olympus, DP-72, Japan) using Cell*A software (Olympus Soft Imaging Solution GmbH, Germany). Sections were coded and examined blindly. Each marker was examined in ten representative fields of stained sections under X400 magnification which corresponds to an area of one square millimeter. In single stained sections each field was photographed two times, one for specific labeled antibody and another for DAPI by switching filters and light intensity. For double-stained sections three images were taken from each field, two for labeled antibodies, and one for DAPI. Stained cells were counted over images with “manual tag” option of the software and results were expressed as the number of positive cells per mm2. For quantifying CD4+ CD25+ Foxp3+ cells, number of CD4- CD25+ cells was subtracted from CD25+ Foxp3+ cells in each sample to confirm that all CD25+ Foxp3+ cells are CD4+ as well. Frequency of Tregs was expressed as a percentage of the total number of CD4+ cells in every sample.
Statistical analysis
The SPSS software was utilized for statistical analysis. Because the parametric conditions were not accomplished in this study, the nonparametric Mann-Whitney U test was used to compare levels of mRNA and protein expression of Foxp3 between groups. Results are expressed as mean ± standard deviation. All tests were two-tailed, and P values of less than 0.05 were considered significant.
RESULTS
Foxp3 expression is elevated in late lesions
Mean expressions of Foxp3 mRNA normalized to expression of GAPDH were 0.53 ± 0.23 and 1.26 ± 0.99 in early and late lesions, respectively, which displayed a significant increase in chronic lesions (P = 0.007) [Figure 1]. As seen in other experiments, samples showed a wide range of Foxp3 mRNA expression especially in chronic lesions. Four chronic patients had received some therapy before taking biopsy; however, this had no meaningful effect on their level of Foxp3 expression.
Figure 1.
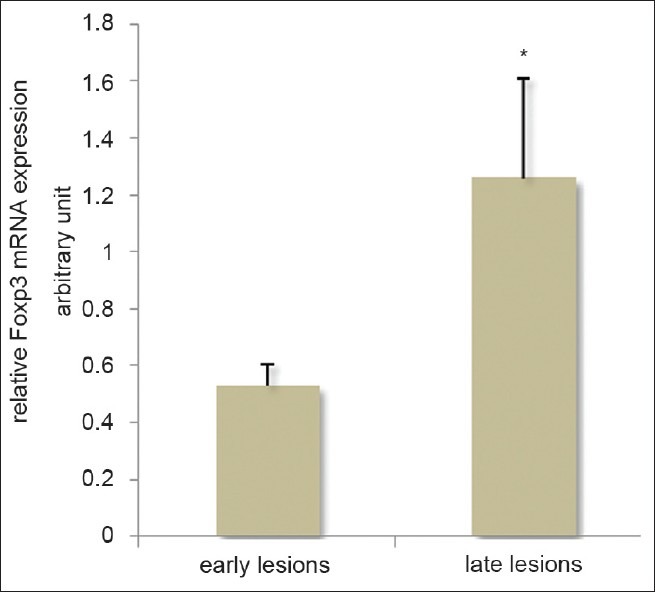
Foxp3 mRNA expression normalized to GAPDH expression in early and late lesions of CL patients, *P < 0.05
Tregs are more frequent in late lesions
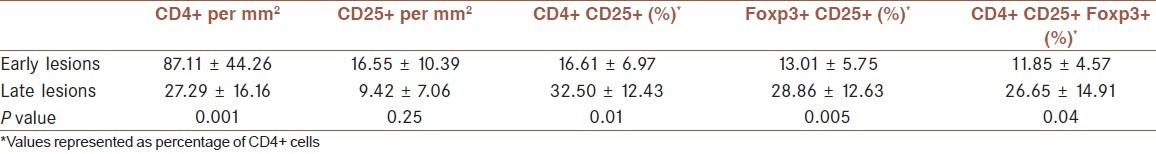
Mean percentages of CD4+ CD25+, CD25+ Foxp3+, and CD4+ CD25+ Foxp3+ cells (calculated as mentioned before) were significantly superior in late lesions than early ones [Table 2]. Samples showed positive staining for all markers but with great variation in number of cells. Late lesions had CD4+ counts significantly lower than early ones but the ratios of Tregs to T helpers were more in these samples than early cases.
Table 2.
Frequency of Treg cell markers in early and late lesions of cutaneous leishmaniasis
In tonsillar sections, we found both CD4- CD25+ and CD25- Foxp3+ cells, but in patient samples CD4- CD25+ cells were minimal (<1%) and CD25- Foxp3+ cells were rarely seen. Because the biopsies were taken from nonulcerated margin of lesions, inflammatory cells were seldom seen in epithelium and most of T-cell accumulations were located in middermis [Figure 2].
Figure 2.
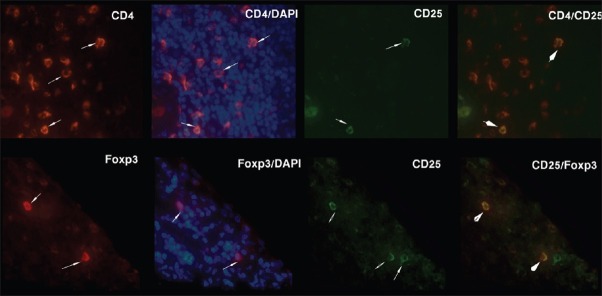
Double staining of CD4/CD25 and CD25/Foxp3 markers in two representative samples. Arrows show positive cells and arrow heads show double positive cells which are stained orange because of merging of red and green colors. Foxp3 stains both cell cytoplasm and nucleus, thus blue-stained cell nuclei appear overlapped with Foxp3 staining. Original magnification: X400
DISCUSSION
In this study, the Foxp3 mRNA and also protein in lesions of patients with L. major infection were identified. Moreover, double immunofluorescent staining of CD4/CD25 and CD25/Foxp3 markers was used to immunophenotype nTregs in patient samples. This method allowed us to easily identify and quantify the cells and has major advantages above single-staining of cutaneous T cells using immunoperoxidase histochemistry.
A 2.3-fold more Foxp3 mRNA expression was found in late CL lesions compared to early ones, proposing a role for nTregs in pathophysiology of chronic human L. major infection. Furthermore, nTreg quantification with IF method validated the prior result (a 2.2-fold increase of nTregsin late lesions).
Bourreau et al. previously reported an eight-fold increase in mRNA expression of Foxp3 in chronic lesions of L. guyanensis infection;[17] however, our results showed a lower difference. Dissimilar parasite species can explain this, as each Leishmania species can trigger special profile of host immune responses. This was confirmed by experimental mouse infections. In L. amazonensis model nTregs had beneficial effects and limited immunopathology,[22] but in L. mexicana infection depletion of CD4+ CD25+ cells did not alter the course of infection, thus nTregs had no effect on this model.[23] L. major has a long history of interaction with humans and well adapted to them, hence is much more different with L. guyanensis and other new world complexes.
Most of nTreg investigations have been directed in murine models of L. major infection. Interestingly, role of nTregs in disease development in these studies depends on both mouse and parasite strains. Belkaid et al. showed that self-cured lesions of C57BL/6 mice inoculated with L. major retain a few viable parasites, mediating long-lasting immunity to secondary challenge. CD4+ CD25+ nTregs were shown to provide a secure environment for these organisms, thus transmission for the parasite and immunity for the host was provided.[24] In another experiment, Infection with resistant strain L. major Sd in C57BL/6 mice caused nonhealing lesions with ulceration and high parasite burden due to accumulation of CD4+ CD25+ Tregs in lesions and suppression of protective Th1 response.[25] In susceptible BALB/c mice nTregs repressed the early Th2 response limiting parasite proliferation and lesion size.[26]
None of these animal models is entirely representative of clinical disease. Moreover, several Treg subsets including inducible Tregs and regulatory cytokines, such as IL10 and TGFβ, may contribute to immune suppression in leishmaniasis.[27] In two separate studies in nonheal models, IL10 derived from CD4+ CD25- effector T cells in late stage of disease had a major contribution to immune suppression in the site of the infection.[28,29] Tregs also use a variety of mechanisms to regulate effector cells; such as direct cell contact, and secretion of suppressor cytokines and molecules; such as IL10, TGFβ, IL35, Fibrinogen-like protein 2 (FGL2), and cyclic AMP[11]
It is not clear to which extend and how nTregs affect acute or chronic leishmaniasis and what is the mechanisms of their regulatory action. Furthermore, the role of other Treg subsets in human disease is not explained. More investigations are crucial to make a clear understanding of regulator–effector balance of immune responses and subsequent use of these perceptions as therapeutic hints in human disease.
The variation detected in both mRNA expression and IF staining of nTregs in patient samples was previously observed in other diseases such as psoriasis, eczematous dermatitis, and grass pollen allergy, as well as in normal skin.[30–33] This may be due to special contact of each person with allergens and pathogens and diversity of disease stage and immune responses.
We found that CD4+ T cells are more frequent in early lesions than late ones. Moreover, DAPI staining of nuclei clearly showed more intact cells in acute samples. This may be as a result of chronic inflammation and consequent fibrosis. The absolute number of nTregs did not differ in two groups, but the ratios of nTregs to CD4+ cells were significantly different. It is now obvious that the balance between nTregs and effector cells determines the outcome of infection; hence, the tendency of this balance toward Tregs in our chronic patients may be the cause of long-lasting disease.
To the best of our knowledge, this is the first experiment on the role of nTregs in human L. major induced leishmaniasis. Although we did not examine the dynamics and function of regulatory cells in the study, this can be a valuable step toward future researches.
ACKNOWLEDGMENTS
Authors gratefully acknowledge Dr M. Mohajeri, Miss Farzaneh Mahmoodi and Mr Mohammad Kardi for their technical help and also thank Parasitology Department and Research Center for Skin Disease and Leishmaniasis, Sedigheh-Tahereh, Isfahan University of Medical Sciences, especially Miss Leila Shirani for her help with patient sampling.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Pearson RD, Sousa AQ. Clinical spectrum of Leishmaniasis. Clin Infect Dis. 1996;22:1–13. doi: 10.1093/clinids/22.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Nylen S, Gautam S. Immunological perspectives of leishmaniasis. J Glob Infect Dis. 2010;2:135–46. doi: 10.4103/0974-777X.62876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hepburn NC. Cutaneous leishmaniasis: An overview. J Postgrad Med. 2003;49:50–4. doi: 10.4103/0022-3859.928. [DOI] [PubMed] [Google Scholar]
- 4.Karamian M, Motazedian MH, Fakhar M, Pakshir K, Jowkar F, Rezanezhad H. Atypical presentation of old-world cutaneous leishmaniasis, diagnosis and species identification by PCR. J Eur Acad Dermatol Venereol. 2008;22:958–62. doi: 10.1111/j.1468-3083.2008.02674.x. [DOI] [PubMed] [Google Scholar]
- 5.Louis J, Himmelrich H, Parra-Lopez C, Tacchini-Cottier F, Launois P. Regulation of protective immunity against Leishmania major in mice. Curr Opin Immunol. 1998;10:459–64. doi: 10.1016/s0952-7915(98)80121-0. [DOI] [PubMed] [Google Scholar]
- 6.Reiner SL, Locksley RM. The regulation of immunity to Leishmania major. Annu Rev Immunol. 1995;13:151–77. doi: 10.1146/annurev.iy.13.040195.001055. [DOI] [PubMed] [Google Scholar]
- 7.Gaafar A, Veress B, Permin H, Kharazmi A, Theander TG, el Hassan AM. Characterization of the local and systemic immune responses in patients with cutaneous leishmaniasis due to Leishmania major. Clin Immunol. 1999;91:314–20. doi: 10.1006/clim.1999.4705. [DOI] [PubMed] [Google Scholar]
- 8.Louzir H, Melby PC, Ben Salah A, Marrakchi H, Aoun K, Ben Ismail R, et al. Immunologic determinants of disease evolution in localized cutaneous leishmaniasis due to Leishmania major. J Infect Dis. 1998;177:1687–95. doi: 10.1086/515297. [DOI] [PubMed] [Google Scholar]
- 9.Ajdary S, Jafari-Shakib R, Riazi-Rad F, Khamesipour A. Soluble CD26 and CD30 levels in patients with anthroponotic cutaneous leishmaniasis. J Infect. 2007;55:75–8. doi: 10.1016/j.jinf.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Jafari-Shakib R, Shokrgozar MA, Nassiri-Kashani M, Malakafzali B, Nikbin B, Khamesipour A. Plasma sCD26 and sCD30 levels in cutaneous leishmaniasis. Acta Trop. 2009;109:61–3. doi: 10.1016/j.actatropica.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Shalev I, Schmelzle M, Robson SC, Levy G. Making sense of regulatory T cell suppressive function. Semin Immunol. 2011;23:282–92. doi: 10.1016/j.smim.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belkaid Y, Sun CM, Bouladoux N. Parasites and immunoregulatory T cells. Curr Opin Immunol. 2006;18:406–12. doi: 10.1016/j.coi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Belkaid Y, Rouse BT. Natural regulatory T cells in infectious disease. Nat Immunol. 2005;6:353–60. doi: 10.1038/ni1181. [DOI] [PubMed] [Google Scholar]
- 14.Sacks D, Anderson C. Re-examination of the immunosuppressive mechanisms mediating non-cure of Leishmania infection in mice. Immunol Rev. 2004;201:225–38. doi: 10.1111/j.0105-2896.2004.00185.x. [DOI] [PubMed] [Google Scholar]
- 15.Campanelli AP, Roselino AM, Cavassani KA, Pereira MS, Mortara RA, Brodskyn CI, et al. CD4 + CD25 + T cells in skin lesions of patients with cutaneous leishmaniasis exhibit phenotypic and functional characteristics of natural regulatory T cells. J Infect Dis. 2006;193:1313–22. doi: 10.1086/502980. [DOI] [PubMed] [Google Scholar]
- 16.Bourreau E, Ronet C, Darsissac E, Lise MC, Marie DS, Clity E, et al. In leishmaniasis due to Leishmania guyanensis infection, distinct intralesional interleukin-10 and Foxp3 mRNA expression are associated with unresponsiveness to treatment. J Infect Dis. 2009;199:576–9. doi: 10.1086/596508. [DOI] [PubMed] [Google Scholar]
- 17.Bourreau E, Ronet C, Darcissac E, Lise MC, Sainte Marie D, Clity E, et al. Intralesional regulatory T-cell suppressive function during human acute and chronic cutaneous leishmaniasis due to Leishmania guyanensis. Infect Immun. 2009;77:1465–74. doi: 10.1128/IAI.01398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasereddin A, Jaffe CL. Rapid diagnosis of old world leishmaniasis by high-resolution melting analysis of the 7SL RNA gene. J Clin Microbiol. 2010;48:2240–2. doi: 10.1128/JCM.00553-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berglund SR, Schwietert CW, Jones AA, Stern RL, Lehmann J, Goldberg Z. Optimized methodology for sequential extraction of RNA and protein from small human skin biopsies. J Invest Dermatol. 2007;127:349–53. doi: 10.1038/sj.jid.5700557. [DOI] [PubMed] [Google Scholar]
- 20.Miyagawa Y, Kiyokawa N, Ochiai N, Imadome K, Horiuchi Y, Onda K, et al. Ex vivo expanded cord blood CD4 T lymphocytes exhibit a distinct expression profile of cytokine-related genes from those of peripheral blood origin. Immunology. 2009;128:405–19. doi: 10.1111/j.1365-2567.2009.03122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 22.Ji J, Masterson J, Sun J, Soong L. CD4 + CD25 + regulatory T cells restrain pathogenic responses during Leishmania amazonensis infection. J Immunol. 2005;174:7147–53. doi: 10.4049/jimmunol.174.11.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas BN, Buxbaum LU. FcgammaRIII mediates immunoglobulin G-induced interleukin-10 and is required for chronic Leishmania mexicana lesions. Infect Immun. 2008;76:623–31. doi: 10.1128/IAI.00316-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belkaid Y, Piccirillo CA, Mendez S, Shevach EM, Sacks DL. CD4 + CD25 + regulatory T cells control Leishmania major persistence and immunity. Nature. 2002;420:502–7. doi: 10.1038/nature01152. [DOI] [PubMed] [Google Scholar]
- 25.Anderson CF, Mendez S, Sacks DL. Nonhealing infection despite Th1 polarization produced by a strain of Leishmania major in C57BL/6 mice. J Immunol. 2005;174:2934–41. doi: 10.4049/jimmunol.174.5.2934. [DOI] [PubMed] [Google Scholar]
- 26.Aseffa A, Gumy A, Launois P, MacDonald HR, Louis JA, Tacchini-Cottier F. The early IL-4 response to Leishmania major and the resulting Th2 cell maturation steering progressive disease in BALB/c mice are subject to the control of regulatory CD4 + CD25 + T cells. J Immunol. 2002;169:3232–41. doi: 10.4049/jimmunol.169.6.3232. [DOI] [PubMed] [Google Scholar]
- 27.Wohlfert E, Belkaid Y. Role of endogenous and induced regulatory T cells during infections. J Clin Immunol. 2008;28:707–15. doi: 10.1007/s10875-008-9248-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson CF, Oukka M, Kuchroo VJ, Sacks D. CD4 (+)CD25 (-)Foxp3 (-) Th1 cells are the source of IL-10-mediated immune suppression in chronic cutaneous leishmaniasis. J Exp Med. 2007;204:285–97. doi: 10.1084/jem.20061886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagase H, Jones KM, Anderson CF, Noben-Trauth N. Despite increased CD4 + Foxp3 + cells within the infection site, BALB/c IL-4 receptor-deficient mice reveal CD4 + Foxp3-negative T cells as a source of IL-10 in Leishmania major susceptibility. J Immunol. 2007;179:2435–44. doi: 10.4049/jimmunol.179.4.2435. [DOI] [PubMed] [Google Scholar]
- 30.Radulovic S, Jacobson MR, Durham SR, Nouri-Aria KT. Grass pollen immunotherapy induces Foxp3-expressing CD4 + CD25 + cells in the nasal mucosa. J Allergy Clin Immunol. 2008;121:1467–72. doi: 10.1016/j.jaci.2008.03.013. 72 e1. [DOI] [PubMed] [Google Scholar]
- 31.Bovenschen HJ, van Vlijmen-Willems IM, van de Kerkhof PC, van Erp PE. Identification of lesional CD4 + CD25 + Foxp3 + regulatory T cells in Psoriasis. Dermatology. 2006;213:111–7. doi: 10.1159/000093849. [DOI] [PubMed] [Google Scholar]
- 32.de Boer OJ, van der Loos CM, Teeling P, van der Wal AC, Teunissen MB. Immunohistochemical analysis of regulatory T cell markers FOXP3 and GITR on CD4 + CD25 + T cells in normal skin and inflammatory dermatoses. J Histochem Cytochem. 2007;55:891–8. doi: 10.1369/jhc.6A7119.2007. [DOI] [PubMed] [Google Scholar]
- 33.Fujimura T, Okuyama R, Ito Y, Aiba S. Profiles of Foxp3 + regulatory T cells in eczematous dermatitis, psoriasis vulgaris and mycosis fungoides. Br J Dermatol. 2008;158:1256–63. doi: 10.1111/j.1365-2133.2008.08504.x. [DOI] [PubMed] [Google Scholar]


