Abstract
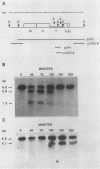
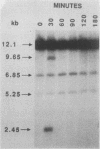
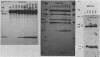
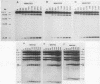
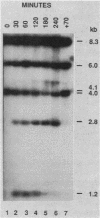
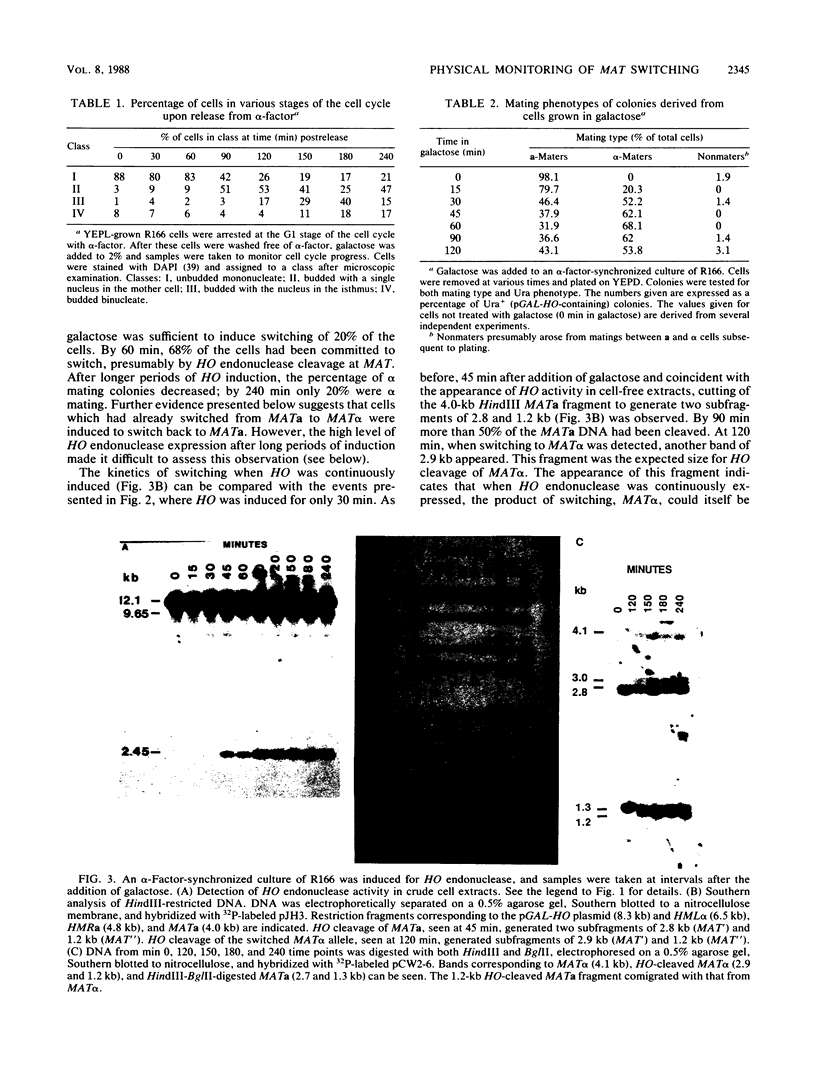
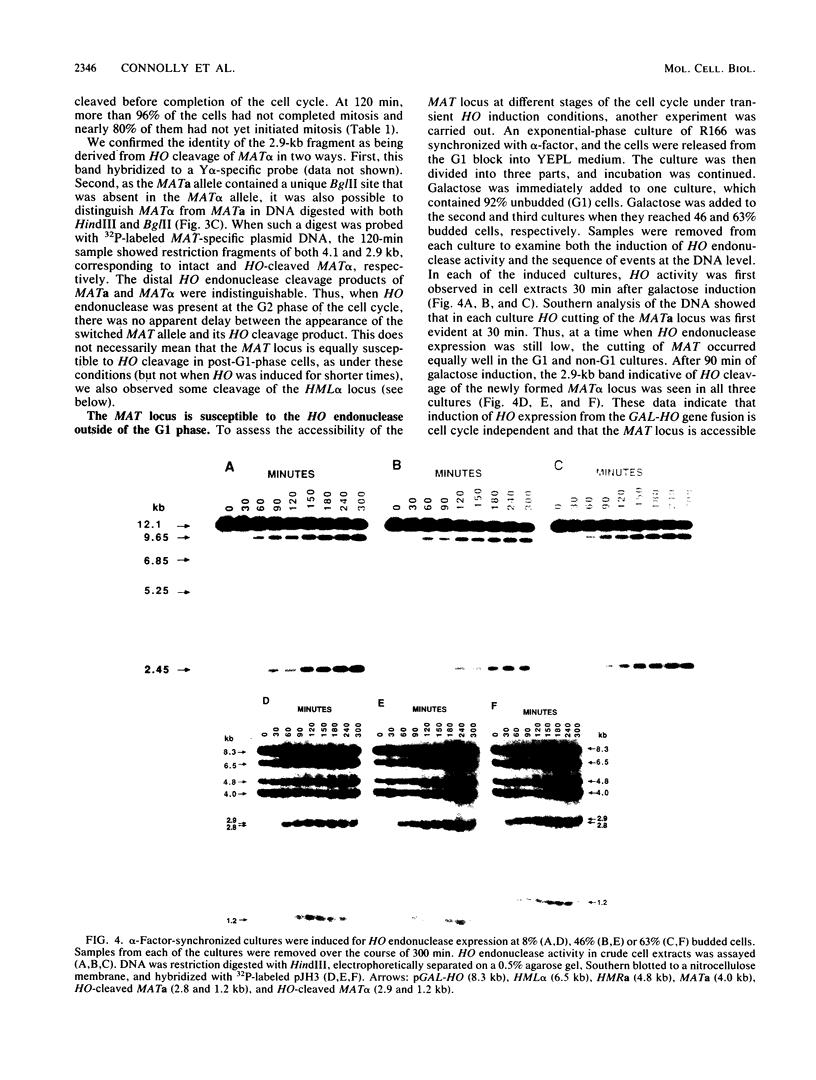
The kinetics of mating type switching in Saccharomyces cerevisiae can be followed at the DNA level by using a galactose-inducible HO (GAL-HO) gene to initiate the event in synchronously growing cells. From the time that HO endonuclease cleaves MAT a until the detection of MAT alpha DNA took 60 min. When unbudded G1-phase cells were induced, switched to the opposite mating type in "pairs." In the presence of the DNA synthesis inhibitor hydroxyurea, HO-induced cleavage occurred but cells failed to complete switching. In these blocked cells, the HO-cut ends of MATa remained stable for at least 3 h. Upon removal of hydroxyurea, the cells completed the switch in approximately 1 h. The same kinetics of MAT switching were also seen in asynchronous cultures and when synchronously growing cells were induced at different times of the cell cycle. Thus, the only restriction that confined normal homothallic switching to the G1 phase of the cell cycle was the expression of HO endonuclease. Further evidence that galactose-induced cells can switch in the G2 phase of the cell cycle was the observation that these cells did not always switch in pairs. This suggests that two chromatids, both cleaved with HO endonuclease, can interact independently with the donors HML alpha and HMRa.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreadis A., Hsu Y. P., Kohlhaw G. B., Schimmel P. Nucleotide sequence of yeast LEU2 shows 5'-noncoding region has sequences cognate to leucine. Cell. 1982 Dec;31(2 Pt 1):319–325. doi: 10.1016/0092-8674(82)90125-8. [DOI] [PubMed] [Google Scholar]
- Astell C. R., Ahlstrom-Jonasson L., Smith M., Tatchell K., Nasmyth K. A., Hall B. D. The sequence of the DNAs coding for the mating-type loci of Saccharomyces cerevisiae. Cell. 1981 Nov;27(1 Pt 2):15–23. doi: 10.1016/0092-8674(81)90356-1. [DOI] [PubMed] [Google Scholar]
- Barford J. P., Hall R. J. Estimation of the length of cell cycle phases from asynchronous cultures of Saccharomyces cerevisiae. Exp Cell Res. 1976 Oct 15;102(2):276–284. doi: 10.1016/0014-4827(76)90043-4. [DOI] [PubMed] [Google Scholar]
- Bücking-Throm E., Duntze W., Hartwell L. H., Manney T. R. Reversible arrest of haploid yeast cells in the initiation of DNA synthesis by a diffusible sex factor. Exp Cell Res. 1973 Jan;76(1):99–110. doi: 10.1016/0014-4827(73)90424-2. [DOI] [PubMed] [Google Scholar]
- Carter B. L., Jagadish M. N. Control of cell division in the yeast Saccharomyces cerevisiae cultured at different growth rates. Exp Cell Res. 1978 Mar 15;112(2):373–383. doi: 10.1016/0014-4827(78)90220-3. [DOI] [PubMed] [Google Scholar]
- Chaleff D. T., Tatchell K. Molecular cloning and characterization of the STE7 and STE11 genes of Saccharomyces cerevisiae. Mol Cell Biol. 1985 Aug;5(8):1878–1886. doi: 10.1128/mcb.5.8.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole G. M., Schild D., Lovett S. T., Mortimer R. K. Regulation of RAD54- and RAD52-lacZ gene fusions in Saccharomyces cerevisiae in response to DNA damage. Mol Cell Biol. 1987 Mar;7(3):1078–1084. doi: 10.1128/mcb.7.3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S., Herskowitz I. The yeast STE12 product is required for expression of two sets of cell-type specific genes. Cell. 1985 Oct;42(3):923–930. doi: 10.1016/0092-8674(85)90288-0. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J Cell Biol. 1980 Jun;85(3):811–822. doi: 10.1083/jcb.85.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J. B., Herskowitz I. Interconversion of Yeast Mating Types I. Direct Observations of the Action of the Homothallism (HO) Gene. Genetics. 1976 Jun;83(2):245–258. doi: 10.1093/genetics/83.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadish M. N., Carter B. L. Effects of temperature and nutritional conditions on the mitotic cell cycle of Saccharomyces cerevisiae. J Cell Sci. 1978 Jun;31:71–78. doi: 10.1242/jcs.31.1.71. [DOI] [PubMed] [Google Scholar]
- Jagadish M. N., Carter B. L. Genetic control of cell division in yeast cultured at different growth rates. Nature. 1977 Sep 8;269(5624):145–147. doi: 10.1038/269145a0. [DOI] [PubMed] [Google Scholar]
- Jensen R. E., Herskowitz I. Directionality and regulation of cassette substitution in yeast. Cold Spring Harb Symp Quant Biol. 1984;49:97–104. doi: 10.1101/sqb.1984.049.01.013. [DOI] [PubMed] [Google Scholar]
- Jensen R., Sprague G. F., Jr, Herskowitz I. Regulation of yeast mating-type interconversion: feedback control of HO gene expression by the mating-type locus. Proc Natl Acad Sci U S A. 1983 May;80(10):3035–3039. doi: 10.1073/pnas.80.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J., Hicks J. B., Strathern J. N. Directionality of yeast mating-type interconversion. Cell. 1982 Mar;28(3):551–561. doi: 10.1016/0092-8674(82)90210-0. [DOI] [PubMed] [Google Scholar]
- Klar A. J., Strathern J. N., Abraham J. A. Involvement of double-strand chromosomal breaks for mating-type switching in Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1984;49:77–88. doi: 10.1101/sqb.1984.049.01.011. [DOI] [PubMed] [Google Scholar]
- Kostriken R., Heffron F. The product of the HO gene is a nuclease: purification and characterization of the enzyme. Cold Spring Harb Symp Quant Biol. 1984;49:89–96. doi: 10.1101/sqb.1984.049.01.012. [DOI] [PubMed] [Google Scholar]
- Kostriken R., Strathern J. N., Klar A. J., Hicks J. B., Heffron F. A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell. 1983 Nov;35(1):167–174. doi: 10.1016/0092-8674(83)90219-2. [DOI] [PubMed] [Google Scholar]
- Malone R. E., Esposito R. E. The RAD52 gene is required for homothallic interconversion of mating types and spontaneous mitotic recombination in yeast. Proc Natl Acad Sci U S A. 1980 Jan;77(1):503–507. doi: 10.1073/pnas.77.1.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A. Molecular genetics of yeast mating type. Annu Rev Genet. 1982;16:439–500. doi: 10.1146/annurev.ge.16.120182.002255. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A repetitive DNA sequence that confers cell-cycle START (CDC28)-dependent transcription of the HO gene in yeast. Cell. 1985 Aug;42(1):225–235. doi: 10.1016/s0092-8674(85)80118-5. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. At least 1400 base pairs of 5'-flanking DNA is required for the correct expression of the HO gene in yeast. Cell. 1985 Aug;42(1):213–223. doi: 10.1016/s0092-8674(85)80117-3. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Molecular analysis of a cell lineage. Nature. 1983 Apr 21;302(5910):670–676. doi: 10.1038/302670a0. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. The determination of mother cell-specific mating type switching in yeast by a specific regulator of HO transcription. EMBO J. 1987 Jan;6(1):243–248. doi: 10.1002/j.1460-2075.1987.tb04745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D. W., Jensen R., Zoller M. J., Burke J., Errede B., Smith M., Herskowitz I. Structure of the Saccharomyces cerevisiae HO gene and analysis of its upstream regulatory region. Mol Cell Biol. 1986 Dec;6(12):4281–4294. doi: 10.1128/mcb.6.12.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater M. L. Effect of reversible inhibition of deoxyribonucleic acid synthesis on the yeast cell cycle. J Bacteriol. 1973 Jan;113(1):263–270. doi: 10.1128/jb.113.1.263-270.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J. N., Klar A. J., Hicks J. B., Abraham J. A., Ivy J. M., Nasmyth K. A., McGill C. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell. 1982 Nov;31(1):183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- Strathern J. N., Spatola E., McGill C., Hicks J. B. Structure and organization of transposable mating type cassettes in Saccharomyces yeasts. Proc Natl Acad Sci U S A. 1980 May;77(5):2839–2843. doi: 10.1073/pnas.77.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980 Jul;10(2):157–166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]
- Weiffenbach B., Haber J. E. Homothallic mating type switching generates lethal chromosome breaks in rad52 strains of Saccharomyces cerevisiae. Mol Cell Biol. 1981 Jun;1(6):522–534. doi: 10.1128/mcb.1.6.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson D. H., Fennell D. J. The use of fluorescent DNA-binding agent for detecting and separating yeast mitochondrial DNA. Methods Cell Biol. 1975;12:335–351. doi: 10.1016/s0091-679x(08)60963-2. [DOI] [PubMed] [Google Scholar]