Abstract
Mucosal innate lymphoid cell (ILC) subsets promote immune responses to pathogens by producing distinct signature cytokines in response to changes in the cytokine microenvironment. We previously identified human ILC3 distinguished by interleukin-22 (IL-22) secretion. Here we characterized a human ILC1 subset that produced Interferon-γ in response to IL-12 and IL-15, had a unique integrin profile, intraepithelial location, hallmarks of TGF-β imprinting and a memory-activated phenotype. Because tissue-resident memory CD8+ T cells share this profile, intraepithelial ILC1 may be their innate counterparts. In mice, intraepithelial ILC1 were distinguished by CD160 expression and required Nfil3-and Tbx21-encoded transcription factors for development, but not IL-15 receptor-α, indicating that intraepithelial ILC1 are distinct from conventional NK cells. Intraepithelial ILC1 were amplified in Crohn’s disease patients and contributed to pathology in the anti-CD40-induced colitis model in mice. Thus, intraepithelial ILC1 may initiate interferon-γ responses against pathogens, but contribute to pathology when dysregulated.
Keywords: innate lymphoid cells, NK cells, epithelium, tonsil, intestine, CD160, IL-15, Crohn’s disease
Introduction
Recent studies have shown that both aggregated and diffuse lymphoid tissues of the oral and gastrointestinal mucosa harbor various innate lymphoid cell (ILC) subsets that contribute to early immune responses against pathogens by secreting different cytokines (Cherrier et al., 2012; Guo et al., 2012; Spits et al., 2013). Remarkably, these ILC subsets resemble the discrete T cell effectors Th1, Th2, Th17 and Th22 cells in terms of the signature cytokines and transcription factors that determine their development. We recently identified a unique type of human ILC marked by the natural killer (NK) cell surface molecule NKp44, the signature cytokine IL-22, expression of the receptors for IL-1 (IL-1R), IL-7 (IL-7Rα) and IL-23 (IL-23Rα), and the chemokine receptor CCR6 (Cella et al., 2009; Cella et al., 2010). We refer to these cells and their murine counterparts as type 3 ILC (ILC3) (Spits et al., 2013). Acting on epithelial cells, IL-22 induces the release of antimicrobial peptides that protect the integrity of the mucosal barrier from pathogens (Ouyang et al., 2011; Sonnenberg et al., 2011). IL-22 release is triggered by IL-23 and enhanced by IL-1, both of which are produced by antigen presenting cells (APC) in response to pathogens or other inflammatory stimuli.
ILC3 development requires IL-7 signaling and the transcription factors Id2, RORγt (Cherrier et al., 2012; Spits et al., 2013), AHR (Kiss et al., 2011; Lee et al., 2012; Qiu et al., 2012) and Notch (Lee et al., 2012; Possot et al., 2011). Human ILC3 maintain a certain degree of functional plasticity, as they can produce IFN-γ when cultured in vitro with IL-7+IL-2, IL-1β+IL-2 or IL-7+IL-1β+IL-23 (Cella et al., 2010).
Additional cells expressing NKp44 or other NK cell markers have been described in the human oral and gastrointestinal mucosa, including tonsilar NKp44+CD103+ NK cells (Cella et al., 2010), small intestine CD7+CD3− intraepithelial lymphocytes (Eiras et al., 2000), colon lamina propria NKp46+NKp44− cells (Takayama et al., 2010), and rectal intraepithelial and lamina propria NKp46+ cells (Sips et al., 2012). Cells with NK cell activity were also reported in the murine intestinal epithelium and lamina propria (Carman et al., 1986; Tagliabue et al., 1982). While these reports suggest further phenotypic and functional heterogeneity of mucosal ILC, the functions and responsiveness to pathogenic stimuli of these ILC subsets are poorly understood. It is also unclear whether these cells are related to ILC3 or classical NK cells. In this study, we demonstrated that the human tonsil and intestinal mucosa contain a unique NKp44+CD103+ cell subset that produced IFN-γ in response to IL-12 and IL-15. These cells were different from classical NK cells in that they were selectively located within the epithelium, expressed the chemokine receptor CXCR6 and reflected features of TGF-β imprinting. Moreover, they had an integrin profile and a memory-activated phenotype similar to that previously observed in tissue resident memory CD8 T cells (Trm) cells, which are virus-specific cells that persist in the mucosae and skin after the resolution of viral infections (Casey et al., 2012; Gebhardt et al., 2009; Jiang et al., 2012; Sathaliyawala et al., 2013; Wakim et al., 2012, reviewed in Cauley and Lefrancois, 2013). Intraepithelial NKp44+CD103+ cell numbers were increased in Crohn’s disease patients, suggesting that they contribute to inflammatory bowel disease (IBD).
We further identified the murine counterparts of intraepithelial NKp44+CD103+ cells, which were distinguished by expression of CD160 as well as NKp46 and NK1.1. We found that these cells were strongly reduced in mice lacking the transcription factors Nfil3 (also known as E4bp4) (Male et al., 2012) and Tbx21, but not in Il15ra-/- mice, corroborating that intraepithelial NKp46+NK1.1+CD160+ cells represent an ILC1 subset distinct from classical NK cells. Murine intraepithelial ILC1 promptly produced IFN-γ in the anti-CD40-induced colitis model and contributed to intestinal inflammation. Thus, intraepithelial ILC1 may facilitate immune responses during exposure to infections, but contribute to IBD when dysregulated.
Results
Human mucosal-associated lymphoid tissues comprise diverse ILC subsets
We sought to delineate the diversity of human ILC in mucosal-associated lymphoid tissues. We isolated CD56+ cells from human tonsils and analyzed them by flow cytometry for the expression of NKp44, CD103 and CD3. Within the CD3− compartment, we identified two cell subsets, NKp44+ and NKp44− cells (Fig. 1A). NKp44+ cells could be further divided into CD103+ and CD103− cells. The NKp44− population was mainly CD103− but included small numbers of CD103lo cells, which varied considerably among individuals and therefore were not further examined.
Figure 1. see also Figure S1. Mucosal NKp44+CD103+ and NKp44−CD103− cells specialize in IFN-γ production in response to IL-12 and IL-15.
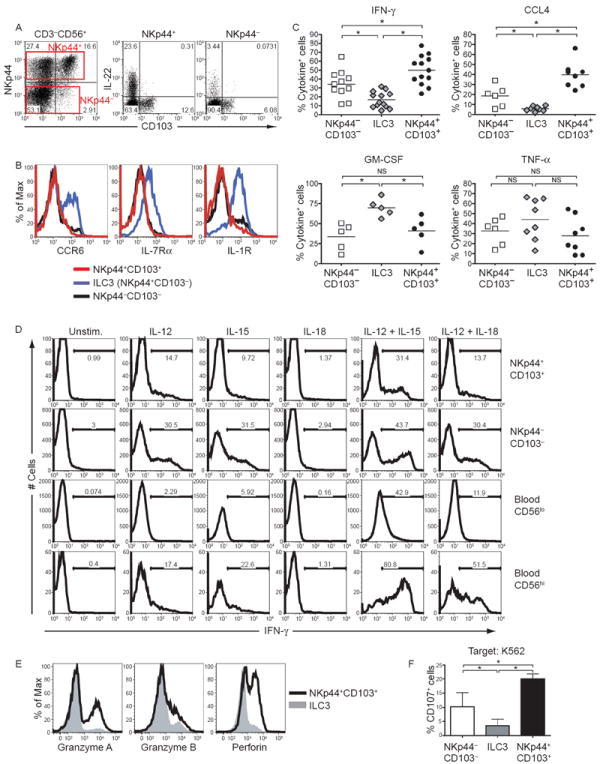
CD56+ cells were enriched from tonsils and analyzed for their surface markers and functions. (A) Left, expression of NKp44 and CD103 on tonsil CD56+ cells, gated on live CD3−CD19− cells. Right, tonsil CD56+ cells were stimulated with IL-23 and stained for intracellular IL-22. Cells were gated on the NKp44+ and the NKp44− fractions. (B) Surface markers of tonsilar NKp44+CD103+ cells (red lines), ILC3 (NKp44+CD103−, blue lines), and NKp44−CD103− cells (black lines). (C) After stimulation in vitro with PMA and ionomycin, cytokine content of tonsil CD56+ cells was analyzed by flow cytometry. Data displayed are the frequencies of cytokine-positive cells within NKp44+CD103+ cells, ILC3 and NKp44−CD103− cells obtained from 5 to 13 different donors. (D) Tonsil and peripheral blood CD56+ cells were cultured with IL-12, IL-15, IL-18, or with combinations of these cytokines. After 9 hours, tonsil NKp44+CD103+ cells, tonsil NKp44−CD103− cells, blood CD56lo (CD3−CD16+CD56lo) and blood CD56hi (CD3−CD16−CD56hi) cells were analyzed for IFN-γ content. (E) Cytotoxic potential of NKp44+ cells was analyzed by intracellular staining for granzymes and perforin. Gray profiles indicate the ILC3 subset; black lines denote NKp44+CD103+ cells. (F) Frequency of CD107a+ cells following co-culture of ILC subsets with K562. Shown are combined results obtained from 7 individual donors. Data are represented as mean +/- SD.
We next determined which of these subsets produced IL-22. CD56+ cells isolated from tonsils were stimulated with IL-23 and analyzed for intracellular content of IL-22 as well as surface expression of NKp44, CD103 and the prototypic ILC3 cell surface markers, including CCR6, IL-1R and IL-7Rα. NKp44+CD103− cells produced IL-22 (Fig. 1A) and many expressed CCR6, IL-1R and IL-7Rα (Fig. 1B) and therefore corresponded to ILC3. In contrast, NKp44+CD103+ cells and NKp44−CD103− cells produced no or little IL-22 (Fig. 1A) and did not express CCR6, IL-1R or IL-7Rα (Fig. 1B).
Mucosal NKp44+CD103+ and NKp44−CD103− cells are type 1 ILC
To evaluate the function of NKp44+CD103+ and NKp44−CD103− cells, we initially assessed cytokine production by intracellular staining following stimulation with PMA and ionomycin. Both NKp44+CD103+ and NKp44−CD103− cells produced more IFN-γ and CCL4 than ILC3, but less GM-CSF (Fig. 1C). We then asked which physiological stimuli induce IFN-γ in NKp44+CD103+ and NKp44−CD103− cells. Typically, ILC produce signature cytokines when challenged with other cytokines, independent of signaling through immunoreceptors (Guo et al., 2012; Soudja et al., 2012). Accordingly, both NKp44+CD103+ and NKp44−CD103− cells produced IFN-γ when stimulated in vitro with IL-12 or IL-15 (Fig. 1D). Moreover, IL-12 and IL-15 had a strong synergistic effect. IL-23, which stimulates ILC3, was ineffective (data not shown). IFN-γ production by NKp44−CD103− cells was particularly efficient in a subset expressing CD27 (Fig. S1), similar to what has been shown for blood NK cells (Silva et al., 2008).
Interestingly, the magnitude of IFN-γ production by NKp44+CD103+ and NKp44−CD103− cells was similar to that of peripheral blood CD56hi NK cells (Fig. 1D), which are known producers of IFN-γ in response to cytokines (Cooper et al., 2001), and superior to that of peripheral blood CD56lo NK cells (Fig. 1D), which specialize in the release of lytic granules (Cooper et al., 2001). However, although CD56hi NK cells were responsive to IL-18, NKp44+CD103+ and NKp44− CD103− cells produced equal amounts of IFN-γ following stimulation with either IL-12+IL-18 or IL-12 alone, demonstrating that these cells do not respond to IL-18 (Fig. 1D). Based on their ability to rapidly produce IFNγ in response to IL-12 and IL-15, we conclude that NKp44+CD103+ cells and NKp44-CD103- cells belong to type 1 ILC (ILC1), which are the innate counterparts of Th1 CD4 T cells and include blood NK cells, particularly the CD56hi subset.
NKp44+CD103+ and NKp44−CD103− ILC1 not only produced IFN-γ but approximately 50% of these cells also contained intracellular perforin and granzymes (Fig. 1E and data not shown), consistent with potential cytotoxic activity. In NKp44−CD103− ILC1, granzyme production precisely paralleled IFN-γ production (Fig. S1). To corroborate the lytic activity of NKp44+CD103+ and NKp44−CD103− ILC1, we monitored degranulation of all ILC subsets by measuring cell surface expression of CD107a in the presence of the tumor cell line K562. Both NKp44+CD103+ and NKp44−CD103− ILC1 expressed more CD107a than ILC3 (Fig. 1F). We conclude that NKp44+CD103+ and NKp44− CD103− ILC1 not only produce IFN-γ but can also release lytic mediators when activating receptors are engaged by cognate ligands expressed on target cells.
NKp44+CD103+ ILC1 have an intraepithelial location
CD103 promotes the interaction of lymphocytes with epithelial cells by binding E-cadherin (Cepek et al., 1994) and is a marker of intraepithelial T lymphocytes (Cheroutre et al., 2011; Sheridan and Lefrancois, 2010). The majority of NKp44+CD103+ ILC1 expressed additional intraepithelial lymphocyte markers, such as CD160, which has been recently shown to bind to HVEM on epithelial cells promoting epithelial cell defense mechanisms (Shui et al., 2012) (Fig. 2A), as well as CD101 (also known as EWI-101) (Allez et al., 2002) (Fig. 2A). Neither ILC3 nor NKp44−CD103− ILC1 expressed substantial amounts of CD160 or CD101. Thus, we hypothesized that NKp44+CD103+ ILC1 are intraepithelial. We investigated serial sections of human tonsils for NKp44 and CD103 expression by immunohistochemistry. We found that numerous NKp44+ cells localized in the epithelia overlaying the crypts and the surface of the tonsil, particularly in “lymphoepithelial” areas where lymphocytes infiltrate the epithelium (Fig. S2). Fewer NKp44+ cells were found in the interfollicular areas, and none in the follicles (Fig. S2 and data not shown). CD103 was found on a fraction of NKp44+ cells, and two-color staining with CD103 and CD3 confirmed the presence of CD103+ cells in tonsil epithelia that were CD3− and thus distinct from T cells (Fig. S2). We conclude that the large majority of NKp44+CD103+ ILC1 have an intraepithelial location.
Figure 2. see also Figure S2. Expression of adhesion molecules, markers of TGF-β imprinting and activation in NKp44+CD103+ILC1.
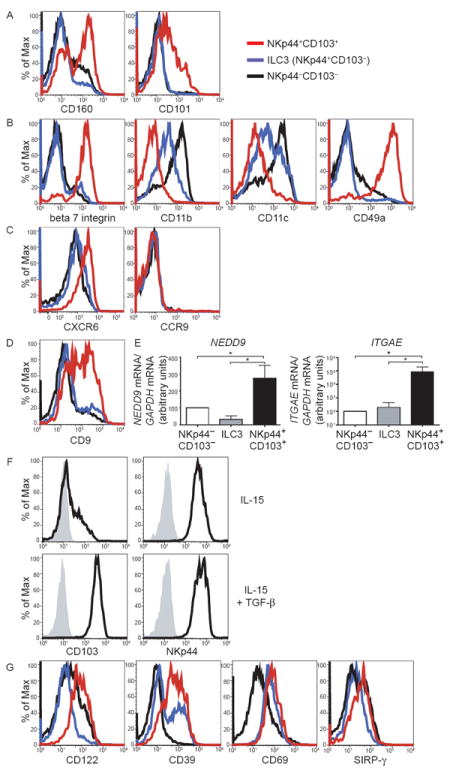
(A-D) CD56+ cells enriched from tonsils were analyzed for surface markers gating on live CD3− CD19− cells. Red lines indicate NKp44+CD103+ ILC1, blue lines denote ILC3 (NKp44+CD103−), and black lines indicate NKp44−CD103− cells. Representative data from experiments with 3 or more individual tonsil samples are shown. (E) Tonsil CD56+ cells were sorted into the three subsets described in (A-D), and mRNA content for NEDD9 and ITGAE (encoding CD103) was analyzed by qRT-PCR. Displayed are normalized data from qRT-PCR experiments with 3 (NEDD9), and 5 (ITGAE) individual donors. Data are represented as mean +/- SD. (F) NKp44+CD103+ cells were sorted from tonsils, cultured in IL-15 with or without the addition of TGF-β for 9 days and analyzed for CD103 and NKp44 expression. (G) Tonsil CD56+ cells were analyzed for surface expression of activation markers as in (A-D).
Intraepithelial ILC1 have the integrin profile of CD8 Trm cells
We asked whether CD103 is part of a unique integrin profile that distinguishes NKp44+CD103+ ILC1 from other ILC subsets within tonsils. We found that NKp44+CD103+ ILC1 selectively express high amounts of beta7 integrin, which associates with CD103 to form αEβ7 heterodimers (Fig. 2B). Conversely, NKp44+CD103+ ILC1 expressed low amounts of CD11b (alpha M integrin) and CD11c (alpha X integrin). Additionally, NKp44+CD103+ ILC1 expressed high levels of the collagen-binding integrin CD49a (also known as alpha 1 integrin or VLA-1) (Fig. 2B). Notably, expression of αEβ7 and CD49a is characteristic of CD8 Trm, which are also preferentially located within the epithelium and poised for prompt effector functions (Cauley and Lefrancois, 2013). In contrast to NKp44+CD103+ ILC1, NKp44−CD103− ILC1 and ILC3 expressed the integrins CD11b and CD11c and lacked αEβ7 and CD49a.
We also assessed the chemokine receptor profile of NKp44+CD103+ ILC1. Unexpectedly, NKp44+CD103+ ILC1 did not express CCR9, which has been shown to be a major chemokine receptor that drives lymphocytes into the epithelium (Fig. 2C). In contrast, they expressed CXCR6, a chemokine receptor that was previously found on liver NK cells with memory-like features (Paust et al., 2010) and that mediates lymphoepithelial adhesion in the gut (Hase et al., 2006). Thus, intraepithelial ILC1 express a unique repertoire of homing receptors that promote retention in the epithelia.
Intraepithelial ILC1 bear hallmarks of TGF-β imprinting and activation
It has been shown that CD103 denotes a cellular response to TGF-β (Keskin et al., 2007). Indeed, NKp44+CD103+ ILC1 expressed additional markers of TGF-β exposure including the tetraspanin CD9 (Keskin et al., 2007) and the integrin-signaling molecule NEDD9 (Giampieri et al., 2009) (Fig. 2D, E). Corroborating the influence of TGF-β in NKp44+CD103+ ILC1 development, we found that tonsil NKp44+CD103+ cells cultured in vitro with TGF-β together with either IL-2 or IL-15 maintained CD103 expression, whereas in the presence of IL-2 and IL-15 alone cells survived and proliferated but lost CD103 expression over time (Fig. 2F). By regulating the expression of integrins and integrin signaling molecules, TGF-β may promote adhesion of ILC1 to epithelial cells (Giampieri et al., 2009). Interestingly, blood NK cells only engender few NKp44+CD103+ ILC1 cells when cultured with either IL-2 or IL-15 and TGF-β or supernatant from intestinal epithelial cell lines (data not shown), suggesting that, although related to conventional NK cells, NKp44+CD103+ ILC1 cells may have a unique developmental pathway.
Intraepithelial NKp44+CD103+ ILC1 expressed higher amounts of the IL-2 receptor subunit beta (IL-2Rβ or CD122) as well as the activation markers CD39, CD69 and SIRP-γ than did NKp44−CD103− ILC1 (Fig. 2G). NK cell receptors, such as NKp46, NKp30, CD244, CD94, NKG2A, NKG2C and NKG2D, were similar in NKp44+CD103+ ILC1 and NKp44−CD103− ILC1 (Fig. S2). KIR expression was very low on both cell subsets as it was on ILC3 (Fig. S2). CD57, which was previously noted on rectal mucosa intraepithelial NK cells (Sips et al., 2012), was not observed on any subset (Fig. S2). Considering the entire phenotypic analysis of ILC subsets, we conclude that intraepithelial ILC1 express hallmarks of TGF-β imprinting and activation. The expression of activation markers on intraepithelial ILC1 is reminiscent of the activated phenotype seen on CD8 Trm (Cauley and Lefrancois, 2013).
NKp44+CD103+ ILC1 express T-bet and EOMES
To gain insight into the development of intraepithelial ILC1, we compared all ILC subsets for expression of transcription factors that play a key role in ILC development by intracellular staining and RT-PCR. Both NKp44+CD103+ and NKp44−CD103− ILC1 expressed T-bet and EOMES (Fig. 3A, B), which have been implicated in the development and function of conventional NK cells (Gordon et al., 2012), but not RORγt or AHR, which drive ILC3 cell differentiation (Spits et al., 2013). Foxp3 was undetectable in all subsets. Interestingly, NKp44+CD103+ ILC1 expressed higher levels of IKZF3 transcript than other ILC. IKZF3 encodes Aiolos, a member of the Ikaros family of transcription factors that regulates B and T cell development (Cortes et al., 1999) but heretofore has not been implicated in ILC development or function.
Figure 3. NKp44+CD103+cells express T-bet, Eomes and Aiolos.
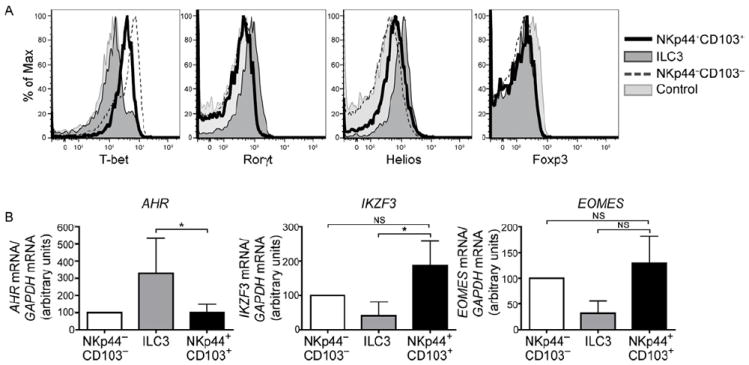
(A) Tonsil CD56+ cells were stained for intracellular T-bet, RORγt, Helios, and FoxP3. Dark gray profiles indicate ILC3, black lines represent NKp44+CD103+ ILC1, and dotted lines indicate NKp44−CD103− cells. Light gray profiles indicate staining with isotype-matched control antibody. Representative data from 3 individual tonsil samples are shown. (B) Tonsil CD56+ cells were sorted into the three subsets described in (A), and mRNA content for AHR, IKZF3 (encoding Aiolos) and EOMES was analyzed by qRT-PCR. Displayed are normalized data from 4 (AHR), 9 (IKZF3), and 5 (EOMES) donors. Data are represented as mean +/- SD.
Intraepithelial ILC1 respond to disparate danger signals
Because NKp44+CD103+ ILC1 are intraepithelial, they may produce IFN-γ in response to signals produced by epithelial cells. To test this hypothesis, we challenged an epithelial cell line with different TLR agonists. We then added CD56+ cells isolated from human tonsils to the cultures and analyzed intracellular IFN-γ content in NKp44+CD103+ ILC1 by flow cytometry. NKp44+CD103+ ILC1 produced more IFN-γ when incubated with epithelial cells stimulated with the TLR2 agonist Pam3CSK4 than with epithelial cells stimulated with the TLR3 agonist polyI:C or unstimulated epithelial cells (Fig. 4A, B). Similarly, elevated IFN-γ levels were detected in the supernatants from FACS-sorted NKp44+CD103+ ILC1 co-cultured with Pam3CSK4-stimulated epithelial cells (Fig. 4C, D). TLR2 agonists had no direct effect on ILC1 (data not shown). These results demonstrate that intraepithelial ILC1 are responsive to danger signals produced by epithelial cells.
Figure 4. Activation of NKp44+CD103+cells by co-culture with epithelial cells and myeloid cells stimulated with TLR agonists.
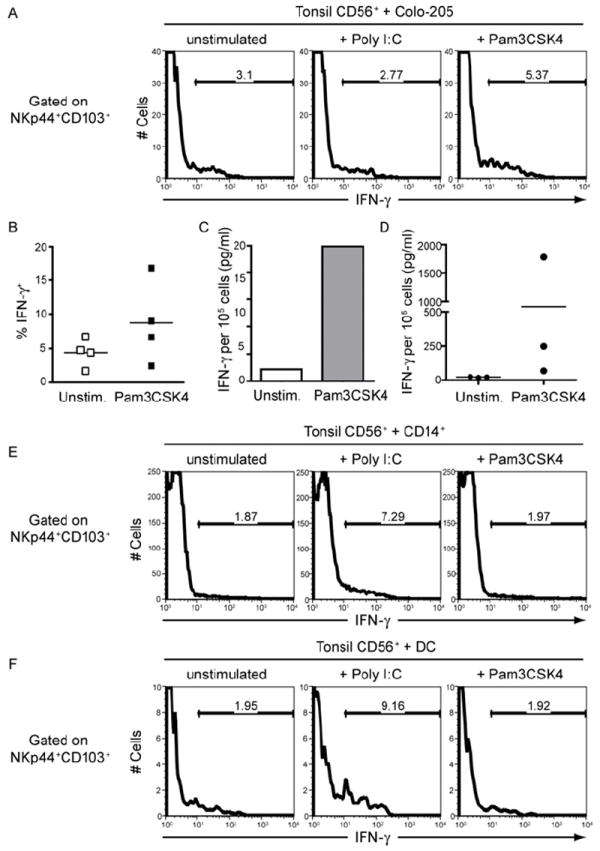
(A-B) Tonsil CD56+ cells were co-cultured for 9 hours with the colon carcinoma cell line Colo-205 that was pre-treated with poly I:C or Pam3CSK4. IFN-γ content of NKp44+CD103+ ILC1 was determined by intracellular staining. (A) Staining from one representative experiment. (B) Compiled data from four experiments, showing the percentage of IFN-γ cells within the NKp44+CD103+ ILC1. (C-D) Tonsil NKp44+CD103+ cells were FACS sorted and co-cultured for 48 hours with Colo-205 cells that were left unstimulated or treated with Pam3CSK4. IFN-γ levels in culture supernatants were determined by CBA. (C) Representative results from one experiment. (D) Summary of experiments with 3 donors, showing IFN-γ levels produced by NKp44+CD103+ cells of individual donors. (E-F) Tonsil CD56+ cells were co-cultured for 9 hours with peripheral blood CD14+ monocytes (E) or monocyte-derived dendritic cells (F) pre-treated with poly I:C or Pam3CSK4. IFN-γ content of NKp44+CD103+ cells was determined by intracellular staining.
Because IL-12 induces IFN-γ production by intraepithelial ILC1 (see Fig. 1) and is secreted by inflammatory monocytes, macrophages and DC (Trinchieri, 2003), we asked whether NKp44+CD103+ ILC1 produce IFN-γ in response to signals generated by monocytes and monocyte-derived DC. We challenged monocytes and monocyte-derived DC with different TLR agonists. We then added CD56+ cells isolated from human tonsils to the cultures and analyzed the IFN-γ content in ILC1 by flow cytometry. ILC1 produced more IFN-γ when incubated with polyI:C-stimulated monocytes and DC than with Pam3CSK4-stimulated monocytes and DC (Fig. 4E, F). Altogether these results indicate that intraepithelial ILC1 respond to danger signals originating from both epithelial cells and myeloid cells. It is likely that epithelial cells transmit the dominant danger signals when pathogenic attack is limited to the mucosal surface, whereas monocytes and DC stimulation of intraepithelial ILC1 might prevail when the epithelial barrier is breached, leading to DC activation in the underlying tissue.
Intraepithelial ILC1 expand during Crohn’s disease
We sought to extend our analysis of NKp44+CD103+ ILC1 to the intestinal mucosa. We separated intraepithelial lymphocytes (IEL) and lamina propria lymphocytes (LPL) from non-inflamed areas of human small intestine specimens that had been surgically removed to treat IBD. NKp44+CD103+ ILC1 were found primarily in the IEL and only in low numbers within LPL, corroborating our hypothesis that NKp44+CD103+ ILC1 are mainly intraepithelial lymphocytes (Fig. 5A). In contrast, ILC3 were mainly found in the lamina propria. NKp44+CD103+ ILC1 from the small intestine expressed most prototypic markers noted on their tonsilar counterparts, including NKp44, NKp46, CD39 (Fig. 5A), CD101, CD49a and CD160 (data not shown). The only exception was CD94, which was poorly expressed on small intestine NKp44+CD103+ ILC1 (Fig. 5A). NKp44+CD103+ ILC1 were also present in the epithelium of the colon (Fig. 5B) but were not detected in non-mucosal tissues, such as axillary lymph nodes (Fig. S3) or peripheral blood (not shown). These results confirmed that NKp44+CD103+ ILC1 are selectively located within the mucosal epithelium.
Figure 5. see also Figure S3. NKp44+CD103+ILC1 in normal and CD intestinal mucosa.
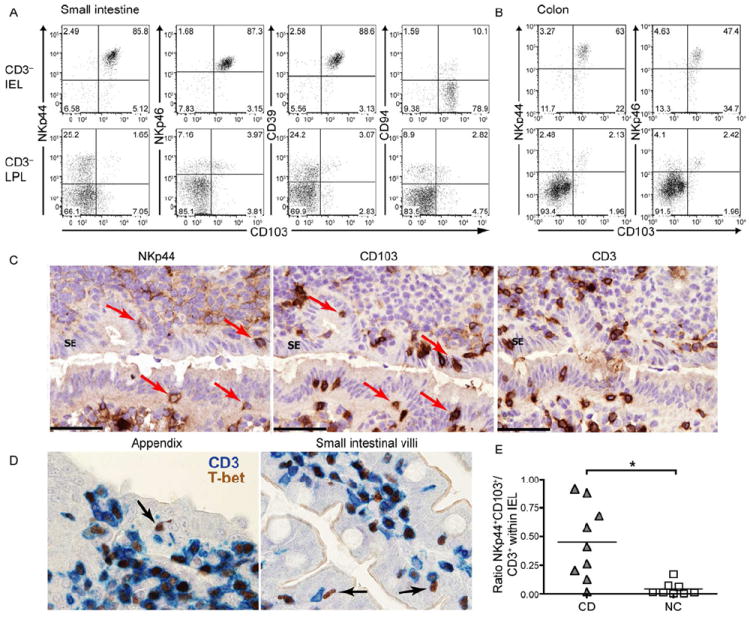
(A-B) Intraepithelial lymphocytes (IEL) and lamina propria lymphocytes (LPL) of human small intestine (A) and colon (B) were stained for CD103 and several ILC1 markers. Cells were gated on live CD45+CD3− lymphocytes. Shown are representative data from 2-3 different individuals. (C) Serial frozen sections from human appendix were stained for NKp44 (brown, left panel), CD103 (brown, middle panel), and CD3 (brown, right panel). Cells expressing both NKp44+, CD103+ within the surface epithelium (SE) are indicated (arrows). Sections are counterstained with haematoxylin. (D) Fixed sections from human appendix (left) and small intestinal villi (right) were stained for CD3 (blue) and T-bet (brown). T-bet+CD3− are indicated (arrows). (E) Samples from ileal resections of CD patients (CD) and non-IBD controls (NC) were analyzed for the ratio of NKp44+CD103+ cells versus CD3+ T cells within CD45+ IEL.
Immunohistochemical analysis of NKp44, CD103 and CD3 in the appendix and intestinal villi showed that while the majority of NKp44+ cells are found in the lamina propria, some cells are located within the epithelium (Fig. 5C and data not shown). Among numerous CD103+ cells detectable within the epithelium, some were CD3−, indicating that the intestinal epithelium harbors a small but detectable subset of NKp44+CD103+ ILC1 among numerous intraepithelial T cells. Immunohistochemical analysis of T-bet and CD3 distribution in appendix and intestinal villi confirmed the presence in the intestinal epithelium of a small but detectable number of CD3−T-bet+ ILC amid numerous CD3+T-bet+ T cells (Fig. 5D).
During IBD, dysregulated T cell production of IFN-γ and IL-17 causes excessive inflammation, tissue damage and loss of barrier function (Geremia et al., 2011; Strober and Fuss, 2011). Because of their ability to secrete IFN-γ and CCL4, intraepithelial NKp44+CD103+ ILC1 might contribute to the pathogenesis of IBD if inappropriately activated. We compared the frequencies of these cells in samples of small intestine surgically excised from Crohn’s disease (CD) patients and non-IBD controls. Remarkably, we detected higher frequencies of NKp44+CD103+ ILC1 within the intraepithelial lymphocytes in CD patients compared to non-IBD controls (Fig. 5E). Although low cell yields from these small tissue samples prevented functional studies, their increase during inflammation suggested that NKp44+CD103+ ILC1 are involved in the pathogenesis of CD.
Murine ILC1 are largely independent of IL-15 receptor α
We sought to identify the murine counterpart of intraepithelial NKp44+CD103+ ILC1. To facilitate their recognition, we isolated IEL from Rag1-/- mice. NKp44 is only encoded in humans, and CD103 was unexpectedly inadequate as CD103+ cells were hardly detectable within CD3− IEL (data not shown). In contrast, CD160 was an optimal marker of intraepithelial ILC1, as we detected a distinct population of CD160+NKp46+NK1.1+ cells within the IEL of Rag1-/- mice (Fig. 6A). Spleen NK cells did not express CD160 in steady state, corroborating the specificity of this marker for intraepithelial ILC1 (Fig. S4). Stimulation of IEL of Rag1-/- and C57BL/6 mice with IL-12 and IL-15 induced rapid IFN-γ production by CD3−CD160+NKp46+NK1.1+ cells, mirroring the human ILC1 response (Fig. 6B, C). In contrast, IL-23 did not induce IFN-γ production (Fig. 6C). The identification of the phenotype of murine intraepithelial ILC1 gave us the opportunity to investigate the transcription factors required for their development. Intraepithelial ILC1 were conserved in Rorc-/- and Ahr-/- mice, which lack ILC3 (Fig. 6D). In contrast, these cells were dramatically reduced in the IEL of mice with knock-out mutations of Nfil3 or Tbx21, which encode two transcription factors previously shown to be master regulators of NK cell development (Gordon et al., 2012; Male et al., 2012) (Fig. 6D). Normal numbers of ILC1 were present in the IEL of germ-free mice and slightly reduced numbers of ILC1 were found in neonatal mice (Fig. 6E). Altogether, these results indicated that intraepithelial ILC1 can develop independently of the intestinal microbiota and are more related to the NK cell lineage than to ILC3.
Figure 6. see also Figure S4. Phenotype, function and developmental requirements of murine intraepithelial ILC1.
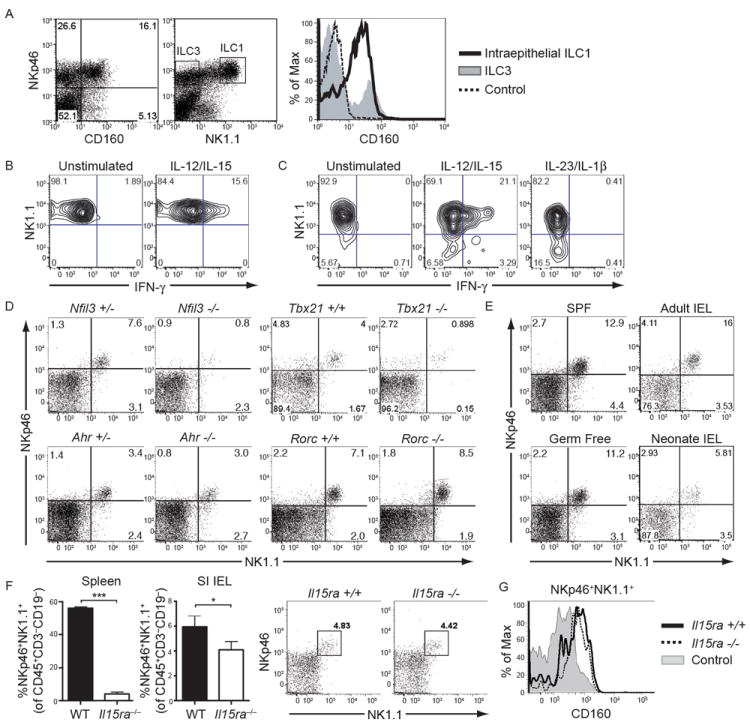
(A) Small intestine IEL of Rag-1-/- mice were analyzed for the expression of NKp46, NK1.1 and CD160. The majority of NKp46+NK1.1+ IEL (histogram, black line) express CD160, while NKp46+NK1.1− ILC3 (histogram, gray profile) do not. Control staining of NKp46+NK1.1+ cells is indicated by a dotted line. Cells in the dot plots were gated on live lymphocytes. (B) IEL from Rag-1-/- mice were stimulated in vitro with IL-12 and IL-15 and analyzed for intracellular IFN-γ. Cells were gated on CD45+NK1.1+ cells. (C) IEL from C57BL/6 mice were stimulated in vitro with a combination of IL-12 and IL-15, or with IL-23 and IL-1β, and analyzed for intracellular IFN-γ. Cells were gated on CD45+CD3−NKp46+ cells. (D) Frequencies of intraepithelial ILC1 in the small intestine of Nfil3-/-, Ahr-/-, Tbx21-/- and Rorc-/- mice and littermate controls. Cells were gated on CD45+CD3−CD19−. (E) Small intestinal IEL of conventionally housed (SPF) or germ-free C57BL/6 mice, as well as of adult and neonate C57BL/6 mice, were analyzed for the presence of NKp46+NK1.1+ cells. Cells were gated on CD45+CD3−CD19−. (F) ILC1 within IEL are largely preserved in Il15ra-/- mice. Left, frequencies of NKp46+NK1.1+ cells within spleen and intestinal IEL (gated on CD45+CD3−CD19−) from wild-type and IL15ra-/- mice from experiments with four mice each. Data are represented as mean +/- SD. Right, representative dot plots from IEL from wild-type and IL15ra-/- mice. (G) ILC1 from small intestinal epithelium of wild-type (black line) and IL15ra-/- mice (dotted line) express similar levels of CD160. Cells were gated on CD45+CD3−CD19−, followed by gating on NKp46+NK1.1+ cells. The gray profile indicates CD160 on wild-type splenic NKp46+NK1.1+ cells.
To gain more insights into the developmental relationship between intraepithelial ILC1 and conventional NK cells, we analyzed these cells in Il15ra-/- mice. As previously reported (Lodolce et al., 1998), these mice showed an almost complete absence of splenic NK cells (Fig. 6F). However, intraepithelial ILC1 were minimally affected by lack of IL-15 receptor α (IL-15Rα) (Fig. 6F) and maintained expression of CD160 (Fig. 6G). These data corroborate that intraepithelial ILC1 are a unique ILC1 subset distinct from conventional NK cells.
Intraepithelial ILC1 contribute to inflammation in a model of colitis
Because the numbers of intraepithelial ILC1 are increased in CD and may have pro-inflammatory functions, we evaluated the presence and function of intraepithelial ILC1 in a mouse model of IBD caused by injection of anti-CD40 in Rag1-/- mice (Buonocore et al., 2010; Uhlig et al., 2006). Following injection of anti-CD40, Rag1-/- mice rapidly lost weight and developed colitis as previously described (Uhlig et al., 2006). Analysis of IEL isolated from the small intestine 36 hrs after injection of anti-CD40 revealed a robust production of IFN-γ by CD160+NKp46+NK1.1+ cells (Fig. 7A), corroborating their early pro-inflammatory function in this model. IFN-γ was also produced by a small percentage of NKp46+NK1.1− cells, which correspond to ILC3 (Spits et al., 2013). This observation was consistent with our previous demonstration that human ILC3 are functionally plastic. ILC3 can produce IFN-γ when cultured in vitro with IL-7+IL-2, IL-1β+IL-2 or IL-7+IL-1β+IL-23, acquire responsiveness to IL-12 upregulate the expression of T-bet and downregulate that of RORγt ((Cella et al., 2010) and Fig. S5). In vivo, the functional conversion of ILC3 into IFN-γ-producing cells may occur when there is increased production of IL-15, IL-23 or IL-2 due to excessive activation of APC or T cells in IBD.
Figure 7. see also Figure S5. Intraepithelial ILC1 produce IFN-γ and contribute to pathology during anti-CD40-induced colitis.
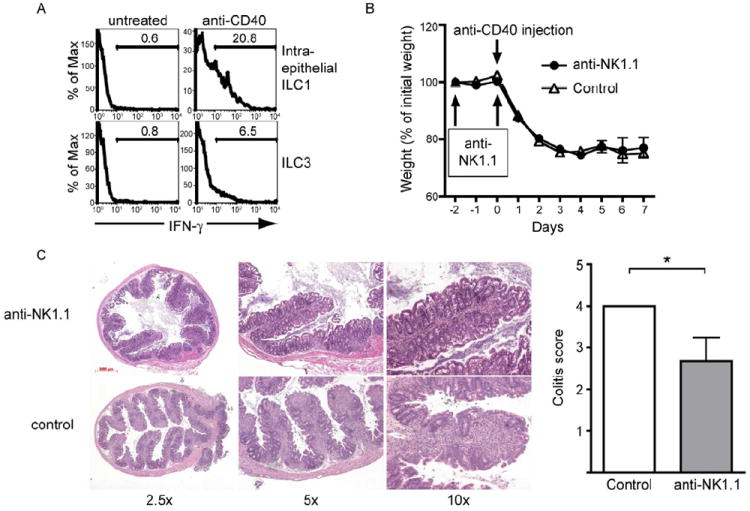
(A) Rag-1-/- mice were left untreated or injected with anti-CD40 to induce colitis. 36 hours after injection, small intestinal IEL were isolated and IFN-γ content of ILC1 (top panel) ILC3 cells (bottom panel), gated as shown in Figure 6A, was determined by intracellular staining. (B-C) ILC1 contribute to the intestinal pathology during anti-CD40-induced colitis. Rag-1-/- mice were treated with anti-NK1.1 to deplete intraepithelial ILC1, and colitis was induced with anti-CD40. Intestinal tissue pathology in the proximal colon was analyzed on day 7 after anti-CD40 treatment. Control, mice injected with anti-CD40 without anti-NK1.1 treatment. (B) Weight of mice recorded as the percentage of initial weight. Data are represented as mean +/- SD. (C) Left, H&E staining of proximal colon sections at day 7 after anti-CD40 injection, showing more severe cellular infiltration in control mice compared to anti-NK1.1-treated mice. Right, colitis score at day 7 determined from H&E staining of proximal colon samples. Data are represented as mean +/- SD.
To determine whether CD160+NKp46+NK1.1+ ILC1 directly contribute to the pathogenesis of anti-CD40 induced colitis in Rag1-/- mice, we depleted intraepithelial ILC1 and conventional NK cells with anti-NK1.1. Anti-NK1.1 treatment did not affect weight loss over a 7 day time period (Fig. 7B), suggesting that depletion of ILC1 or NK cells does not affect the systemic wasting disease. However, histochemical analysis of tissue sections of proximal colon revealed significantly less cellular inflammatory infiltrates and epithelial damage in NK1.1 depleted than control mice (Fig. 7C). These results suggest that intraepithelial ILC1 may play a pathogenic role in IBD through IFN-γ secretion.
Discussion
Our study extends the diversity of human ILC within the oral and gastrointestinal mucosa, characterizing a subset of human IFN-γ-producing ILC designated intraepithelial ILC1. While related to NK cells, intraepithelial ILC1 were unique in that they were located in the mucosal epithelium, shared multiple phenotypic and functional features with CD8 Trm and had several hallmarks of TGF-β imprinting. We also identified the murine intraepithelial ILC1 and found that they were largely independent of IL-15Rα, corroborating that these ILC are distinct from conventional NK cells, which require IL-15Rα for development. Although sharing the NKp44 marker with human ILC3, ILC1 also clearly differed from ILC3 in terms of phenotype, function and transcription factors involved in development. In addition to intraepithelial ILC1, we also identified a subset of human IFN-γ-producing NKp44-CD103- ILC1 that may be related to intraepithelial ILC1 and perhaps can acquire similar characteristics when transiting from the lamina propria to the epithelium.
ILC produce signature cytokines in response to cytokine stimulation (Guo et al., 2012). Optimal production requires stimulation by a STAT activator and an NF-κB inducer, most commonly an IL-1 family member. For example, IFN-γ-producing CD56hi NK cells respond to the IL-1 family member IL-18 and the STAT4 activator IL-12. However, mucosal ILC1 differed from conventional NK cells because they required a STAT4 (IL-12) and a STAT5 (IL-15) activator, whereas the NF-κB activator IL-18 was not effective alone or in combination with IL-12 or IL-15. Given that IL-15 has been shown to be an important inflammatory mediator in gastrointestinal diseases such as celiac disease (Jabri and Sollid, 2009), the responsiveness of intraepithelial ILC1 to IL-15 may be an important pathogenic factor in intestinal inflammation.
Interestingly, intraepithelial ILC1 shared multiple phenotypic and functional features with CD8 Trm, which persist in the gastrointestinal tract, lung mucosae and skin long after a primary infection has cleared and are very effective at preventing re-infection (Cauley and Lefrancois, 2013). Given this, we envision that intraepithelial ILC1 are the innate counterparts of non-migratory CD8 Trm. Epstein-Barr virus (Strowig et al., 2008) and human cytomegalovirus infection of tonsils as well as viral infections of the small intestine (Carman et al., 1986) may result in an accumulation or activation of intraepithelial ILC1, which are retained in the epithelium to prevent reinfection. It will be important to explore the relationship between intraepithelial ILC1 and the recently reported memory NK cells (Cooper et al., 2009; Romee et al., 2012; Sun and Lanier, 2011). Like intraepithelial CD8 T cells, ILC1 also expressed CD160 and therefore may be the major innate trigger of HVEM-mediated epithelial anti-microbial response (Shui et al., 2012).
Intraepithelial ILC1 were unique in that they were located in the mucosal epithelium and showed several hallmarks of TGF-β imprinting, particularly expression of CD103, CD9 and NEDD9, most likely reflecting close interaction with the epithelium that produces TGF-β. TGF-β imprinting was previously observed in decidual NK cells, which are in close contact with the fetal trophoblast and presumably contribute to maternal-fetal tolerance (Koopman et al., 2003). However, in contrast to decidual NK cells, intraepithelial ILC1 had an activated-memory phenotype and secreted IFN-γ as well as lytic mediators, consistent with a proinflammatory function. By inducing a unique profile of adhesion and integrin signaling molecules, TGF-β may be important for maintaining the adhesiveness of ILC1 to epithelial cells rather than inducing a tolerogenic function.
In our study we identified the murine counterpart of intraepithelial ILC1 as CD3− IEL that co-express CD160, NKp46 and NK1.1. This observation enabled us to examine the developmental pathway of intraepithelial ILC1 in mice with targeted mutations of master transcription factors, demonstrating the requirement of Nfil3 and Tbx21, factors involved in the development of conventional NK cells. In contrast, Rorc and Ahr were dispensable. While these results, together with the abundance of T-bet protein and EOMES transcript in human intraepithelial ILC1, suggested a relationship of intraepithelial ILC1 with conventional NK cells, we found that intraepithelial ILC1 were largely independent of IL-15Rα, indicating that intraepithelial ILC1 are developmentally distinct from conventional NK cells. It is possible that intraepithelial ILC1 are capable of exploiting other cytokines for their development, such as IL-7 or IL-2. Moreover, ILC1 expressed the transcription factor Aiolos, which may contribute to some of the unique developmental and phenotypic features of these cells.
We found that frequencies of intraepithelial ILC1 were increased in Crohn’s disease patients. We also observed that intraepithelial ILC1 produced high amounts of IFN-γ in Rag1-/- mice treated with anti-CD40, a model of colitis characterized by wasting syndrome and severe intestinal inflammation. Although it was previously shown that IL-12 and IL-23 play a major role in the wasting syndrome and colon inflammation respectively (Buonocore et al., 2010; Uhlig et al., 2006), we found that depletion of intraepithelial ILC1 ameliorated proximal colon inflammation. Together, these results indicate that intraepithelial ILC1 are poised for robust IFN-γ production, which may be important for orchestrating innate responses to infectious agents and preventing reinfection after elimination of the inciting pathogen. However, they may contribute to IBD when dysregulated.
Experimental Procedures
Antibodies and flow cytometry
The commercial sources of the antibodies used for flow cytometric analysis are indicated in Supplemental Experimental Procedures. Stained cells were analyzed on a FACS Calibur or FACS Canto using CellQuest and Diva programs, respectively. Dead cells were excluded with propidium iodide (Sigma Aldrich) or 7-AAD (BD Biosciences). Anti-CD45 was included in flow cytometric analysis of small intestinal lymphocytes to gate on live CD45+ cells. Flow cytometric analyses were performed using the FlowJo software (Tree Star).
Mice
All mice were maintained in a pathogen-free facility and animal protocols were approved by the Washington University School of Medicine (WUSM) animal studies committee. Ahr-/- mice were described before (Lee et al., 2012). Rorc-/-and Rag-1-/- mice were obtained from Jackson laboratories. Nfil3-/-, Tbx21-/-, Il15ra-/- and germ-free adult female mice were kindly provided by Drs Paul Rothman (University of Iowa), Takeshi Egawa (WUSM), Anthony R. French (WUSM) and Jeffrey I. Gordon (WUSM), respectively. All mice were on a C57BL/6 background. Mice were used between 6 and 8 weeks of age, with the exception of those used to study neonate IEL from the small intestine, which were 10 day-old.
Tissues and cell isolation
Tonsils, intestinal samples, and peripheral blood products were obtained under the approval of the WUSM institutional review board. Tissue sources are described in Supplemental Experimental Procedures. Tonsilar lymphocyte suspensions and peripheral blood mononuclear cells (PBMC) were prepared as previously described (Cella et al., 2009; Cella et al., 2010). IEL and LPL were prepared as previously described (Lefrancois and Lycke, 2001). CD56+ cells were enriched from tonsil lymphocytes by magnetic cell sorting using CD56 MicroBeads (Miltenyi Biotec). ILC subsets were sorted from CD56+ cells as previously reported (Cella et al., 2010) and described in Supplemental Experimental Procedures.
ILC culture for analysis of cytokines and degranulation
For cytokine analyses, CD56-enriched tonsil cells were stimulated with 10-7 M PMA and 1 μg/ml ionomycin (both from Sigma Aldrich) for 6 hours at 37°C. 2 μM monensin (Sigma Aldrich) was added during the final 4 hours of culture. Alternatively, human tonsil CD56+ cells or mouse IEL were cultured for 9 hours in the presence of 50 ng/ml human IL-15 and 10 ng/ml human or mouse IL-12, or with 100 ng/ml human or mouse IL-23 (all from Peprotech). Cells were stained for surface markers, followed by fixation, intracellular cytokine staining and analysis by flow cytometry. IFN-γ content in supernatants of CD56+ cells cultured with epithelial cells was measured by Cytometric Bead Array (CBA) (BD Biosciences). For degranulation assays, CD56+ cells were co-cultured with K562 tumor cells in the presence of anti-CD107a for 5 hours. Co-cultures are described in detail in Supplemental Experimental Procedures.
Tissue immunohistochemistry and immunofluorescence
Immunohistochemistry and immunofluorescence staining of frozen or paraffin-embedded tissues are described in Supplemental Experimental Procedures.
qRT-PCR
RNA and cDNA were prepared from sorted ILC as previously described (Cella et al., 2009). Quantitative PCR was performed using SYBRGreen (BIORAD). Results were normalized to the housekeeping gene GAPDH and displayed as [mRNA (gene of interest) per mRNA (GAPDH)] × 106, using the following formula: 2-[Ct (gene of interest) − Ct (GAPDH)] × 106. Primers are listed in Supplemental Experimental Procedures.
Anti-CD40-induced colitis model
Rag-1-/- mice were injected ip with 50 μl of anti-CD40 ascites (kind gift of Dr. Antonius Rolink, University of Basel). To deplete ILC1, mice were injected ip with 200 μl anti-NK1.1 ascites (PK136) at days -2 and 0 of anti-CD40 treatment. Mice were weighed daily and sacrificed at day 7 after anti-CD40 injection to analyze intestinal pathology as previously described (Uhlig et al., 2006). To evaluate IFN-γ production by IEL, mice were sacrificed 36 hours after anti-CD40 injection. Small intestinal IEL were cultured for 4 hours in the presence of monensin and then analyzed for intracellular IFN-γ content.
Supplementary Material
Acknowledgments
We would like to thank Paul Rothman, Antony R. French, Kenneth M. Murphy, Jeffrey I. Gordon and Takeshi Egawa for mouse lines; Antonius Rolink for antibodies; William E. Gillanders and Isaiah Turnbull for tissue specimens. This work was supported by NIH grants R01 DE021255-01 and U01 AI095542-01 to M. Co. W. V. was supported by PRIN (CKARAL, Ministero dell’Istruzione dell’Università e della Ricerca, 2009) and AIRC (Associazione Italiana per la ricerca sul cancro, 2010, IG 11924). J. S. L. was supported by the Ruth L. Kirschstein National Research Service Award Training Grant; R. D. N. was supported by NIH grant R01DK064798. The Washington University DDRCC Biobank core is supported by NIH grant P30DK052574.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allez M, Brimnes J, Dotan I, Mayer L. Expansion of CD8+ T cells with regulatory function after interaction with intestinal epithelial cells. Gastroenterology. 2002;123:1516–1526. doi: 10.1053/gast.2002.36588. [DOI] [PubMed] [Google Scholar]
- Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ, Powrie F. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman PS, Ernst PB, Rosenthal KL, Clark DA, Befus AD, Bienenstock J. Intraepithelial leukocytes contain a unique subpopulation of NK-like cytotoxic cells active in the defense of gut epithelium to enteric murine coronavirus. J Immunol. 1986;136:1548–1553. [PubMed] [Google Scholar]
- Casey KA, Fraser KA, Schenkel JM, Moran A, Abt MC, Beura LK, Lucas PJ, Artis D, Wherry EJ, Hogquist K, et al. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J Immunol. 2012;188:4866–4875. doi: 10.4049/jimmunol.1200402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauley LS, Lefrancois L. Guarding the perimeter: protection of the mucosa by tissue-resident memory T cells. Mucosal Immunol. 2013;6:14–23. doi: 10.1038/mi.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK, Doherty JM, Mills JC, Colonna M. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M, Otero K, Colonna M. Expansion of human NK-22 cells with IL-7, IL-2, and IL-1beta reveals intrinsic functional plasticity. Proc Natl Acad Sci U S A. 2010;107:10961–10966. doi: 10.1073/pnas.1005641107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepek KL, Shaw SK, Parker CM, Russell GJ, Morrow JS, Rimm DL, Brenner MB. Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the alpha E beta 7 integrin. Nature. 1994;372:190–193. doi: 10.1038/372190a0. [DOI] [PubMed] [Google Scholar]
- Cheroutre H, Lambolez F, Mucida D. The light and dark sides of intestinal intraepithelial lymphocytes. Nat Rev Immunol. 2011;11:445–456. doi: 10.1038/nri3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrier M, Ohnmacht C, Cording S, Eberl G. Development and function of intestinal innate lymphoid cells. Curr Opin Immunol. 2012;24:277–283. doi: 10.1016/j.coi.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Cooper MA, Elliott JM, Keyel PA, Yang L, Carrero JA, Yokoyama WM. Cytokine-induced memory-like natural killer cells. Proc Natl Acad Sci U S A. 2009;106:1915–1919. doi: 10.1073/pnas.0813192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- Cortes M, Wong E, Koipally J, Georgopoulos K. Control of lymphocyte development by the Ikaros gene family. Curr Opin Immunol. 1999;11:167–171. doi: 10.1016/s0952-7915(99)80028-4. [DOI] [PubMed] [Google Scholar]
- Eiras P, Leon F, Camarero C, Lombardia M, Roldan E, Bootello A, Roy G. Intestinal intraepithelial lymphocytes contain a CD3- CD7+ subset expressing natural killer markers and a singular pattern of adhesion molecules. Scand J Immunol. 2000;52:1–6. doi: 10.1046/j.1365-3083.2000.00761.x. [DOI] [PubMed] [Google Scholar]
- Gebhardt T, Wakim LM, Eidsmo L, Reading PC, Heath WR, Carbone FR. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol. 2009;10:524–530. doi: 10.1038/ni.1718. [DOI] [PubMed] [Google Scholar]
- Geremia A, Arancibia-Carcamo CV, Fleming MP, Rust N, Singh B, Mortensen NJ, Travis SP, Powrie F. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J Exp Med. 2011;208:1127–1133. doi: 10.1084/jem.20101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giampieri S, Manning C, Hooper S, Jones L, Hill CS, Sahai E. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol. 2009;11:1287–1296. doi: 10.1038/ncb1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, Lindsten T, Reiner SL. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36:55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Junttila IS, Paul WE. Cytokine-induced cytokine production by conventional and innate lymphoid cells. Trends Immunol. 2012;33:598–606. doi: 10.1016/j.it.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase K, Murakami T, Takatsu H, Shimaoka T, Iimura M, Hamura K, Kawano K, Ohshima S, Chihara R, Itoh K, et al. The membrane-bound chemokine CXCL16 expressed on follicle-associated epithelium and M cells mediates lympho-epithelial interaction in GALT. J Immunol. 2006;176:43–51. doi: 10.4049/jimmunol.176.1.43. [DOI] [PubMed] [Google Scholar]
- Jabri B, Sollid LM. Tissue-mediated control of immunopathology in coeliac disease. Nat Rev Immunol. 2009;9:858–870. doi: 10.1038/nri2670. [DOI] [PubMed] [Google Scholar]
- Jiang X, Clark RA, Liu L, Wagers AJ, Fuhlbrigge RC, Kupper TS. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature. 2012;483:227–231. doi: 10.1038/nature10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin DB, Allan DS, Rybalov B, Andzelm MM, Stern JN, Kopcow HD, Koopman LA, Strominger JL. TGFbeta promotes conversion of CD16+ peripheral blood NK cells into CD16- NK cells with similarities to decidual NK cells. Proc Natl Acad Sci U S A. 2007;104:3378–3383. doi: 10.1073/pnas.0611098104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, Diefenbach A. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334:1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- Koopman LA, Kopcow HD, Rybalov B, Boyson JE, Orange JS, Schatz F, Masch R, Lockwood CJ, Schachter AD, Park PJ, Strominger JL. Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J Exp Med. 2003;198:1201–1212. doi: 10.1084/jem.20030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, Mantovani A, Kopan R, Bradfield CA, Newberry RD, Colonna M. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2012;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois L, Lycke N. Isolation of mouse small intestinal intraepithelial lymphocytes, Peyer’s patch, and lamina propria cells. Curr Protoc Immunol. 2001;Chapter 3(Unit 3):19. doi: 10.1002/0471142735.im0319s17. [DOI] [PubMed] [Google Scholar]
- Lodolce JP, Boone DL, Chai S, Swain RE, Dassopoulos T, Trettin S, Ma A. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity. 1998;9:669–676. doi: 10.1016/s1074-7613(00)80664-0. [DOI] [PubMed] [Google Scholar]
- Male V, Nisoli I, Gascoyne DM, Brady HJ. E4BP4: an unexpected player in the immune response. Trends Immunol. 2012;33:98–102. doi: 10.1016/j.it.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol. 2011;29:71–109. doi: 10.1146/annurev-immunol-031210-101312. [DOI] [PubMed] [Google Scholar]
- Paust S, Gill HS, Wang BZ, Flynn MP, Moseman EA, Senman B, Szczepanik M, Telenti A, Askenase PW, Compans RW, von Andrian UH. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol. 2010;11:1127–1135. doi: 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possot C, Schmutz S, Chea S, Boucontet L, Louise A, Cumano A, Golub R. Notch signaling is necessary for adult, but not fetal, development of RORgammat(+) innate lymphoid cells. Nat Immunol. 2011;12:949–958. doi: 10.1038/ni.2105. [DOI] [PubMed] [Google Scholar]
- Qiu J, Heller JJ, Guo X, Chen ZM, Fish K, Fu YX, Zhou L. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 2012;36:92–104. doi: 10.1016/j.immuni.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romee R, Schneider SE, Leong JW, Chase JM, Keppel CR, Sullivan RP, Cooper MA, Fehniger TA. Cytokine activation induces human memory-like NK cells. Blood. 2012;120:4751–4760. doi: 10.1182/blood-2012-04-419283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathaliyawala T, Kubota M, Yudanin N, Turner D, Camp P, Thome JJ, Bickham KL, Lerner H, Goldstein M, Sykes M, et al. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity. 2013;38:187–197. doi: 10.1016/j.immuni.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan BS, Lefrancois L. Intraepithelial lymphocytes: to serve and protect. Curr Gastroenterol Rep. 2010;12:513–521. doi: 10.1007/s11894-010-0148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shui JW, Larange A, Kim G, Vela JL, Zahner S, Cheroutre H, Kronenberg M. HVEM signalling at mucosal barriers provides host defence against pathogenic bacteria. Nature. 2012 doi: 10.1038/nature11242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A, Andrews DM, Brooks AG, Smyth MJ, Hayakawa Y. Application of CD27 as a marker for distinguishing human NK cell subsets. Int Immunol. 2008;20:625–630. doi: 10.1093/intimm/dxn022. [DOI] [PubMed] [Google Scholar]
- Sips M, Sciaranghella G, Diefenbach T, Dugast AS, Berger CT, Liu Q, Kwon D, Ghebremichael M, Estes JD, Carrington M, et al. Altered distribution of mucosal NK cells during HIV infection. Mucosal Immunol. 2012;5:30–40. doi: 10.1038/mi.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, Fouser LA, Artis D. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. doi: 10.1038/ni.2025. [DOI] [PubMed] [Google Scholar]
- Soudja SM, Ruiz AL, Marie JC, Lauvau G. Inflammatory Monocytes Activate Memory CD8(+) T and Innate NK Lymphocytes Independent of Cognate Antigen during Microbial Pathogen Invasion. Immunity. 2012;37:549–562. doi: 10.1016/j.immuni.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, et al. Innate Lymphoid Cells – a proposal for a uniform nomenclature. Nat Rev Immunol. 2013 doi: 10.1038/nri3365. In press. [DOI] [PubMed] [Google Scholar]
- Strober W, Fuss IJ. Proinflammatory cytokines in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1756–1767. doi: 10.1053/j.gastro.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strowig T, Brilot F, Arrey F, Bougras G, Thomas D, Muller WA, Munz C. Tonsilar NK cells restrict B cell transformation by the Epstein-Barr virus via IFN-gamma. PLoS Pathog. 2008;4:e27. doi: 10.1371/journal.ppat.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Lanier LL. NK cell development, homeostasis and function: parallels with CD8(+) T cells. Nat Rev Immunol. 2011;11:645–657. doi: 10.1038/nri3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliabue A, Befus AD, Clark DA, Bienenstock J. Characteristics of natural killer cells in the murine intestinal epithelium and lamina propria. J Exp Med. 1982;155:1785–1796. doi: 10.1084/jem.155.6.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama T, Kamada N, Chinen H, Okamoto S, Kitazume MT, Chang J, Matuzaki Y, Suzuki S, Sugita A, Koganei K, et al. Imbalance of NKp44(+)NKp46(-) and NKp44(-)NKp46(+) natural killer cells in the intestinal mucosa of patients with Crohn’s disease. Gastroenterology. 2010;139:882–892. 892 e881–883. doi: 10.1053/j.gastro.2010.05.040. [DOI] [PubMed] [Google Scholar]
- Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- Uhlig HH, McKenzie BS, Hue S, Thompson C, Joyce-Shaikh B, Stepankova R, Robinson N, Buonocore S, Tlaskalova-Hogenova H, Cua DJ, Powrie F. Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity. 2006;25:309–318. doi: 10.1016/j.immuni.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Wakim LM, Woodward-Davis A, Liu R, Hu Y, Villadangos J, Smyth G, Bevan MJ. The molecular signature of tissue resident memory CD8 T cells isolated from the brain. J Immunol. 2012;189:3462–3471. doi: 10.4049/jimmunol.1201305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


