Abstract
Epithelial cells reside on specialized extracellular matrices that provide instructive cues to regulate and support cell function. The authors have previously demonstrated that substrate topography with dimensions similar to the native extracellular matrix (submicrometer and nanoscale features) significantly impacts corneal epithelial proliferation and migration. In this work, synthetic hydrogels were modified with both topographic and biochemical cues, where specified peptide ligands were immobilized within nanopatterned hydrogels. The efficient, systematic study of multiple instructive cues (peptide, peptide concentration, topographic dimensions), however, is contingent on the development of higher throughput platforms. Toward this goal, the authors developed a hydrogel array platform to systematically and rapidly evaluate combinations of two different peptide motifs and a range of nanoscale topographic dimensions. Specifically, distinct functional pegylated peptide ligands, RGD (GGGRGDSP) and AG73 (GRKRLQVQLSIRT), were synthesized for incorporation into an inert hydrogel network. Elastomeric stencils with arrays of millimeter-scale regions were used to spatially confine hydrogel precursor solutions on elastomeric stamps with nanoscale patterns generated by soft lithography. The resulting topographically and peptide-functionalized hydrogel arrays were used to characterize single cell migration. Epithelial cell migration speed and persistence were governed by both the biochemical and topographical cues of the underlying substrate.
INTRODUCTION
The extracellular matrix (ECM) is a specialized network that supports surrounding cells and also plays a principal role in engineering the local cellular environment by actively sequestering, releasing, and presenting critical biochemical cues. The unique combinations of proteins, proteoglycans, and glycosaminoglycans that constitute the heterogeneous ECM produce the specific chemical and physical cues that regulate and maintain proper cell phenotypes. A challenge in the field of tissue engineering is developing well-defined platforms to systematically and efficiently identify combinations of specific ECM cues that guide and support representative cell behavior.
Numerous ECM cues have previously been investigated as individual independent variables and shown to impact cellular behavior. For example, stiff polymeric substrates with nanometer scale topographic features that mimic the dimensions of pores and fibers of the ECM control cell proliferation,1 migration,2 and differentiation.3 These topographic substrates, however, maintain no control of the presentation of chemical cues as they allow nonspecific protein adsorption. Likewise, others have shown substrate stiffness,4 and presentation and density of biological ligands5, 6 individually impacts cell spreading and proliferation. While these individual cues provide important information about cell–substrate interactions, the intersection of these physical and chemical cues has significant potential in the development and design of cell-instructive biomaterials. Thus, there is a need to develop new synthetic ECMs that can simultaneously control nanometer scale topography and peptide presentation to cells.
Numerous studies have focused on the development of innovative techniques to spatially pattern chemical cues.7 Microcontact printing is a common technique to design chemical patterns on the order of the size of cells (microns) to study, for example, geometric restraints on cell apoptosis8 or viral propagation.9 Other techniques such as e-beam lithography,10, 11 dip pen lithography,12 laser scanning lithography,13 and colloidal lithography14 have been used to pattern chemical cues on the length scales of focal adhesions (nanometers). Chemical patterns on the nanometer scale have been used to explore fundamental cell–substrate interactions, including the limits of focal adhesion spacing. In addition, nanometer length scale patterns have been used to develop patterns for applied applications, for example, patterns that support neuronal cell elongation while preventing the adhesion of astrocytes.10 The chemical patterns have recently advanced to include techniques to spatially pattern multiple chemical cues.11, 13 This paper is not an additional demonstration of the ability to pattern biochemical cues in hydrogels, but rather, we use biomimetic length scale topographic patterns to enable the simultaneous study of biomimetic physical and biologically relevant chemical cues in a soft material platform.
The systematic investigation of combinations of biologically relevant physical and biochemical cues requires the development of screening platforms with increased throughput in order to study the large number of possible inputs. There have been significant advancements in the combinatorial screening of materials for the optimization of cell culture conditions over the last decade. The most prevalent studies investigate the cellular response to an array of chemical parameters including combinations of peptides,15 ECM proteins,16 and acrylated monomers.17 While these studies enable the identification of an optimal surface chemistry for a specific outcome (e.g., clonal growth of pluripotent stem cells), there is little control over other material properties known to influence cell behavior, such as nanometer-scale topography.
In the current work, we simultaneously investigate two different cues by developing strategies to replica mold well-defined cell instructive hydrogel materials. Elastomeric stencils separated different hydrogel precursor solutions into millimeter-scale circular array spots, which were formed on nanometer-scale ridge and groove patterned stamps. When the hydrogels were polymerized and released from the stamp and stencil, the result was an array of peptide functionalized, nanopatterned hydrogels. This approach allowed multiple parameters to be tested within the same culture conditions while also limiting material consumption. These arrays are a facile platform for investigation of multiple biologically relevant cues, namely nanotopography and specific peptide ligands. Using these arrays, we show that human epithelial cell migration was dictated by both the nanoscale topography and the specific peptides presented in the hydrogel substrate. Platforms with increased throughput, like these arrays, are necessary to elucidate key synergistic combinations of instructive physical and chemical cues.
EXPERIMENT
Fabrication of micro- and nanopatterned hydrogels
Ridge and groove patterned silicon surfaces18 (pitch of 400, 1400, or 4000 nm; depth of features was 300 nm; ridge to groove ratio was 1:1) were used as masters to generate polydimethylsiloxane (PDMS) stamps.1 PDMS stamps were used as templates for replication of the patterns in poly(ethylene glycol) diacrylate (PEGDA) hydrogels. A PDMS stencil (rectangular [1.2 mm × 6 mm] or circular [d = 2 mm] arrays) was fabricated via replica molding of a silicon master.19 A stencil was applied to a PDMS stamp with the desired topographic features. Hydrogel precursor solutions of 3.4 kDa PEGDA (Ref. 20) (20 wt. % w/w), the photoinitiator lithium phenyl-2,4,6-trimethylbenzoyl-phosphinate21 (2.2 mM), and acryloyl functionalized pegylated peptides6 (RGD = acryloyl-PEG27-GGGRGDSP; AG73 = acryloyl-PEG27-GRKRLQVQLSIRT) were pipetted within the void spaces of the stencil. The precursor solutions were then covered with glass coverslips that had been treated with 3 -(trichlorosilyl)propyl methacrylate (Sigma Aldrich) and exposed to a UV source (365λ, 8.7 mW/cm2) in a nitrogen environment for 300 s (Fig. 1). Samples were hydrated and maintained in a hydrated state for the duration of the experiment. Hydrogels were sterilized with ethanol (70% v/v) and UV light (265λ) for 15 min and then equilibrated with phosphate buffered saline (1X PBS) prior to cell culture.
Figure 1.
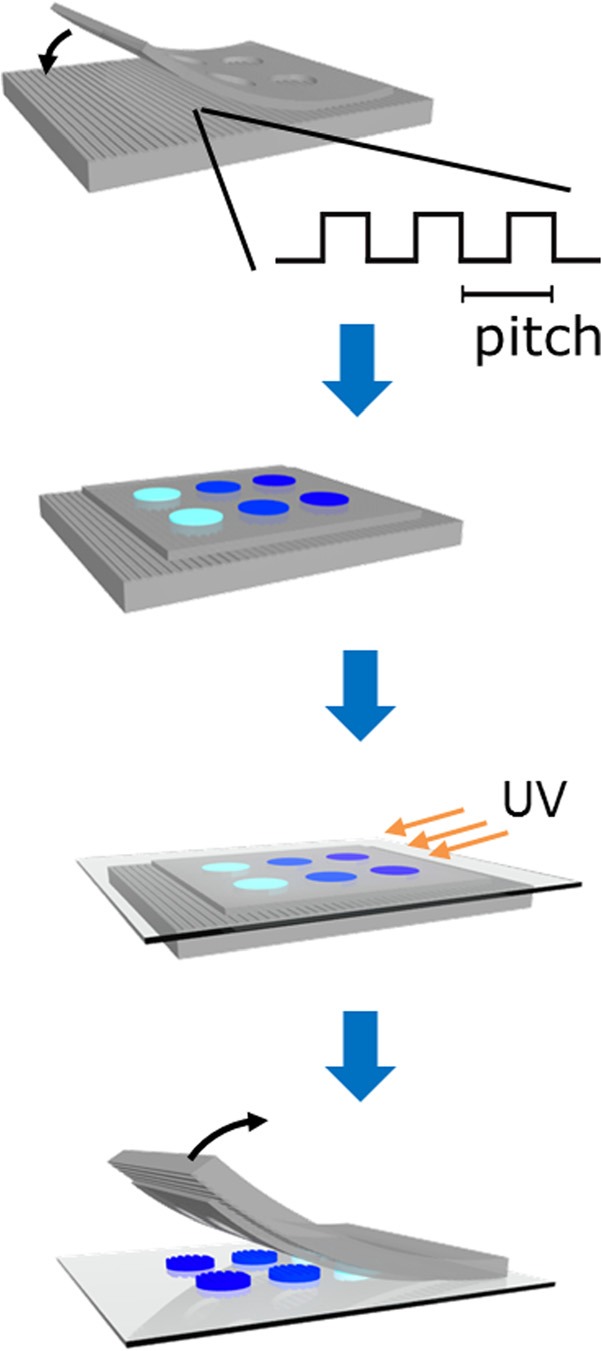
(Color online) Schematic of the molding process demonstrates the use of a PDMS stencil to obtain distinct, topographically patterned hydrogel regions. In the process, PDMS stencils were placed on PDMS stamps with ridge and groove features, where the pitches range from 400 to 4000 nm. Hydrogel precursor solutions were constrained within the void space of the stencil, covered with a treated glass cover slide and polymerized by exposure to UV light source. Discrete arrays of topographically molded hydrogels were then released from the elastomeric stamp and stencil.
Hydrogel characterization
Hydrogels were imaged by atomic force microscopy (AFM) to measure the pitch of the ridge and groove features. Samples were hydrated in deionized water for at least 48 h at room temperature prior to measurements. In a fluid cell, samples were scanned in contact mode using a SNL-10 silicon nitride cantilever with a silicon tip (Veeco Probes, CA) using a Nanoscope IIIa Multimode scanning probe microscope (Veeco Instruments, Inc., CA).
Topographically patterned hydrogels with different concentrations of peptide were imaged with a GE Healthcare Typhoon Tri Variable Mode Imager to qualitatively measure peptide concentration. Hydrogel arrays were equilibrated in 1X PBS and then equilibrated with 0.15 M NaHCO3 buffer solution. An excess of Alexa Fluor 488 5-SDP Ester (Life Technologies) was then added to the buffered solution and allowed to react with any available primary amine incorporated in the hydrogel (AG73 sequence: GRKRLQVQLSIRT) for 1 h. After the reaction, the hydrogel arrays were equilibrated with 1X PBS. Scanned images were colored and analyzed with imagej.
Cell culture
Telomerase-immortalized human corneal epithelial (hTCEpi) cells22 were donated by James Jester from University of California, Irvine. hTCEpi cells were cultured in epithelial medium containing a 3:2 ratio of Ham's F-12:Dulbecco's modified eagle medium (DMEM) (Invitrogen) supplemented with 2.5% fetal bovine serum, 0.4 μg ml−1 hydrocortisone, 8.4 ng ml−1 cholera toxin, 5 μg ml−1 insulin, 24 μg ml−1 adenine, 10 ng ml−1 epidermal growth factor, 100 IU ml−1 penicillin, and 100 μg ml−1 streptomycin.23, 24 Mitomycin-c treated Swiss 3T3 fibroblasts were used as a feeder layer to maintain hTCEpi stocks on 100 mm tissue culture plates. hTCEpi stock plates were incubated at 37 °C and 5% CO2. When hTCEpi colonies reached approximately 70%–80% confluence, hTCEpi cells were plated on hydrogel arrays. hTCEpi cells between passages 45 and 55 were used for experiments.
hTCEpi cell migration analysis
hTCEpi cells plated at a density of 10 000 cells cm−2 on topographically patterned, peptide functionalized hydrogels were incubated for 4 h for initial attachment. An automated Nikon TiE inverted microscope with a stage top incubation system to control temperature and CO2 levels was used to collect 10× images of hTCEpi cells every 12 min for 6 h. The images were analyzed with NIS Elements. A migrating cell was defined as a cell whose center of mass traveled a distance of more than one cell length during the tracking period. Cells that divided, interacted with other cells, or migrated out of the imaging field were excluded from the analysis. Mean squared displacements for the centroid path of each cell were calculated using the method of overlapping intervals using cell_motility software.25 Cell speed was determined by dividing the root mean square displacement by the tracking interval Δt = 12 min. The confinement ratio was calculated by dividing the displacement of the cell from the origin by its total path length. Cell persistence time was fit using a nonlinear least squares regression in matlab by inserting the cell's average speed into a random walk model for cell migration
where ⟨(Δx)2⟩ is the mean square displacement of the cell over time interval (Δt), S is the root mean square cell speed, and P is the persistence time. Error bars represent the 95% confidence interval of the mean.
RESULTS AND DISCUSSION
Design of nanopatterned, peptide functionalized arrays
In the current work, we developed an approach to investigate a combination of two independent variables that mimic instructive cues of the native extracellular matrix of the corneal epithelium. Specifically, peptide cell-adhesion ligands were incorporated into nanoscale patterned hydrogel arrays, enabling the efficient study of peptide type and topographic dimensions. The two peptides selected, RGD and AG73, have significant biological roles in wound healing. RGD, a fragment of fibronectin, interacts with select integrin cell–surface receptors that, when upregulated, promote migration and proliferation.26 AG73, a fragment of laminin-1, interacts with the heparin sulfate domain of syndecan-127 that is found at the leading edge of wounds,28 and the lack of syndecan-1 activation results in a decreased migration rate.29 The tight regulation of both integrins and syndecans in the native cellular environment during wound healing provides sufficient motivation to use RGD and AG73 as the initial chemical cues in the hydrogel arrays described here.
The arrays of topographically and peptide-functionalized hydrogels were generated using elastomeric millimeter-scale stencils on nano- and micron patterned stamps (Fig. 1). The lateral dimensions of the large area PDMS stamps used in this study were 400, 1400, or 4000 nm with feature depths of 300 nm. The lateral dimensions were selected as they span from the biologically relevant nanoscale features of the native extracellular matrices30 to the micron-scale, which is the prevalent length scale for topographic features in previous studies. For confined area replica molding, PDMS stencils with arrays of millimeter-scale rectangles or circles were used to spatially confine and isolate hydrogel precursor solutions. The hydrogel precursor solution consists of a nonfouling PEG polymer and peptides, where both components were functionalized with acrylate groups.6 In the presence of a photoinitiator and UV source, the hydrogel precursor components were copolymerized, creating a cross-linked hydrogel network that retained the nanoscale ridge and groove pattern of the stamp. The hydrogel arrays were hydrated and allowed to swell prior to cell culture, which resulted in an approximate 30% increase in the hydrogel volume. Yanez-Soto et al. demonstrated that hydrogels composed of 20 wt. % PEGDA (MW 3400) retained the topographic features and had anti-fouling properties.31
There was good fidelity between the stamp lateral dimensions and the resulting hydrated hydrogel surface dimensions; replica molded hydrogels had pitches of 400, 1400, or 4000 nm with groove depths ranging from 450 to approximately 200 nm, as measured with AFM (Fig. 2). The primary purpose of the AFM measurements was to demonstrate the retention of the topographic features in hydrated substrates following release from the mold. The feature depths of the substrates were not absolutely known due to the limitations of using contact mode AFM on the soft hydrogel surfaces (Young's Modulus ∼ 100 kPa). We have previously demonstrated that feature depth of topographic features is a significant parameter, where increased groove depth resulted in an increase in the number of cells elongating and aligning in the direction of the grooves.32 It is important to note, however, that differences in feature depths ranging from 200 to 450 nm, as measured by AFM (absolute or not) in this study, would not have elicited significantly different cellular responses.
Figure 2.
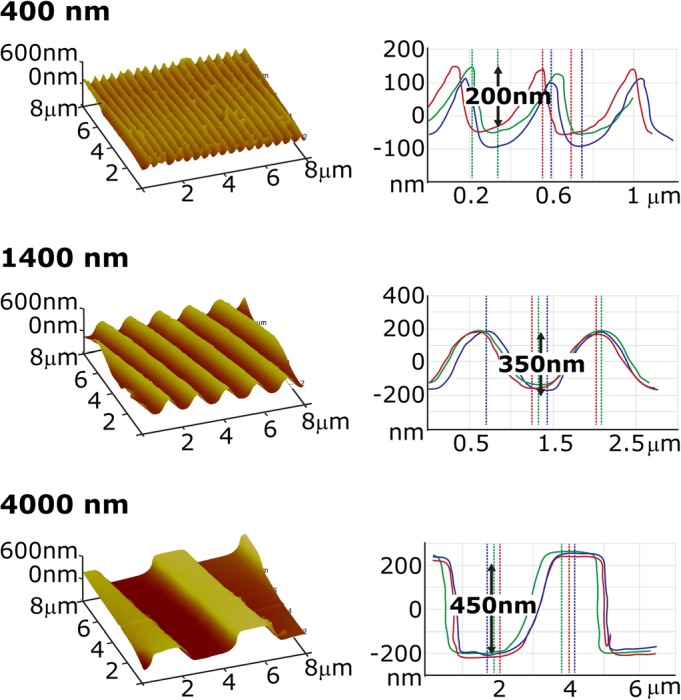
(Color online) AFM measurements of the hydrated samples show the groove and ridge features molded into hydrogels with pitch sizes of 400, 1400, and 4000 nm. The difference in elevation from the ridge and groove is approximately 300 nm.
The hydrogel arrays provide exquisite control of both physical and chemical cues presented to the cells. Previously, we demonstrated that the conjugation of peptides to pure, monodisperse PEG spacers and subsequent acrylate functionalization allows peptides to be incorporated via radical polymerization at high efficiencies, with high reproducibility.6 The peptides are covalently bound via an amide bond to eliminate concerns of hydrolysis of the peptide for long-term studies that usually accompany peptide incorporation schemes like Michael-type addition.5 The hydrolytic cleavage of the PEGDA ester bonds between the aliphatic polymer backbone and the crosslinking unit has been shown by other groups to be limited during the time frame of our experiments, and the swelling ratio has been shown to not significantly change over the course of 6 weeks (where complete degradation is estimated to be on the order of months to years).33
The final arrays featured isolated millimeter-scale hydrogel regions that never contacted adjacent precursor solutions, thus preserving the desired biochemical properties. To demonstrate that the precursor solutions were isolated and retained within the desired regions of the stencil, three different precursor solutions with increasing concentrations of AG73 were dispensed into three adjacent regions. After polymerization, the AG73 peptide was fluorescently labeled and imaged with a fluorescence scanner. Each hydrogel array element had a distinct concentration, as demonstrated by fluorescence intensity, that was unaffected by the proximity of neighboring solutions (Fig. 3). In addition to isolating components, the PDMS stencil also reduced the amount of precursor material consumed and enabled multiple precursor solutions (varying peptide density or combinations of multiple peptides) to be tested on a single coverslip for a given topographic pitch. The resulting arrays, in combination with an automated microscope, were used to effectively monitor cellular dynamics or image fixed cells.
Figure 3.
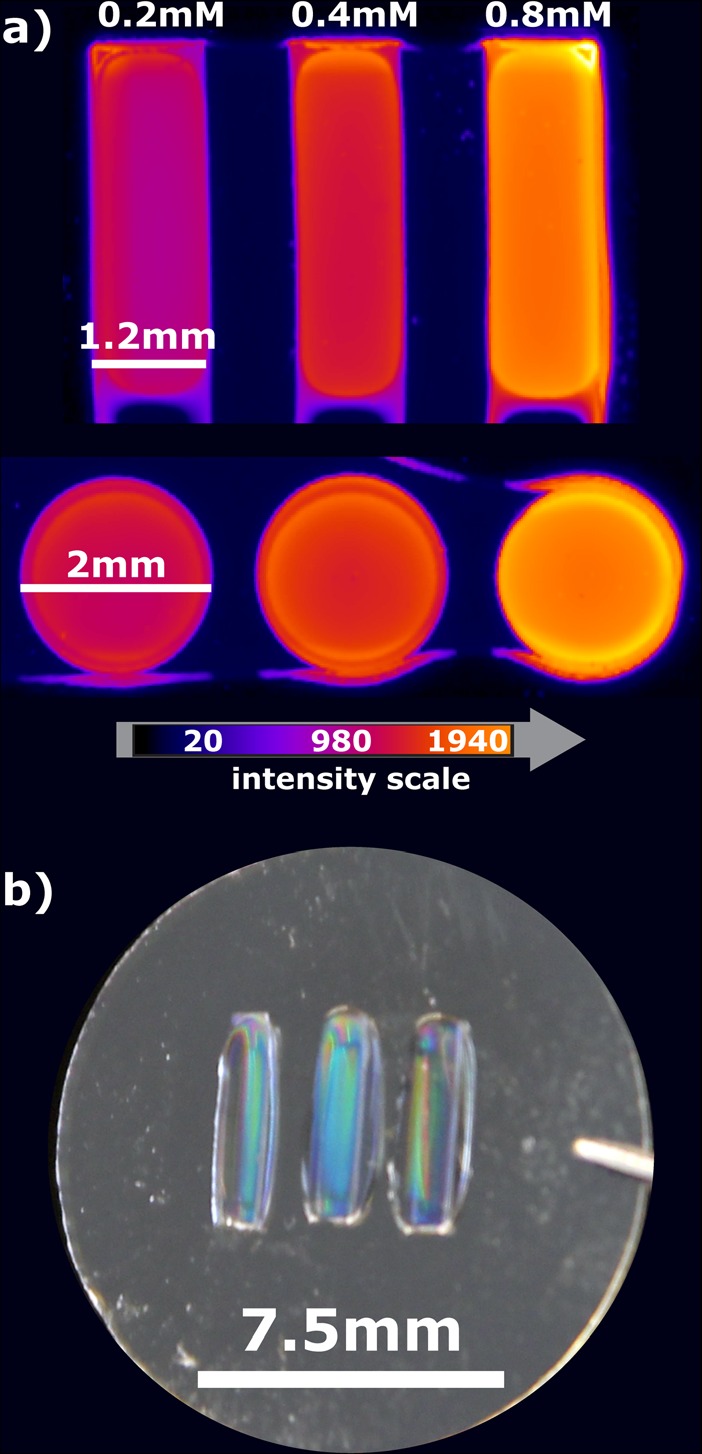
(Color online) (a) Fluorescence image of topographically molded hydrogel arrays functionalized with different concentrations of fluorescently tagged peptide. Rectangular or circular areas were spatially defined by a PDMS stamp, where the individual domains are isolated from the other regions. (b) Photograph of a rectangular topographically molded hydrogel array.
Epithelial cell migration on hydrogel arrays
To highlight this new materials platform and strategy for its use in medium throughput assays, we selected three topographic features and two distinct peptides for cell studies. Note that this platform will also allow the study of many other variables, including peptide density, combinations of many different peptides, different topographic geometries and substrate compliances, which have the potential to significantly alter the cell response. The peptides RGD and AG73, which interact with two distinct cell–surface receptors, were incorporated into patterned hydrogel arrays at concentrations previously shown to support similar cell spreading and proliferation rates. The topography was also varied, including molded ridge and groove patterns of 400, 1400, and 4000 nm pitches and nonpatterned surfaces. The combinatorial effect of topography and biochemical cues on epithelial cell migration was assessed using the aforementioned array platform.
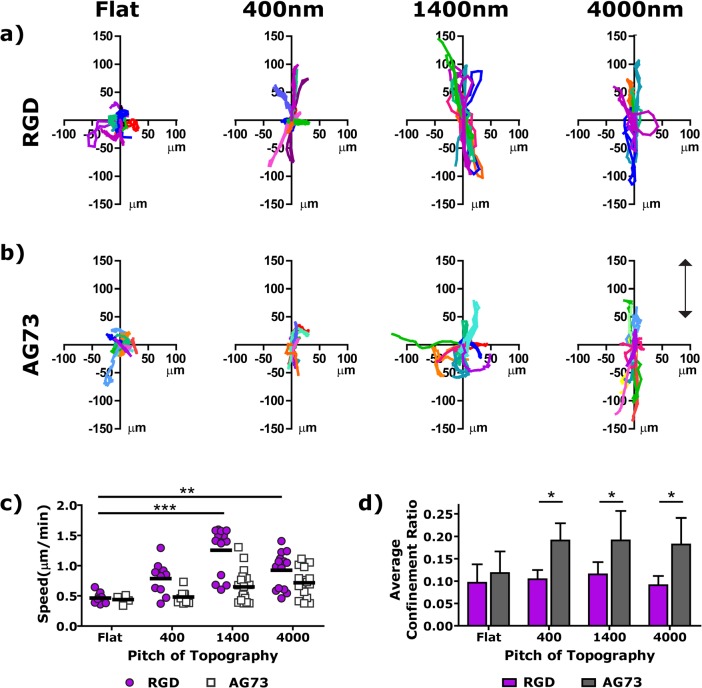
hTCEpi cell migration was analyzed on the array surfaces. hTCEpi cells were selected for the cell experiments to align with our interest in developing synthetic basement membrane constructs for corneal prosthetic devices.18 The experimental format, however, is generalizable for other cell types where control of chemical and physical cues is desirable. For migration experiments, hTCEpi cells were imaged with an automated microscope every 12 min for 6 h. The migration of the cells was analyzed to obtain cell tracks, cell speed, confinement ratios, and persistence times for the eight different conditions. The confinement ratio is the ratio of the average distance from the origin to the total cumulative path length. Persistence time provides an assessment of cell motility, where short persistence times correlate with random movement and long persistence times suggest persistent migration in a particular direction. The migration analysis provides biologically meaningful output to understand the cellular response to the combinatorial sample set.
Corneal epithelial cells migrated parallel to the ridge and groove topographies, where the extent of directional migration was dictated by the peptides incorporated into the hydrogel. Control surfaces, where the PEGDA was not functionalized with a peptide, did not support cell attachment for any of the flat or patterned conditions (data not shown). Cells on RGD functionalized surfaces migrated in the direction of the ridges and grooves for all topographic pitches [Fig. 4a]. The cell tracks were distinct on the topographically molded surfaces when compared to the nonpatterned hydrogels; the cells explored a larger space, migrating on average over 100 μm, and had directed migration parallel to the ridges and grooves. The hTCEpi cell speed on RGD surfaces, however, was dependent on the pitch of the topography. Cells migrated significantly faster on 1400 and 4000 nm pitches than on the nonpatterned surfaces [Fig. 4c]. On the 1400 nm patterned RGD surfaces, a significant fraction of the cell population migrated at a faster speed than on any other surface tested. In addition, the cell population appears to be divided, where one subset migrated at a fast rate, while the other subset migrated at a much slower rate. This distribution of cells is possibly related to a percentage of cells in certain phases of the mammalian cell cycle. The confinement ratio of the cells on both nonpatterned and patterned RGD surfaces was approximately constant [Fig. 4d].
Figure 4.
(Color online) hTCEpi cells migrate parallel to the pattern on both RGD (a) and AG73 (b) functionalized surfaces, where the arrow indicates the direction of the ridges and grooves. (c) The speed of cell migration was dependent on both the pitch of the topography and the biochemical cues of the peptide. (d)Similarly, the confinement ratio was also dependent on the pitch of the topography and the peptide. * is 0.01 ≤ p < 0.05, ** is 0.001 ≤ p < 0.01, and *** is p < 0.001 compared to the flat control (c) or the corresponding RGD gel (d).
Similar to what was observed for RGD surfaces, hTCEpi cells on patterned AG73 surfaces migrated further and had more directed migration as compared to cells on nonpatterned AG73 surfaces. The cell tracks on AG73 surfaces, however, suggest minor differences in cellular migration on AG73 and RGD surfaces. For instance, cells had less directed migration parallel to the pattern on AG73 surfaces on 1400 nm pitches than on the analogous RGD surfaces. Similarly, cells migrated shorter distances from the origin on 400 nm AG73 surfaces as compared to the corresponding RGD surface. There were also significant differences between cell speed trends and confinement ratios on the different peptide functionalized surfaces. The hTCEpi cell speed increased as the pitch of the topography increased on AG73 patterned surfaces. The confinement ratio was larger on topographically patterned AG73 surfaces than on the nonpatterned AG73 surfaces.
These results, namely directional migration and speed of migration on RGD modified surfaces, were in agreement with our previous reports on stiff polyurethane substrates that did not selectively target specific cell receptors. On polyurethane substrates, the epithelial cell migration on a range of topographic features was characterized as a biphasic response, where migration was stimulated in the 1600 nm range when compared to both larger and smaller size topographic pitches.2 This is likely correlated with previous observations on polyurethane substrates that the 1400 nm region was at the transition region where pitches smaller than 1400 nm elicited stronger cell adhesion than on larger pitches.34 Cell migration has previously been shown to be an inverse function of the affinity of cell–substrate interactions.35 Interestingly, this biphasic trend was not observed on AG73 functionalized surfaces. These results suggest that the activated integrin receptor dominates the cell behavior response on nonselective surfaces for corneal epithelial cells.
hTCEpi cells migrated faster and more frequently returned to the origin on RGD patterned hydrogels as compared to the AG73 surfaces. Cells migrated approximately 100 μm in a directed path from the origin during the 6 h of observation on both RGD and AG73 functionalized surfaces; however, the hTCEpi cells on RGD surfaces frequently retraced their migration path (supplementary Fig. 1, Ref. 36). This backtracking phenomenon explains the smaller confinement ratios and shorter persistence times (supplementary Fig. 2, Ref. 36) measured on RGD surfaces as compared to AG73 surfaces. There are numerous factors that contribute to persistent migration;37 external guidance cues such as the topographic cues presented here can geometrically constrain adhesion sites or intrinsic cues that can result in a decrease in the number of peripheral cell protrusions that is correlated with a decrease in Rac1 activity.38 Syndecan-1 mediated cell adhesion (mode of cell–substrate interaction on AG73 surfaces) has previously been linked to rapid changes in activity of several Rho GTP binding proteins.39 We hypothesize that the activity of Rac1 or other GTP binding proteins plays a role in the increased persistent migration on AG73 surfaces. Biomaterials that implement instructive cues, such as guided nanoscale topography or peptides that support persistent migration in a particular direction could have considerable impact on wound healing applications where sustained, directed migration is desirable.
SUMMARY AND CONCLUSIONS
We developed a hydrogel array platform suitable for systematically measuring multiple peptides in combination with different topographic dimensions. The arrays were used to demonstrate that both biochemical and physical cues provide essential signals for epithelial cell migration. The choice of peptide affected speed and persistence of the cell migration on the patterned surfaces, while the pitch of the topography only influenced the speed of migration. Tools, like these arrays, that facilitate the investigation of an increased combinatorial space have great potential for gaining insight on cell–substrate interactions for the rational design of biomaterial substrates. In addition to controlling topography and peptide type, other parameters of this synthetic array can also be modulated, including substrate stiffness, peptide density, and peptide presentation.
ACKNOWLEDGMENTS
The authors would like to thank Eric Nguyen for assistance with fabrication of the PDMS stencils, Dr. Hyo Seon Suh for help designing the schematic for the project, and Anna Kiyanova for providing PDMS molds from the silicon masters.
References
- Liliensiek S. J., Campbell S., Nealey P. F. and Murphy C. J., J. Biomed. Mater. Res. Part A 79A 185 (2006). 10.1002/jbm.a.30744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl K. A., Foley J. D., Nealey P. F., and Murphy C. J., J. Biomed. Mater. Res. Part A 75A, 603 (2005). 10.1002/jbm.a.30467 [DOI] [PubMed] [Google Scholar]
- McMurray R. J. et al. , Nature Mater. 10, 637 (2011). 10.1038/nmat3058 [DOI] [PubMed] [Google Scholar]
- Engler A., Bacakova L., Newman C., Hategan A., Griffin M., and Discher D., Biophys. J. 86, 617 (2004). 10.1016/S0006-3495(04)74140-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbert D. L. and Hubbell J. A., Biomacromolecules 2, 430 (2001). 10.1021/bm0056299 [DOI] [PubMed] [Google Scholar]
- Wilson M. J., Liliensiek S. J., Murphy C. J., Murphy W. L., and Nealey P. F., Soft Matter 8, 390 (2012). 10.1039/c1sm06589k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R. C. and Healy K. E., J. Biomed. Mater. Res. Part A 90A, 1252 (2009). 10.1002/jbm.a.32501 [DOI] [PubMed] [Google Scholar]
- Chen C. S., Mrksich M., Huang S., Whitesides G. M., and Ingber D. E., Science 276, 1425 (1997). 10.1126/science.276.5317.1425 [DOI] [PubMed] [Google Scholar]
- Endler E. E., Duca K. A., Nealey P. F., Whitesides G. M., and Yin J., Biotechnol. Bioeng. 81, 719 (2003). 10.1002/bit.10516 [DOI] [PubMed] [Google Scholar]
- Krsko P., Sukhishvili S., Mansfield M., Clancy R., and Libera M., Langmuir 19, 5618 (2003). 10.1021/la034157r [DOI] [Google Scholar]
- Christman K. L., Schopf E., Broyer R. M., Li R. C., Chen Y., and Maynard H. D., J. Am. Chem. Soc. 131, 521 (2009). 10.1021/ja804767j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. W., Oh B. K., Sanedrin R. G., Salaita K., Fujigaya T., and Mirkin C. A., Adv. Mater. 18, 1133 (2006). 10.1002/adma.200600070 [DOI] [Google Scholar]
- Slater J. H., Miller J. S., Yu S. S., and West J. L., Adv. Funct. Mater. 21, 2876 (2011). 10.1002/adfm.201100297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold M., Cavalcanti-Adam E. A., Glass R., Blummel J., Eck W., Kantlehner M., Kessler H., and Spatz J. P., ChemPhysChem 5, 383 (2004). 10.1002/cphc.200301014 [DOI] [PubMed] [Google Scholar]
- Jung J. P., Moyano J. V., and Collier J. H., Integr. Biol. 3, 185 (2011). 10.1039/c0ib00112k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaim C. J., Teng D., Chien S., and Bhatia S. N., Stem Cells Dev. 17, 29 (2008). 10.1089/scd.2007.0085 [DOI] [PubMed] [Google Scholar]
- Mei Y. et al. , Nature Mater. 9, 768 (2010). 10.1038/nmat2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira A. I., Abrams G. A., Bertics P. J., Murphy C. J., and Nealey P. F., J. Cell Sci. 116, 1881 (2003). 10.1242/jcs.00383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch A., Jo B. H., Hurtado O., Beebe D. J., and Toner M., J. Biomed. Mater. Res. 52, 346 (2000). [DOI] [PubMed] [Google Scholar]
- Lin C. C. and Metters A. T., J. Biomed. Mater. Res. Part A 83A, 954 (2007). 10.1002/jbm.a.31282 [DOI] [PubMed] [Google Scholar]
- Fairbanks B. D., Schwartz M. P., Bowman C. N., and Anseth K. S., Biomaterials 30, 6702 (2009). 10.1016/j.biomaterials.2009.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D. M., Li L., Fisher S., Pearce V. P., Shay J. W., Wright W. E., Cavanagh H. D., and Jester J. V., Invest. Ophthalmol. Visual Sci. 46, 470 (2005). 10.1167/iovs.04-0528 [DOI] [PubMed] [Google Scholar]
- Allen-Hoffmann B. L. and Rheinwald J. G., Proc. Natl. Acad. Sci. U.S.A. 81, 7802 (1984). 10.1073/pnas.81.24.7802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini L. M., Allen-Hoffmann B. L., Warner T. F., and Azen E. A., In Vitro Cell. Dev. Biol. 27, 939 (1991). 10.1007/BF02631121 [DOI] [PubMed] [Google Scholar]
- Martens L., Monsieur G., Ampe C., Gevaert K., and Vandekerckhove J., BMC Bioinf. 7, 289 (2006). 10.1186/1471-2105-7-289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuss J. M. et al. , J. Cell Sci. 108, 2241 (1995). [DOI] [PubMed] [Google Scholar]
- Hoffman M. P., Nomizu M., Rogue E., Lee S., Jung D. W., Yamada Y., and Kleinman H. K., J. Biol. Chem. 273, 28633 (1998). 10.1074/jbc.273.44.28633 [DOI] [PubMed] [Google Scholar]
- Chakravarti R., Sapountzi V., and Adams J. C., Mol. Biol. Cell 16, 3678 (2005). 10.1091/mbc.E04-10-0907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepp M. A. et al. , J. Cell Sci. 120, 2851 (2007). 10.1242/jcs.03480 [DOI] [PubMed] [Google Scholar]
- Abrams G. A., Schaus S. S., Goodman S. L., Nealey P. F., and Murphy C. J., Cornea 19, 57 (2000). 10.1097/00003226-200001000-00012 [DOI] [PubMed] [Google Scholar]
- Yanez-Soto B., Liliensiek S., Murphy C. J., and Nealey P. F., “Biochemically and topographically engineered poly(ethylene glycol) diacrylate hydrogels with biomimetic characteristics as substrates for human corneal epithelial cells,” J. Biomed. Mater. Res. Part A (in press). [DOI] [PMC free article] [PubMed]
- Fraser S. A., Ting Y. H., Mallon K. S., Wendt A. E., Murphy C. J., and Nealey P. F., J. Biomed. Mater. Res. Part A 86A, 725 (2008). 10.1002/jbm.a.31519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning M. B. and Cosgriff-Hernandez E., Biomacromolecules 13, 779 (2012). 10.1021/bm201707z [DOI] [PubMed] [Google Scholar]
- Karuri N. W., Liliensiek S., Teixeira A. I., Abrams G., Campbell S., Nealey P. F., and Murphy C. J., J. Cell Sci. 117, 3153 (2004). 10.1242/jcs.01146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek S. P., Loftus J. C., Ginsberg M. H., Lauffenburger D. A., and Horwitz A. F., Nature 385, 537 (1997). 10.1038/385537a0 [DOI] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1116/1.4762842 for figures that illustrate the migration path of the hTCEpi cells on RGD or AG73 surfaces (Supplemental Fig. 1) and the calculated persistence times for hTCEpi cells on RGD or AG73 functionalized surfaces (Supplemental Fig. 2).
- Petrie R. J., Doyle A. D., and Yamada K. M., Nat. Rev. Mol. Cell Biol. 10, 538 (2009). 10.1038/nrm2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankov R., Endo Y., Even-Ram S., Araki M., Clark K., Cukierman E., Matsumoto K., and Yamada K. M., J. Cell Biol. 170, 793 (2005). 10.1083/jcb.200503152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachy S., Letourneur F., and Rousselle P., J. Cell. Physiol. 214, 238 (2008). 10.1002/jcp.21184 [DOI] [PubMed] [Google Scholar]