Abstract
Background & Aims
HFE and transferrin receptor 2 (TFR2) are each necessary for the normal relationship between body iron status and liver hepcidin expression. In murine Hfe and Tfr2 knockout models of hereditary hemochromatosis (HH), signal transduction to hepcidin via the bone morphogenetic protein 6 (Bmp6)/Smad1,5,8 pathway is attenuated. We examined the effect of dietary iron on regulation of hepcidin expression via the Bmp6/Smad1,5,8 pathway using mice with targeted disruption of Tfr2, Hfe, or both genes.
Methods
Hepatic iron concentrations and mRNA expression of Bmp6 and hepcidin were compared with wild-type mice in each of the HH models on standard or iron-loading diets. Liver phospho-Smad (P-Smad)1,5,8 and Id1 mRNA levels were measured as markers of Bmp/Smad signaling.
Results
While Bmp6 expression was increased, liver hepcidin and Id1 expression were decreased in each of the HH models compared with wild-type mice. Each of the HH models also demonstrated attenuated P-Smad1,5,8 levels relative to liver iron status. Mice with combined Hfe/Tfr2 disruption were most affected. Dietary iron loading increased hepcidin and Id1 expression in each of the HH models. Compared with wild-type mice, HH mice demonstrated attenuated (Hfe knockout) or no increases in P-Smad1,5,8 levels in response to dietary iron loading.
Conclusions
These observations demonstrate that Tfr2 and Hfe are each required for normal signaling of iron status to hepcidin via Bmp6/Smad1,5,8 pathway. Mice with combined loss of Hfe and Tfr2 up-regulate hepcidin in response to dietary iron loading without increases in liver BMP6 mRNA or steady-state P-Smad1,5,8 levels.
Keywords: bone morphogenetic protein 6, Id1
Introduction
Hereditary hemochromatosis (HH) is a genetically heterogeneous hereditary disorder caused by elevated iron absorption from the diet, with consequent iron overload and tissue injury.1, 2 The most common form of HH is caused by mutation in the HFE gene. A much more rare form of HH (type 3) results from mutations in the gene for transferrin receptor 2 (TFR2).3 It is now widely accepted that impaired regulation of hepcidin expression plays a central role in the pathogenesis of HH. Hepcidin acts to downregulate the iron exporter ferroportin on the surface of duodenal enterocytes and macrophages, thereby inhibiting iron release from these cells.4 Human patients and mouse models of TFR2-related5, 6 and HFE-related7-10 HH each demonstrate inappropriately low expression of hepcidin. The mechanisms by which TFR2 and HFE influence hepcidin expression remain unclear. Several observations suggest a model in which TFR1 may participate as well. In this model, as the transferrin saturation increases, diferric transferrin displaces HFE from TFR1, thereby making HFE available to bind to TFR2.11-13 The complex of HFE and TFR2 is then postulated to influence hepcidin expression differently than TfR2 alone14. Mice with inactivating mutations in both Hfe and Tfr2 demonstrate a severe HH phenotype and very low hepcidin expression, raising the possibility that each may serve to regulate hepcidin expression even in the absence of the other15.
A bone morphogenetic protein 6 (BMP6)-dependent signaling pathway has been shown to play a key role in regulation of hepcidin expression.16, 17 BMPs bind to type I and type II serine threonine kinase receptors, which phosphorylate specific intracellular SMAD proteins (SMAD1,5,8). Phosphorylated SMAD1,5,8 (P-SMAD1,5,8) binds to the common mediator SMAD4, and the SMAD complex translocates to the nucleus to affect transcription of target genes such as ID1.18, 19 HAMP (encoding hepcidin) is transcriptionally upregulated by BMPs.20-23 Impaired hepatic Bmp signaling, through mutations in genes encoding either the ligand Bmp6,16, 17 the Bmp co-receptor hemojuvelin (Hjv)24, 25 or Smad4,26 leads to low hepcidin levels and iron overload in mice. Conversely, dietary iron loading increases hepatic Bmp6 mRNA expression in mice concordantly with Hamp1 and Id1 mRNAs.27 Collectively, these data demonstrate that BMP-SMAD signaling is an important regulatory pathway for hepcidin expression and thus iron metabolism. In Hfe knockout mice 28, 29, and in patients with HFE-associated HH 30, 31, the induction of Bmp6 mRNA by iron is intact, but Smad1,5,8 signaling to hepcidin is impaired. Impaired Bmp6 signaling to hepcidin has also been reported in murine models of Tfr2-associated HH 15, 32
The goal of this study was to investigate the Bmp6-Smad-hepcidin signaling pathway in the Tfr2 and Hfe mutant mouse models of HH, under standard iron diets and with dietary iron loading. We observed the expected impaired signaling to hepcidin via the Bmp6/Smad pathway in Tfr2 and Hfe HH mouse models. Signaling to hepcidin via the Bmp/Smad pathway was more impaired in Hfe/Tfr2 mice than in mice with loss of either gene product individually. Dietary iron loading increased hepcidin expression in each of the murine HH model systems. In Tfr2 mice and Hfe/Tfr2 mice, hepcidin up-regulation occurred without an increase in liver P-Smad1,5,8 levels. Taken together, these results indicate that Hfe and Tfr2 are each necessary for normal signaling from Bmp6 to hepcidin, that each can influence hepcidin expression independent of the other, and that mechanisms regulating hepcidin expression in response to dietary iron exist which do not require Hfe or Tfr2.
Methods
Animal care
Hfe knockout mice33 and Tfr2Y245X mice34 were bred to uniformity on an FVB background for greater than 7 generations. The Tfr2Y245X mice have no detectable Tfr2 or truncated form of the protein in hepatocellular membrane preparations and are a functional knockout34. These two mouse lines were crossed with each other and bred to homozygosity for each mutant allele. Colonies were maintained as homozygotes for each allele individually (hereafter referred to as Hfe mice or Tfr2 mice), and as compound mutant homozygotes (Hfe/Tfr2 mice). Mice were fed standard chow (Purina 5001, containing 270 ppm iron) ad libitum after weaning at 21 days. Dietary iron loading was achieved by weaning mice onto a diet containing an additional 25,000 ppm of carbonyl iron. At 5 weeks of age, the mice were sacrificed by exposure to hypercarbia followed by exsanguination, and tissues were harvested. To minimize potential variability related to gender, samples from only male mice were used in subsequent studies. Sample sizes unless otherwise indicated in figure legends were: 13 wild-type on a standard diet, 4 wild type on high iron, 3 Hfe knockout, 3 Hfe knockout on high iron, 5 Tfr2 knockout, 3 Tfr2 knockout on high iron, 5 Hfe/Tfr2, and 3 Hfe/Tfr2 on high iron. The murine studies were performed under protocols approved by the Institutional Animal Care and Use Committee of Saint Louis University, and in accord with the NIH Guide for the Care and Use of Laboratory Animals.
Liver iron content
Liver specimens were homogenized, and a portion was desiccated overnight at room temperature and analyzed for non-heme iron content by the method of Torrance and Bothwell35 Data were expressed as μg iron per g dry weight of liver.
Quantitative real-time RT-PCR
Total RNA was isolated from mouse liver tissue using the RNeasy Mini Kit (Qiagen), with DNAse digestion using the RNase-Free DNase Set (Qiagen). Quantitation of murine Bmp6, Hamp1, Id1 and Rpl19 mRNA transcripts was performed using two-step quantitative real-time RT-PCR as previously described.28 Samples were analyzed in triplicate and expression levels were normalized to the housekeeping gene Rpl19. Additional quantitation of Bmp6, Hamp1, and β-actin mRNA transcripts were performed using one-step quantitative real-time RT-PCR (TaqMan, Applied Biosystems ABI7700) and the following probes and primers: Hamp1 forward: CCTATCTCCATCAACAGGTG, reverse: AACAGATACCACACTGGGAA, and probe: 6FAM-CCCTGCTTTCTTCCCCGTGCAAAGT-TAMRA β-actin forward: CCGTGAAAAGATGACCCAGATCATG, reverse: TCTTCATGAGGTAGTCCGTCAGGTC and probe: 6FAM-TACGAGGGCTATGCTCTCCCTCACGCT-TAMRA. Hamp1 expression relative to β-actin expression was compared across groups using both the delta Ct method, and the method described by Pfaffl et al.36 using REST software (Qiagen). Similar results were obtained using each analytical and real-time PCR method.
Western blot analyses
Liver specimens were homogenized in lysis buffer (1× Tris-buffered saline, 0.1% sodium dodecyl sulfate, 10 μL/mL Triton X-100, 1 g/dL sodium deoxycholate, 2 μL/mL EDTA) containing protease inhibitors (Complete Mini; Roche Diagnostic) and phosphatase inhibitors (Halt Phosphatase Inhibitor Cocktail, Thermo Scientific). Western blots of liver lysates for P-Smad1,5,8 protein (relative to total Smad1 protein and to β-actin) and chemiluminescence quantitation were performed as previously described28. P-Stat3 was quantified by Western blot using the PhosphoPlus Stat3 antibody kit (Cell Signaling) per the manufacturer's instructions, and normalized to β-actin.
Statistical analyses
Statistical analyses across multiple groups were performed by ANOVA with Dunnett&apos:s test comparing each experimental group to each control (wild-type) group, or (in separate experiments) by ANOVA with Neuman-Keuls test, when comparing across each group. For iron loading studies, comparison was made within each genotype between mice on a high-iron diet or standard diet by two-tailed Student's t-test. P<0.05 was considered statistically significant.
Results
Elevated Bmp6 mRNA expression is associated with hepatic iron loading in mice with loss of Hfe and/or Tfr2
Functional loss of Hfe or Tfr2 is known to result in inappropriately low hepatic expression of hepcidin and consequent iron overload. Several lines of evidence suggest that upregulation of Bmp6 contributes to iron-dependent regulation of hepcidin. We measured the hepatic expression of Bmp6 mRNA in Hfe, Tfr2, and Hfe/Tfr2 mice to assess if the decreased hepcidin expression could be attributed to decreased hepatic Bmp6 expression. To provide a comparison group for the degree of hepatic iron loading observed in the murine HH models, wild-type mice were placed on a high-iron (25,000 ppm) diet. As seen in Figure 1, Hfe, Tfr2, and Hfe/Tfr2 mice on a standard iron diet demonstrated the expected elevated liver iron concentrations. Hepatic iron concentrations were highest in the Hfe/Tfr2 mice, although only statistically increased when compared with the Hfe mice and wild-type mice. The wild-type mice placed on a high-iron diet had hepatic iron concentrations comparable to the Hfe/Tfr2 mice, and not statistically different from the other mouse HH models.
Figure 1. Hepatic iron concentrations.
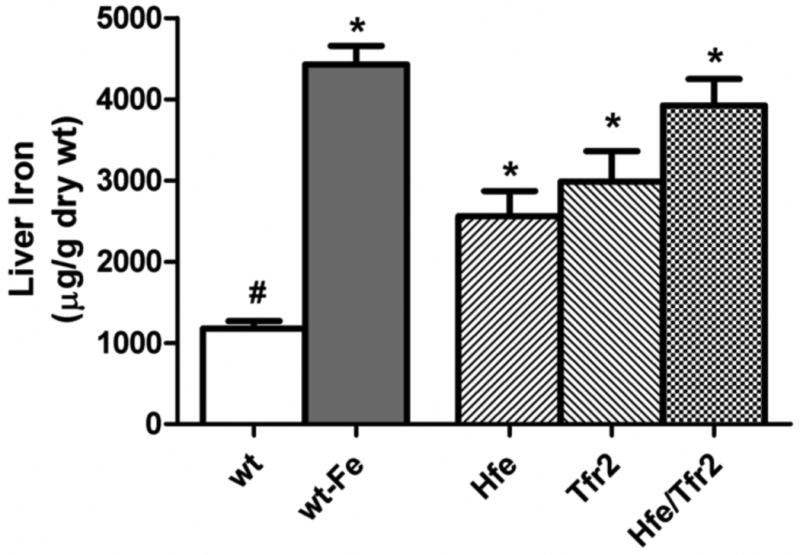
Hepatic non-heme iron concentrations were measured in wild-type (wt) mice and murine models of hemochromatosis (Hfe, Tfr2, and Hfe/Tfr2) on a standard iron diet (270 ppm iron). As an iron-loaded control, wt mice were fed a diet containing 25,000 ppm iron (wt-Fe). ANOVA P<0.0001. *P<0.05 compared to wt. #P<0.05 compared to wt-Fe.
We compared hepatic Bmp6 expression in each of the above described groups. As expected, dietary iron loading of the wild-type mice led to a significant increase in hepatic Bmp6 mRNA (Figure 2). No increase in Bmp6 expression in the small intestine was observed with dietary iron loading (Supplemental Figure 1). Hepatic expression of Bmp6 mRNA in each of the murine HH models was similar to that observed in the iron-loaded wild-type mice, and increased over wild-type mice on the standard diet. While the difference in Bmp6 expression between Hfe knockout and wild type mice on a standard iron diet was not statistically significant in this multigroup analysis, separate analysis of mice from these two groups demonstrated a statistically significant increase in Bmp6 mRNA in the Hfe knockout mice (P<0.05, Student's t-test). This observation is in agreement with observations by other groups29,32. These data indicate that hepatic iron loading is associated with increased hepatic Bmp6 mRNA expression despite the absence of Hfe and/or Tfr2.
Figure 2. Liver Bmp6 mRNA expression.
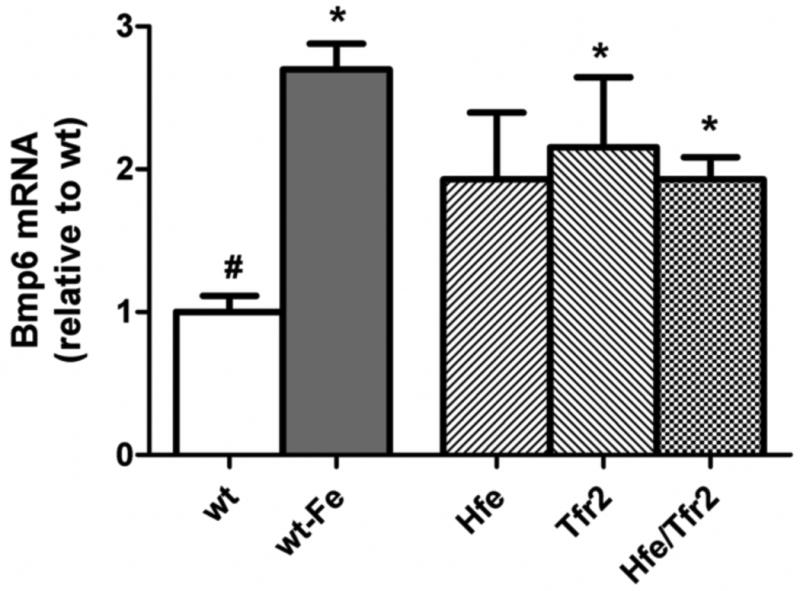
Bmp6 mRNA was quantified by real-time RT-PCR, normalized to Rpl19 mRNA, and expressed relative to the mean value obtained in wild-type (wt) mice on a standard diet. As an iron-loaded control, wt mice were fed a diet containing 25,000 ppm iron (wt-Fe). ANOVA P<0.001. *P<0.05 compared to wt. #P<0.05 compared to wt-Fe.
Impaired relationship between hepatic Bmp6 and hepcidin expression in mice with loss of Hfe and/or Tfr2
The observation that Bmp6 is upregulated in the HH mouse models led us to examine in these mice the relationship between Bmp6 expression and hepcidin expression. Hamp1 mRNA was measured by real-time RT-PCR (Figure 3A) and expressed relative to that in wild-type mice. We observed an absolute decrease in liver Hamp1 mRNA with loss of Hfe or Tfr2 individually as expected. Mice with combined loss of each protein had the lowest liver hepcidin mRNA content; however, in this multigroup analysis, the difference between the Hfe/Tfr2 mice and the single Hfe or Tfr2 mutant mice did not achieve statistical significance. We directly compared the Hfe/Tfr2 mice with mice carrying each mutation individually in a separate set of experiments. We found that indeed the Tfr2/Hfe mice had statistically lower hepcidin expression than did mice with only one of these mutations (Supplemental Figure 2). These observations demonstrate that Tfr2 and Hfe can influence hepcidin expression independently of each other.
Figure 3. Liver hepcidin mRNA expression.
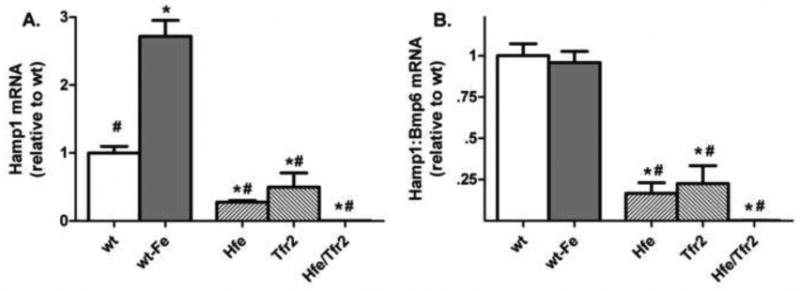
(A) Liver Hamp1 mRNA was quantified by realtime RT-PCR, normalized to Rpl19 mRNA, and expressed relative to the mean value obtained in wild-type (wt) mice on a standard diet. (B) The ratio between Hamp1 and Bmp6 mRNA expression was determined individually for each sample, and expressed relative to the mean ratio obtained from wt mice on a standard diet. Iron-loaded wt mice (wt-Fe) were fed a diet containing 25,000 ppm iron. ANOVA P<0.0001. *P<0.05 compared to wt. #P<0.05 compared to wt-Fe.
We next expressed Hamp1 mRNA relative to Bmp6 mRNA content as a ratio for each of the groups (Figure 3B). The relationship between Bmp6 mRNA and Hamp1 mRNA was unchanged in the wild-type mice placed on an iron-loading diet. However, each of the HH mouse models demonstrated markedly decreased Hamp1 mRNA expression relative to Bmp6 mRNA expression. This effect was most pronounced in the Hfe/Tfr2 mice. These data demonstrate that loss of Hfe and/or Tfr2 negatively affects the relationship between hepatic Bmp6 mRNA and hepcidin mRNA expression.
In order to further characterize the requirement for both Hfe and Tfr2 in the signaling between Bmp6 mRNA and hepcidin, independent analysis of 3 additional Tfr2, Hfe, and Hfe/Tfr2 mice was performed. Comparison of Hamp1:Bmp6 mRNA ratios demonstrated a significant decrease (P<0.05) in the Hfe/Tfr2 mice compared with the Hfe mice alone. While the Hamp1:Bmp6 mRNA ratio was also decreased in the Hfe/Tfr2 mice compared with the Tfr2 mutant mice, this difference was not statistically significance. These observations indicate that Hfe and Tfr2 each contribute to the normal relationship between Bmp6 mRNA and hepcidin mRNA expression.
Attenuated hepatic P-Smad1,5,8 levels in mice with loss of Hfe and/or Tfr2
Bmp6 induces hepcidin expression via phosphorylation of Smad1,5,8. We compared steady-state levels of P-Smad1,5,8 in wild-type mice on standard and iron-loading diets and observed the anticipated increase in response to iron loading (Figure 4). We then compared levels of P-Smad1,5,8 in each of the murine HH models with the iron-loaded wild-type mice. While P-Smad1,5,8 levels were not significantly lower in the HH mouse models compared with wild-type mice, they were significantly less that those observed in the iron-loaded wild-type mice. This was the case despite similar increases in Bmp6 mRNA levels in the HH mouse models and iron-loaded wild-type mice. These data are consistent with an impairment in the relationship of hepatic iron status and Bmp6 mRNA levels to P-Smad1,5,8 levels in the murine HH models.
Figure 4. Liver P-Smad1,5,8 levels.
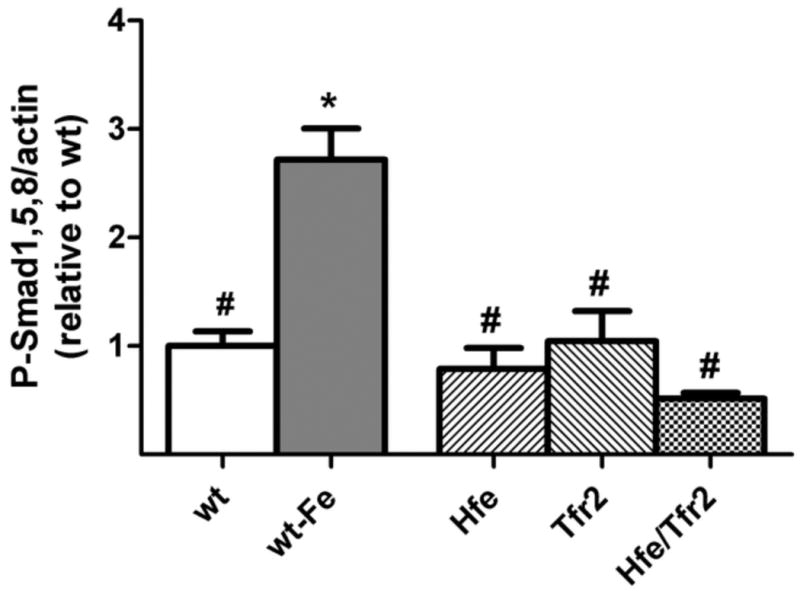
P-Smad1,5,8 levels obtained by Western blot analysis were quantified, normalized to the signal obtained for β-actin, and expressed relative to the mean value obtained from wt mice on a standard diet. As an iron-loaded control, wt mice were fed a diet containing 25,000 ppm iron (wt-Fe). Sample sizes: wt=4, wt-Fe=4, Hfe=3, Tfr2=3, Hfe/Tfr2=5. ANOVA P<0.0001. *P<0.05 compared to wt. #P<0.05 compared to wt-Fe.
Decreased expression of Id1 relative to Bmp6 with loss of Hfe and/or Tfr2
We next examined the expression of Id1, a Bmp-Smad signaling pathway target transcript that has been shown to be modified by dietary iron loading. Dietary iron loading in wild-type mice was associated with the expected increase in Id1 mRNA expression (Figure 5A). However, in each of the murine HH models, Id1 mRNA levels were significantly lower than those observed in the iron-loaded wild-type mice. The relationship between Bmp6 mRNA and Id1 mRNA levels was examined by comparing their ratios in each of the groups (Figure 5B). As seen with hepcidin, the expression of Id1 mRNA relative to Bmp6 mRNA was markedly decreased in each of the HH mouse models.
Figure 5. Liver Id1 mRNA expression.
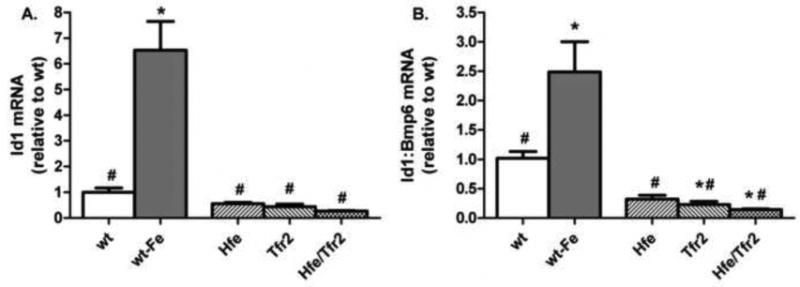
(A) Liver Id1 mRNA was quantified by real-time RT-PCR, normalized to Rpl19 mRNA, and expressed relative to the mean value obtained in wild-type (wt) mice on a standard diet. (B) The ratio between Id1 and Bmp6 mRNA expression was determined individually for each sample, and expressed relative to the mean ratio obtained from wt mice on a standard diet. Iron-loaded wt mice (wt-Fe) were fed a diet containing 25,000 ppm iron. ANOVA P<0.0001. *P<0.05 compared to wt. #P<0.05 compared to wt-Fe.
Upregulation of hepcidin and Id1 in mice with dietary iron loading in mice despite loss of Hfe and/or Tfr2
Previous studies have shown that dietary iron is capable of upregulating hepcidin expression in mice despite loss of Hfe. We performed studies to determine if mice in which Hfe and/or Tfr2 has been disrupted demonstrate a response to dietary iron loading with an increase in Bmp6, hepcidin, or Id1 mRNA expression. We found that there was a modest but not-statistically significant increase in Bmp6 mRNA expression in Hfe and Tfr2 mice with dietary iron loading. Hfe/Tfr2 mice demonstrated no increase in Bmp6 mRNA (Figure 6B). Nonetheless, each of the mutant mouse models, including Hfe/Tfr2 mice, demonstrated an increase in both hepcidin (Figure 6C) and in Id1 (figure 6D) mRNA expression in response to dietary iron loading. While the increase in hepcidin expression in the Tfr2 mice with dietary iron in this sampling was not statistically significant, we have observed statistically significant up-regulation of hepcidin in response to dietary iron in other samplings. Interestingly, the increase in hepcidin and Id1 mRNA expression in the iron-loaded Tfr2 and Hfe/Tfr2 mice was not associated with an increase in P-Smad1,5,8 levels. Indeed, when normalized to β-actin, the P-Smad1,5,8 levels in the iron-loaded Hfe/Tfr2 mice were statistically lower than observed in these mice on a standard iron diet (Figure 7). Normalization of these data to total Smad1 rather than β-actin resulted in overall similar findings; however, the decrease in P-Smad1,5,8 in the iron-loaded Hfe/Tfr2 mice was not statistically significant. These observations suggest that in Hfe/Tfr2 mice hepcidin upregulation with dietary iron loading can occur independently of changes in steady-state P-Smad1,5,8 levels. The Jak/STAT pathway mediates an increase in hepcidin expression in response to IL6-mediated inflammatory stimuli. To determine if this pathway was responsible for the increase in hepcidin mRNA in the iron-loaded mutant mice, we measured liver P-Stat3 levels in selected groups by Western blot analyses, with normalization to β-actin. There was no change in liver P-Stat3 levels in the iron-loaded wild-type mice compared with those on a standard iron diet (0.93 fold, p=0.71). Moreover, there was no change in P-Stat3 expression in the Hfe/Tfr2 mice on the high-iron diet compared with a standard diet (0.89 fold, p=0.78).
Figure 6. Effect of dietary iron loading in murine HH models.
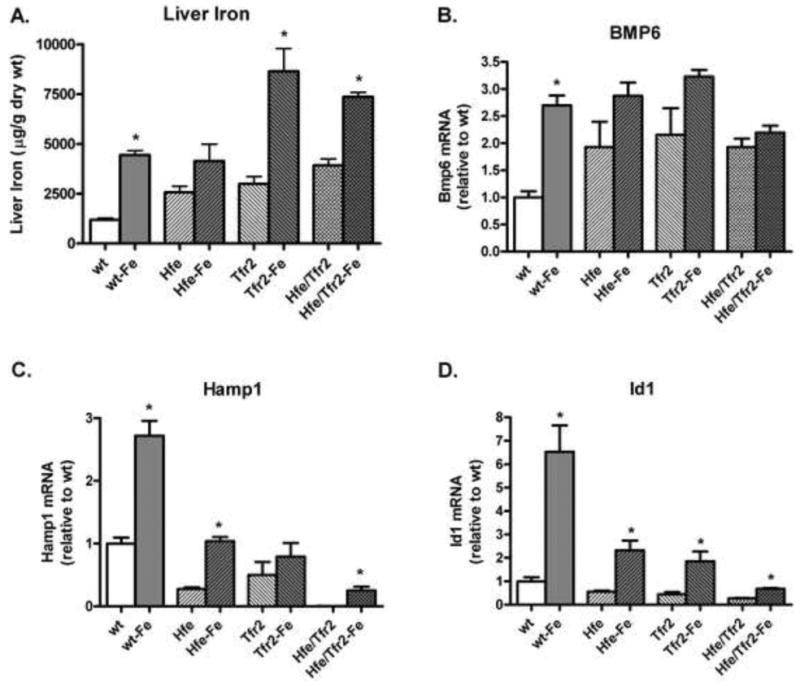
(A) Liver non-heme iron levels were measured after 2 weeks on a standard iron or high-iron (Fe) diet. (B-D) mRNA levels of the identified gene were quantified by real-time RT-PCR, normalized to Rpl19 mRNA, and expressed relative to the mean value obtained in wild-type (wt) mice on a standard diet. *P<0.05 comparing mean values from mice on a standard diet to mice of like genotype on a high-iron diet.
Figure 7. Effect of dietary iron loading on hepatic P-Smad1,5,8 levels in murine HH models.
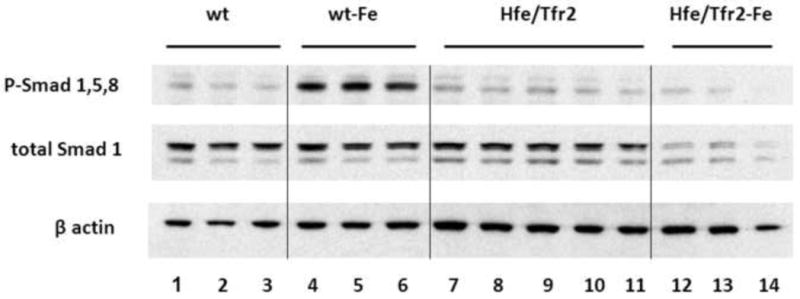
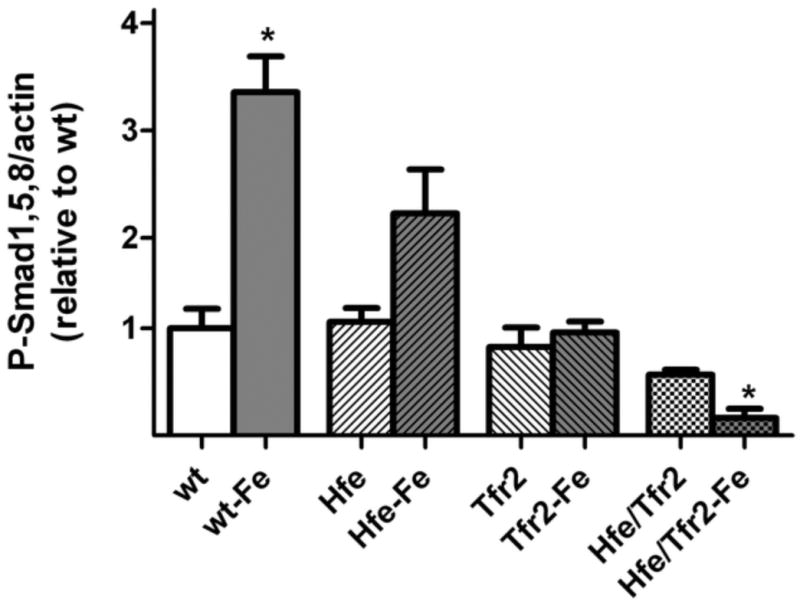
(A) Representative Western blot of P-Smad1,5,8 in total liver homogenates from 3 wild-type (wt) mice on a standard diet (lanes 1-3), 3 wt mice on a high-iron diet (lanes 4-6), 5 Hfe/Tfr2 mice (lanes 7-11), and 3 Hfe/Tfr2 mice on a high iron diet (lanes 12-14). Blots were also reacted with antibodies to detect total Smad1 protein or β-actin. (B) P-Smad1,5,8 levels were quantified by Western blot analyses and normalized to expression of β-actin, and expressed relative to the mean value obtained from the wild-type (wt) mice on a standard iron diet. Sample sizes: wt=3, Hfe=3, Hfe-Fe=3, Tfr2=3 Tfr2-Fe=3, Hfe/Tfr2=5, Hfe/Tfr2-Fe=3. ANOVA P<0.0001. *P<0.05 comparing mean values from mice on a standard diet to mice of like genotype on a high-iron diet.
Discussion
The normal relationship between body iron status and liver hepcidin expression requires the action of several identified genes, including HFE, TFR2, HJV, BMP6, and SMAD4.26, 37 Mutations in any one of these genes generates the classic HH phenotype, i.e., excess dietary iron absorption, elevated serum iron concentrations, hepatocellular iron loading, and macrophage iron sparing.1, 2 TFR2 is the only one of these gene products known to directly interact with an iron-containing protein, i.e., its ligand ferri-transferrin.38 As such, TFR2 has been proposed to be an essential component of a hepatocellular iron sensor. Recent observations support a model in which elevated body iron stores normally upregulate BMP6 expression, leading to increased hepatocellular hepcidin expression via an intracellular pathway that involves that SMAD signaling.37 While some evidence suggested the possibility that the intestine is the relevant source for iron-regulated Bmp6 expression39, our studies (Figure S1) and others40 implicate a hepatic rather than intestinal source.
In agreement with recent reports, our studies support a role for Tfr2 and for Hfe in the regulation of hepcidin downstream of the regulation of Bmp6 mRNA. It has been proposed that HFE and TFR2 form a complex with the BMP receptor and HJV on the hepatocellular surface41, possibly affecting the interaction of BMP6 with its receptor. As another possibility, HFE or TFR2 might influence the function of genes responsible for the relative distribution of hepatocellular HJV. While membrane-associated HJV serves as a co-receptor that augments BMP signaling, soluble HJV (released from the cell surface) serves instead to inhibit BMP signaling.42 The release of HJV from the hepatocellular surface appears to be mediated at least in part by the serine protease matriptase2.43, 44 Whether hepatic matriptase2 activity is influenced by loss of HFE or TFR2 is unknown.
Several lines of evidence suggest that HFE and TFR2 function as a complex,11, 14 and that the ability of TFR2 to regulate hepcidin via transferrin requires HFE13. Most recently, Gao et al. used an adeno-associated viral delivery system to express Hfe or Tfr2 in vivo in the livers of mice in which the complimentary gene was disrupted.45 They found that Tfr2 overexpression had no effect on iron parameters in Hfe mice or wild-type mice. They moreover found that Hfe overexpression had no effect in Tfr2 mice, but did in wild-type mice. Such observations suggest that Tfr2 is functionally dependent on Hfe in the regulation of hepcidin. The studies we present here examine instead the effects of expression of Tfr2 and Hfe at endogenous levels when compared with mice in which neither is expressed. The Hfe/Tfr2 mice had lower hepcidin mRNA expression than did mice with loss of either Hfe or Tfr2, consistent with the previously reported severe phenotypic abnormalities in Hfe/Tfr2 mice15 and in patients with mutations in both HFE and TFR2 genes.46 The observation that Hfe/Tfr2 mice have lower hepatic hepcidin mRNA expression than mice with either gene individually knocked out is in agreement with our studies on serum hepcidin levels 47. We thus find that Hfe and Tfr2 can each influence hepcidin expression in the absence of the other molecule. Our studies do not examine whether the functional properties of Tfr2 or Hfe might be enhanced by the presence of other molecule, or whether they function as a complex when both are present. Indeed, observations by Gao et al. in cell culture13 and in vivo45 suggest that the level of expression of Hfe might be a limiting factor in the regulation of hepcidin by Tfr2. Nonetheless, our observations indicate that Tfr2 retains functional activity in the regulation of hepcidin in the absence of Hfe. Moreover, Hfe influences liver hepcidin expression without a requirement for Tfr2.
We observe that dietary iron loading increases hepcidin and Id1 expression in each of the murine models tested, although the level of liver hepcidin expression in the Hfe/Tfr2 mice on an iron loading diet remains a small fraction of that observed in wild-type mice. The increase in hepcidin and Id1 expression in the mice without Hfe or Tfr2 occurred without an increase in Bmp6 expression or P-Smad1,5,8 levels. The mechanism(s) by which hepcidin is increased in this mouse model has yet to be elucidated. However, multiple hepcidin signaling pathways have been identified, not each of which would be expected to require an change in P-Smad1,5,8 levels. These additional regulatory pathways include TWSG148, TGF-β26, SMAD749, members of the MAPK pathway50,15, the IL-6 receptor-Jak-STAT pathway 51, 52 and IL-153. The transcription factor CREB-H mediates an increase in hepcidin expression in response to endoplasmic reticulum (ER) stress.54 Signaling to hepcidin via one or more of these pathways might have been influenced by the very high iron concentrations in the iron-loaded Hfe/Tfr2 mice., Substantial cross-communication may exist between these hepcidin regulatory signaling pathways and the iron-regulatory BMP/Smad1,5,8 signaling pathway.
The effects of the HH gene mutations on expression of hepcidin were similar to those observed for Id1. However, they were not identical. For example, the Id1:Bmp6 mRNA ratio of the iron-loaded wild-type mice was higher than the ratio observed in the standard-diet wild-type mice. On the contrary, the hepcidin:Bmp6 mRNA ratios of iron-loaded wild-type mice and standard-diet wild-type mice were not different from each other. These observations suggest that changes in BMP6 expression have greater influence on Id1 transcription compared with hepcidin, and/or that iron loading increases Id1 transcription by mechanisms in addition to the BMP/SMAD pathway. Supporting these concepts, loss of Hfe and/or Tfr2 had a greater effect on hepcidin expression than on Id1 expression.
In summary, our findings support a model in which Tfr2 and Hfe each modulate the signal between iron status and hepcidin expression via the Bmp/Smad pathway. The effects of these molecules on Bmp signaling occur downstream of the regulation of Bmp6 mRNA by iron, and influence the relative abundance of hepatocellular P-Smad1,5,8. Moreover, mice with combined loss of Hfe and Tfr2 demonstrate no increase in hepatic steady-state P-Smad1,5,8 levels with dietary iron loading, yet demonstrate an increase in hepcidin expression. The mechanism by which iron regulates liver Bmp6 mRNA expression remains to be elucidated, but does not require Hfe or Tfr2.
Supplementary Material
Acknowledgments
We thank Rosemary O'Neill for excellent technical assistance.
Grant support: This work was supported by National Institutes of Health grants (R01 DK063016, R.E.F.; K08 DK075846, J.L.B.; R01 DK069533 and R01 DK071837, H.Y.L.) and a Claflin Distinguished Scholar Award from the Massachusetts General Hospital (J.L.B).
Abbreviations
- Bmp6
bone morphogenetic protein 6
- HH
hereditary hemochromatosis
- Hjv
hemojuvelin
- Tfr2
transferrin receptor 2
- wt
wild-type
Footnotes
Disclosures: The authors have no conflicts to disclose.
Author contributions: E.C. (study concept and design; acquisition of data; analysis and interpretation of data; drafting of the manuscript); M.R. (acquisition of data; analysis and interpretation of data); D.M. (acquisition of data; analysis and interpretation of data; critical revision of the manuscript for important intellectual content); A.O. (acquisition of data; analysis and interpretation of data); H.Y.L. (study concept and design; critical revision of the manuscript for important intellectual content; funding); Q.F. (acquisition of data; analysis and interpretation of data); M.C.M. (acquisition of data; analysis and interpretation of data); R.S.B. (analysis and interpretation of data; critical revision of the manuscript for important intellectual content); J.L.B. (study concept and design; analysis and interpretation of data; drafting of the manuscript; funding); R.E.F. (study concept and design; analysis and interpretation of data; drafting of the manuscript; statistical analysis; funding)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pietrangelo A. Hereditary hemochromatosis--a new look at an old disease. N Engl J Med. 2004;350:2383–2397. doi: 10.1056/NEJMra031573. [DOI] [PubMed] [Google Scholar]
- 2.Fleming RE, Britton RS, Waheed A, Sly WS, Bacon BR. Pathophysiology of hereditary hemochromatosis. Semin Liver Dis. 2005;25:411–419. doi: 10.1055/s-2005-923313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camaschella C, Roetto A, Cali A, De Gobbi M, Garozzo G, Carella M, Majorano N, Totaro A, Gasparini P. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet. 2000;25:14–15. doi: 10.1038/75534. [DOI] [PubMed] [Google Scholar]
- 4.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 5.Kawabata H, Fleming RE, Gui D, Moon SY, Saitoh T, O&apos:Kelly J, Umehara Y, Wano Y, Said JW, Koeffler HP. Expression of hepcidin is down-regulated in Tfr2 mutant mice manifesting a phenotype of hereditary hemochromatosis. Blood. 2005;105:376–381. doi: 10.1182/blood-2004-04-1416. [DOI] [PubMed] [Google Scholar]
- 6.Nemeth E, Roetto A, Garozzo G, Ganz T, Camaschella C. Hepcidin is decreased in TFR2 hemochromatosis. Blood. 2005;105:1803–1806. doi: 10.1182/blood-2004-08-3042. [DOI] [PubMed] [Google Scholar]
- 7.Ahmad KA, Ahmann JR, Migas MC, Waheed A, Britton RS, Bacon BR, Sly WS, Fleming RE. Decreased liver hepcidin expression in the Hfe knockout mouse. Blood Cells Mol Dis. 2002;29:361–366. doi: 10.1006/bcmd.2002.0575. [DOI] [PubMed] [Google Scholar]
- 8.Nicolas G, Viatte L, Lou DQ, Bennoun M, Beaumont C, Kahn A, Andrews NC, Vaulont S. Constitutive hepcidin expression prevents iron overload in a mouse model of hemochromatosis. Nat Genet. 2003;34:97–101. doi: 10.1038/ng1150. [DOI] [PubMed] [Google Scholar]
- 9.Muckenthaler M, Roy CN, Custodio AO, Minana B, deGraaf J, Montross LK, Andrews NC, Hentze MW. Regulatory defects in liver and intestine implicate abnormal hepcidin and Cybrd1 expression in mouse hemochromatosis. Nat Genet. 2003;34:102–107. doi: 10.1038/ng1152. [DOI] [PubMed] [Google Scholar]
- 10.Bridle KR, Frazer DM, Wilkins SJ, Dixon JL, Purdie DM, Crawford DH, Subramaniam VN, Powell LW, Anderson GJ, Ramm GA. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis. Lancet. 2003;361:669–673. doi: 10.1016/S0140-6736(03)12602-5. [DOI] [PubMed] [Google Scholar]
- 11.Goswami T, Andrews NC. Hereditary hemochromatosis protein, HFE, interaction with transferrin receptor 2 suggests a molecular mechanism for mammalian iron sensing. J Biol Chem. 2006;281:28494–28498. doi: 10.1074/jbc.C600197200. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt PJ, Toran PT, Giannetti AM, Bjorkman PJ, Andrews NC. The transferrin receptor modulates Hfe-dependent regulation of hepcidin expression. Cell Metab. 2008;7:205–214. doi: 10.1016/j.cmet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao JW, Chen JX, Kramer M, Tsukamoto H, Zhang AS, Enns CA. Interaction of the Hereditary Hemochromatosis Protein HFE with Transferrin Receptor 2 Is Required for Transferrin-Induced Hepcidin Expression. Cell Metabolism. 2009;9:217–227. doi: 10.1016/j.cmet.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waheed A, Britton RS, Grubb JH, Sly WS, Fleming RE. HFE association with transferrin receptor 2 increases cellular uptake of transferrin-bound iron. Arch Biochem Biophys. 2008;474:193–197. doi: 10.1016/j.abb.2008.02.041. [DOI] [PubMed] [Google Scholar]
- 15.Wallace DF, Summerville L, Crampton EM, Frazer DM, Anderson GJ, Subramaniam VN. Combined Deletion of Hfe and Transferrin Receptor 2 in Mice Leads to Marked Dysregulation of Hepcidin and Iron Overload. Hepatology. 2009;50:1992–2000. doi: 10.1002/hep.23198. [DOI] [PubMed] [Google Scholar]
- 16.Andriopoulos B, Corradini E, Xia Y, Faasse SA, Chen SZ, Grgurevic L, Knutson MD, Pietrangelo A, Vukicevic S, Lin HY, Babitt JL. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nature Genetics. 2009;41:482–487. doi: 10.1038/ng.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meynard D, Kautz L, Darnaud V, Canonne-Hergaux F, Coppin H, Roth MP. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nature Genetics. 2009;41:478–481. doi: 10.1038/ng.320. [DOI] [PubMed] [Google Scholar]
- 18.Miyazono K, Miyazawa K. Id: a target of BMP signaling. Sci STKE. 2002;2002:e40. doi: 10.1126/stke.2002.151.pe40. [DOI] [PubMed] [Google Scholar]
- 19.Korchynskyi O, ten DP. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 20.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, Andrews NC, Lin HY. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 21.Babitt JL, Huang FW, Xia Y, Sidis Y, Andrews NC, Lin HY. Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J Clin Invest. 2007;117:1933–1939. doi: 10.1172/JCI31342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Truksa J, Peng H, Lee P, Beutler E. Bone morphogenetic proteins 2, 4, and 9 stimulate murine hepcidin 1 expression independently of Hfe, transferrin receptor 2 (Tfr2), and IL-6. Proc Natl Acad Sci USA. 2006;103:10289–10293. doi: 10.1073/pnas.0603124103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casanovas G, Mleczko-Sanecka K, Altamura S, Hentze MW, Muckenthaler MU. Bone morphogenetic protein (BMP)-responsive elements located in the proximal and distal hepcidin promoter are critical for its response to HJV/BMP/SMAD. Journal of Molecular Medicine-Jmm. 2009;87:471–480. doi: 10.1007/s00109-009-0447-2. [DOI] [PubMed] [Google Scholar]
- 24.Huang FW, Pinkus JL, Pinkus GS, Fleming MD, Andrews NC. A mouse model of juvenile hemochromatosis. J Clin Invest. 2005;115:2187–2191. doi: 10.1172/JCI25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niederkofler V, Salie R, Arber S. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload. J Clin Invest. 2005;115:2180–2186. doi: 10.1172/JCI25683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang RH, Li C, Xu X, Zheng Y, Xiao C, Zerfas P, Cooperman S, Eckhaus M, Rouault T, Mishra L, Deng CX. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 27.Kautz L, Meynard D, Monnier A, Darnaud V, Bouvet R, Wang RH, Deng C, Vaulont S, Mosser J, Coppin H, Roth MP. Iron regulates phosphorylation of Smad1/5/8 and gene expression of Bmp6, Smad7, Id1, and Atoh8 in the mouse liver. Blood. 2008;112:1503–1509. doi: 10.1182/blood-2008-03-143354. [DOI] [PubMed] [Google Scholar]
- 28.Corradini E, Garuti C, Montosi G, Ventura P, Andriopoulos B, Lin HY, Pietrangelo A, Babitt JL. Bone Morphogenetic Protein Signaling Is Impaired in an Hfe Knockout Mouse Model of Hemochromatosis. Gastroenterology. 2009;137:1489–1497. doi: 10.1053/j.gastro.2009.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kautz L, Meynard D, Besson-Fournier C, Darnaud V, Al Saati T, Coppin H, Roth MP. BMP/Smad signaling is not enhanced in Hfe-deficient mice despite increased Bmp6 expression. Blood. 2009;114:2515–2520. doi: 10.1182/blood-2009-02-206771. [DOI] [PubMed] [Google Scholar]
- 30.Ryan JD, Ryan E, Fabre A, Lawless MW, Crowe J. Defective Bone Morphogenic Protein Signaling Underlies Hepcidin Deficiency in HFE Hereditary Hemochromatosis. Hepatology. 2010;52:1266–1273. doi: 10.1002/hep.23814. [DOI] [PubMed] [Google Scholar]
- 31.Bolondi G, Garuti C, Corradini E, Zoller H, Vogel W, Finkenstedt A, Babitt JL, Lin HY, Pietrangelo A. Altered hepatic BMP signaling pathway in human HFE hemochromatosis. Blood Cells Molecules and Diseases. 2010;45:308–312. doi: 10.1016/j.bcmd.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roetto A, Di Cunto F, Pellegrino RM, Hirsch E, Azzolino O, Bondi A, Defilippi I, Carturan S, Miniscalco B, Riondato F, Cilloni D, Silengo L, Altruda F, Camaschella C, Saglio G. Comparison of 3 Tfr2-deficient murine models suggests distinct functions for Tfr2-alpha and Tfr2-beta isoforms in different tissues. Blood. 2010;115:3382–3389. doi: 10.1182/blood-2009-09-240960. [DOI] [PubMed] [Google Scholar]
- 33.Zhou XY, Tomatsu S, Fleming RE, Parkkila S, Waheed A, Jiang J, Fei Y, Brunt EM, Ruddy DA, Prass CE, Schatzman RC, O'Neill R, Britton RS, Bacon BR, Sly WS. HFE gene knockout produces mouse model of hereditary hemochromatosis. Proc Natl Acad Sci USA. 1998;95:2492–2497. doi: 10.1073/pnas.95.5.2492. see comments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fleming RE, Ahmann JR, Migas MC, Waheed A, Koeffler HP, Kawabata H, Britton RS, Bacon BR, Sly WS. Targeted mutagenesis of the murine transferrin receptor-2 gene produces hemochromatosis. Proc Natl Acad Sci USA. 2002;99:10653–10658. doi: 10.1073/pnas.162360699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torrance JD, Bothwell TH. Tissue Iron. Methods Hematol. 1:90–115. [Google Scholar]
- 36.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST (c)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Research. 2002;30 doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camaschella C. BMP6 orchestrates iron metabolism. Nature Genetics. 2009;41:386–388. doi: 10.1038/ng0409-386. [DOI] [PubMed] [Google Scholar]
- 38.Kawabata H, Yang R, Hirama T, Vuong PT, Kawano S, Gombart AF, Koeffler HP. Molecular cloning of transferrin receptor 2. A new member of the transferrin receptorlike family. J Biol Chem. 1999;274:20826–20832. doi: 10.1074/jbc.274.30.20826. [DOI] [PubMed] [Google Scholar]
- 39.Arndt S, Maegdefrau U, Dorn C, Schardt K, Hellerbrand C, Bosserhoff AK. Iron-Induced Expression of Bone Morphogenic Protein 6 in Intestinal Cells Is the Main Regulator of Hepatic Hepcidin Expression In Vivo. Gastroenterology. 2010;138:372–382. doi: 10.1053/j.gastro.2009.09.048. [DOI] [PubMed] [Google Scholar]
- 40.Kautz L, Besson-Fournier C, Meynard D, Latour C, Roth MP, Coppin H. Iron overload induces BMP6 expression in the liver but not in the duodenum. Haematologica. 2011;96:199–203. doi: 10.3324/haematol.2010.031963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ganz T. Iron homeostasis: fitting the puzzle pieces together. Cell Metab. 2008;7:288–290. doi: 10.1016/j.cmet.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 42.Lin L, Goldberg YP, Ganz T. Competitive regulation of hepcidin mRNA by soluble and cell-associated hemojuvelin. Blood. 2005;106:2884–2889. doi: 10.1182/blood-2005-05-1845. [DOI] [PubMed] [Google Scholar]
- 43.Silvestri L, Pagani A, Nai A, De Domenico I, Kaplan J, Camaschella C. The Serine Protease Matriptase-2 (TMPRSS6) Inhibits Hepcidin Activation by Cleaving Membrane Hemojuvelin. Cell Metabolism. 2008;8:502–511. doi: 10.1016/j.cmet.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Truksa J, Gelbart T, Peng HF, Beutler E, Beutler B, Lee P. Suppression of the hepcidin-encoding gene Hamp permits iron overload in mice lacking both hemojuvelin and matriptase-2/TMPRSS6. British Journal of Haematology. 2009;147:571–581. doi: 10.1111/j.1365-2141.2009.07873.x. [DOI] [PubMed] [Google Scholar]
- 45.Gao J, Chen J, De DI, Koeller DM, Harding CO, Fleming RE, Koeberl DD, Enns CA. Hepatocyte-targeted HFE and TFR2 control hepcidin expression in mice. Blood. 2010;115:3374–3381. doi: 10.1182/blood-2009-09-245209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pietrangelo A, Caleffi A, Henrion J, Ferrara F, Corradini E, Kulaksiz H, Stremmel W, Andreone P, Garuti C. Juvenile hemochromatosis associated with pathogenic mutations of adult hemochromatosis genes. Gastroenterology. 2005;128:470–479. doi: 10.1053/j.gastro.2004.11.057. [DOI] [PubMed] [Google Scholar]
- 47.Harold TLC, van Swelm R, Theurl M, Theurl I, Kemna E, van der Burgt Y, Venselaar H, Dutilh B, Russel F, Weiss G, Masereeuw R, Fleming RE, Swinkels D. Mass spectrometry analysis of hepcidin peptides in experimental mouse models. PLoS ONE. doi: 10.1371/journal.pone.0016762. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanno T, Porayette P, Sripichai O, Noh SJ, Byrnes C, Bhupatiraju A, Lee YT, Goodnough JB, Harandi O, Ganz T, Paulson RF, Miller JL. Identification of TWSG1 as a second novel erythroid regulator of hepcidin expression in murine and human cells. Blood. 2009;114:181–186. doi: 10.1182/blood-2008-12-195503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mleczko-Sanecka K, Casanovas G, Ragab A, Breitkopf K, Muller A, Boutros M, Dooley S, Hentze MW, Muckenthaler MU. SMAD7 controls iron metabolism as a potent inhibitor of hepcidin expression. Blood. 2010;115:2657–2665. doi: 10.1182/blood-2009-09-238105. [DOI] [PubMed] [Google Scholar]
- 50.Poli M, Luscieti S, Gandini V, Maccarinelli F, Finazzi D, Silvestri L, Roetto A, Arosio P. Transferrin receptor 2 and HFE regulate furin expression via mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/Erk) signaling. Implications for transferrin-dependent hepcidin regulation. Haematologica. 2010;95:1832–40. doi: 10.3324/haematol.2010.027003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nemeth E, Rivera S, Gabayan V, Keller C, Taudorf S, Pedersen BK, Ganz T. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest. 2004;113:1271–1276. doi: 10.1172/JCI20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pietrangelo A, Dierrsen U, Valli L, Garuti C, Rump A, Corradini E, Klein C, Trautwein C. STAT3 is required for IL-6-GP130-dependent activation of hepcidin in vivo. Gastroenterology. 2007;132:294–300. doi: 10.1053/j.gastro.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 53.Lee P, Peng H, Gelbart T, Wang L, Beutler E. Regulation of hepcidin transcription by interleukin-1 and interleukin-6. Proc Natl Acad Sci USA. 2005;102:1906–1910. doi: 10.1073/pnas.0409808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vecchi C, Montosi G, Zhang KZ, Lamberti I, Duncan SA, Kaufman RJ, Pietrangelo A. ER Stress Controls Iron Metabolism Through Induction of Hepcidin. Science. 2009;325:877–880. doi: 10.1126/science.1176639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


