Abstract
Background & Aims
Interferon-alfa (IFN)-related cytopenias are common and may be dose-limiting. We performed a genome wide association study on a well-characterized genotype 1 HCV cohort to identify genetic determinants of peginterferon-α (peg-IFN)-related thrombocytopenia, neutropenia, and leukopenia.
Methods
1604/3070 patients in the IDEAL study consented to genetic testing. Trial inclusion criteria included a platelet (Pl) count ≥80 × 109/L and an absolute neutrophil count (ANC) ≥ 1500/mm3. Samples were genotyped using the Illumina Human610-quad BeadChip. The primary analyses focused on the genetic determinants of quantitative change in cell counts (Pl, ANC, lymphocytes, monocytes, eosinophils, and basophils) at week 4 in patients >80% adherent to therapy (n = 1294).
Results
6 SNPs on chromosome 20 were positively associated with Pl reduction (top SNP rs965469, p = 10−10). These tag SNPs are in high linkage disequilibrium with 2 functional variants in the ITPA gene, rs1127354 and rs7270101, that cause ITPase deficiency and protect against ribavirin (RBV)-induced hemolytic anemia (HA). rs1127354 and rs7270101 showed strong independent associations with Pl reduction (p = 10−12, p = 10−7) and entirely explained the genome-wide significant associations. We believe this is an example of an indirect genetic association due to a reactive thrombocytosis to RBV-induced anemia: Hb decline was inversely correlated with Pl reduction (r = −0.28, p = 10−17) and Hb change largely attenuated the association between the ITPA variants and Pl reduction in regression models. No common genetic variants were associated with pegIFN-induced neutropenia or leucopenia.
Conclusions
Two ITPA variants were associated with thrombocytopenia; this was largely explained by a thrombocytotic response to RBV-induced HA attenuating IFN-related thrombocytopenia. No genetic determinants of pegIFN-induced neutropenia were identified.
Keywords: GWAS, ITPA, Thrombocytopenia, Hepatitis C, Neutropenia, IL28B
Introduction
Chronic infection with hepatitis C virus (HCV) affects up to 170 million individuals worldwide [1] and may lead to progressive hepatic fibrosis and cirrhosis with risk of liver failure and hepato-cellular carcinoma. HCV-related liver disease is currently the most common indication for liver transplantation in North America. Antiviral therapy with pegylated-interferon-alfa (pegIFN) plus ribavirin (RBV) may be curative, but is poorly tolerated by many patients.
Bone marrow suppression is an important adverse effect of pegIFN therapy, leading to neutropenia and thrombocytopenia, with risk of secondary sepsis and bleeding, respectively [2,3]. Dose reduction may be required potentially compromising treatment outcome, as rates of viral clearance are significantly reduced in patients who cannot be maintained on at least 80% of their pegIFN and ribavirin dosage for the duration of treatment [4]. Identifying patients at greatest risk for such complications would be clinically useful for selecting patients for therapy, as well as planning the frequency of monitoring and likely need for growth factor support on treatment. Patients with advanced hepatic fibrosis are at highest risk [5], but bone marrow suppression remains prevalent in patients with early stage fibrosis and there is a need for more accurate biomarkers. A genetic biomarker for predicting risk of IFN-related bone marrow suppression would be particularly useful as a pre-treatment test.
A number of lines of evidence suggest that genetic variants may be associated with IFN-induced cytopenia. Firstly, persistently low neutrophil counts are more commonly observed in persons of African American ancestry compared to Caucasians (‘benign ethnic neutropenia’) [6], and this has recently been linked to a regulatory variant in the Duffy Antigen Receptor for Chemokines gene (DARC) [7]. The relevance of this variant to drug-induced neutropenia is not known. Secondly, polymorphism in the region of the interleukin 28B gene (IL28B), coding for IFN-lambda(λ)-3, has recently been identified to be strongly associated with viral clearance following pegIFN plus RBV therapy [8–11]. Although the mechanism remains unclear, the polymorphism is believed to regulate sensitivity to the antiviral effects of IFN. Whether IL28B polymorphism is relevant to other IFN-mediated effects has not been evaluated. Finally, functional variants in the inosine triphosphatase gene (ITPA) causing inosine triphosphatase (ITPase) deficiency, previously recognized as a benign red cell enzymopathy, have recently been identified to protect against RBV-induced hemolytic anemia [12,13]. RBV depletes red cell GTP levels, leading in turn to ATP depletion, oxidative stress, and hemolysis. The protective ITPA variants are associated with red cell inosine triphosphate (ITP) accumulation, and it has been shown that ITP is able to substitute for GTP in the biosynthesis of ATP, thereby protecting against RBV-hemolysis [14].
In this study we have performed genome-wide analyses for determinants of treatment-related bone marrow suppression in a large, well characterized cohort of genotype 1 HCV patients treated with pegIFN plus RBV in the IDEAL study. We have focused primarily on treatment-induced neutropenia and thrombocytopenia.
Materials and methods
Patient and control population
1604/3070 patients in the IDEAL study [15] consented to collection of DNA samples for genetic testing (ClinicalTrials.gov number, NCT00081770). Clinical and laboratory data were collected as described previously [15,16]. All patients included in this study were treatment-naïve and infected with genotype 1 HCV [15]. Patients were treated with either pegIFN-alfa-2b (1.0 or 1.5 μg/kg/week) or pegIFN-alfa-2a (180 μg/week) plus weight-based RBV (800–1400 mg for peg-IFN-alfa-2b, and 1000–1200 mg for pegIFN-alfa-2a) [15]. For all patients, the protocol-specified treatment duration was 48 weeks, with an additional 24 weeks follow-up. All patients had a full blood count performed at baseline, weeks 2, 4, 8, 12, 18, 24, 30, 36, 42, and 48 of therapy and at weeks 4, 12, and 24 post-treatment. Inclusion criteria for the parent study required an absolute neutrophil count (ANC) ≥ 1500/mm3 and platelet count (Pl) ≥80 × 109/L. All patients had compensated liver disease. Protocol specified dose reduction of pegIFN was indicated for ANC <750/mm3 or Pl <50 × 109/L, and discontinuation of both pegIFN and RBV was required for ANC <500/mm3 or Pl <25 × 109/L. The use of growth factor support for neutropenia or thrombocytopenia was not permitted. Detailed records of drug compliance were kept for all patients on-treatment. Only patients who were more than 80% adherent to pegIFN to week 4 of treatment were included in the primary analyses (26 patients were excluded from analysis).
Genotyping
A total of 1604 DNA samples were genotyped in the context of a previously reported study of anti-HCV treatment response, using the Illumina Human610-quad BeadChip (Illumina, San Diego, CA, USA) as previously described [8]. Quality control steps are described in Supplementary Material I. Genotyping of the two ITPA variants, rs1127354 and rs7270101, was performed using the ABI TaqMan allelic discrimination kit (Applied Biosystems, Carlsbad, CA, USA) in a previous study of RBV-induced hemolytic anemia [12,17].
Definition of clinical endpoints
The primary analyses focused on the genetic determinants of quantitative change in (i) platelet, and (ii) leukocyte counts, at week 4 of treatment in adherent patients. The following leukocyte sub-populations were separately analyzed: absolute neutrophil count (ANC), lymphocytes, monocytes, basophils, and eosinophils. Week 4 was chosen as a time point to minimize confounding by dose modification of pegIFN and RBV, or confounding by the use of erythropoietin supplementation.
Statistical analysis
The primary association tests involved single-marker genotype trend tests performed in three independent groups (European-Americans, African-Americans, Hispanics), using a linear regression model. Association tests were implemented in the PLINK software [18], correcting for the relevant clinical covariates baseline cell count (Pl, leukocyte cell lines), age, gender, body mass index, liver fibrosis stage (METAVIR F0–2 vs. F3–4), pegIFN dose (binary variable: pegIFN-α2b 1.0 μg/kg/week vs. pegIFN- α2b 1.5 μg/kg/week and RBV dose (mg/kg). The association signals (p values) were then combined using Stouffer's weighted Z-method [19], adjusting for sample sizes, effect sizes and effect directions in each population. This combined p value was then reported as the main result, along with the p values in each ethnic group. A series of quality control steps resulted in 565,759 polymorphisms being included in the association tests. Methods to assess copy number variants were applied and the relation between copy number variants and reduction of Pl/leukocyte cell lines was tested. To control for the possibility of spurious associations resulting from population stratification, we used a modified EIGENSTRAT method [20] and corrected for population ancestry within each group. We assessed significance with a Bonferroni correction (Pcutoff= 4.4 × 10−8).
Results
Interferon-alfa-mediated thrombocytopenia
We performed a genome-wide association study (GWAS) of genetic determinants of IFN-related thrombocytopenia at week 4 in compliant genotype 1 HCV patients from the IDEAL study. Following quality control steps, 1284 individuals (984 European-Americans, 201 African-Americans, 99 Hispanics) were included in the analysis (patient characteristics are summarized in Table 1). Baseline Pl counts were not significantly different between the 3 populations (p = 0.8977, Table 1). We tested each of 565,759 single nucleotide polymorphisms (SNPs) passing quality control measures in a linear regression model incorporating the relevant clinical covariates: age, gender, body mass index (BMI), hepatic fibrosis stage, pegIFN dose (binary: pegIFN-alfa-2b 1.0 μg/kg/week vs. pegIFN-alfa-2b 1.5 μg/kg/week or pegIFN-alfa-2a 180 μg/week), RBV dose (mg/kg) and baseline Pl count.
Table 1. Patient characteristics.
| European Americans | African Americans | Hispanics | p value | |
|---|---|---|---|---|
| No (platelet analysis) | 984 | 201 | 99 | |
| Gender (n, %) | 608 (62%) | 121 (60%) | 63 (64%) | 0.8387 |
| Age, yrs* | 48 (44-52) | 50 (47-54) | 46 (39-51) | <0.0001 |
| BMI, kg/m2 | 27.4 (24.8-30.4) | 29.3 (26.6-32.6) | 28.6 (25.1-32.8) | <0.0001 |
| METAVIR fibrosis stage (n, %) | 873 (89%) | 183 (91%) | 85 (86%) | 0.3886 |
| Minimal (F0-2) | 111 (11%) | 18 (9%) | 14 (14%) | |
| Advanced (F3-4) | ||||
| RBV starting dose, mg/kg | 13.2 (12.4-14.1) | 12.8 (12.1-13.7) | 13.6 (12.5-14.7) | 0.0004 |
| RBV starting dose (n, %) | 0.0065 | |||
| 800 mg | 86 (9%) | 4 (2%) | 6 (6%) | |
| 1000 mg | 373 (38%) | 65 (32%) | 41 (41%) | |
| 1200 mg | 463 (47%) | 118 (59%) | 44 (44%) | |
| 1400 mg | 62 (6%) | 14 (7%) | 8 (8%) | |
| PegIFN starting dose (n, %) | ||||
| PegIFN-α-2b 1.0 | 332 (34%) | 71 (35%) | 31 (31%) | 0.8532 |
| PegIFN-α-2b 1.5 | 321 (33%) | 62 (31%) | 37 (37%) | |
| PegIFN-α-2a | 331 (34%) | 68 (34%) | 31 (31%) | |
| Baseline Pl count (×109/L) | 225 (184-269) | 228 (184-273) | 230 (186-275) | 0.8977 |
| Baseline Pl count <100×109/L | 17 (1.7%) | 2 (1%) | 1 (1%) | 0.6724 |
| Wk 4 Pl reduction (×109/L) | 37 (11-72) | 28 (0-61) | 26 (2-65) | 0.0052 |
| Wk 4 Pl count (n, %) | ||||
| <75×109/L | 24 (2%) | 4 (2%) | 0 (0%) | 0.2796 |
| <50×109/L | 2 (<1%) | 0 (0%) | 0 (0%) | 0.7369 |
| <25×109/L | 0 (0%) | 0 (0%) | 0 (0%) | 1.0000 |
| No (ANC analysis) | 991 | 203 | 98 | |
| Baseline ANC count (/mm3) | 3.65 (2.96-4.68) | 3.04 (2.14-4.04) | 3.36 (2.77-4.24) | <0.0001 |
| Week 4 ANC reduction (/mm3) | 2.0 (1.34-2.68) | 1.22 (0.61-1.97) | 1.72 (1.0-2.38) | <0.0001 |
| <1.0/mm3 (n, %) | 124 (13%) | 26 (13%) | 12 (12%) | 0.9892 |
| <0.75/mm3 (n, %) | 30 (3%) | 8 (4%) | 1 (1%) | 0.3816 |
| <0.5/mm3 (n, %) | 2 (<1%) | 2 (1%) | 0 (0%) | 0.1588 |
Continuous data are presented as median (25th – 75th centile).
6 SNPs on chromosome 20 were significantly associated with Pl reduction at week 4 (top SNP rs965469, p = 9.02 × 10−10 in European Americans, Fig. 1 and Table 2). These SNPs have previously been shown to co-segregate with 2 functional variants in the ITPA gene on chromosome 20, rs1127354 and rs7270101 (Supplementary Material III), that are each independently associated with reduced ITPase activity and protect against RBV-induced hemolytic anemia (HA) [12]. rs1127354 is a mis-sense variant in exon 2 of the ITPA gene (P32T), and rs7270101 is splicing-altering variant located in the second intron (IVS2). Neither variant was included on the genome-wide array but they had been genotyped in the context of a previous GWAS [12]. These 2 functional variants showed strong independent associations with week 4 Pl reduction (rs1127354, overall p = 10−12 and rs7270101 p = 10−7, respectively, Table 2). The level of ITPase activity may be predicted according to an individual's ITPA genotype, based on previous functional studies (Supplementary Material III), and a combined low activity allele made up of either functional variant may be used to define an ITPase deficiency variable [21–25]. This ITPase deficiency variable was more strongly associated with Pl reduction (p = 10−20). Furthermore, when the two functional ITPA variants were incorporated into a regression model, they were found to entirely explain the genome-wide significant association between rs965469 and Pl reduction (European American patients: p value fell from p = 10−10 to p = 0.9204 after adjustment for the 2 functional variants, Table 2). The functional ITPA variants remained strongly associated with Pl reduction in this model.
Fig. 1. The Manhattan plot shows a genome-wide view of the p values [2log10(P)] for association between SNPs tested and week 4 platelet reduction in patients of European American ancestry.
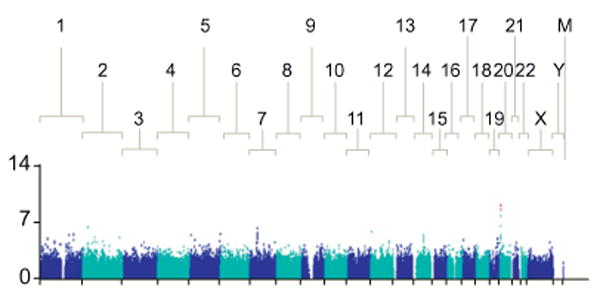
The SNPs that show genome-wide significant association with quantitative reduction in Pl levels are marked in red. [This figure appears in color on the web.]
Table 2. (A) Six variants in the 20p13 were associated with Pl reduction at the genome-wide significant level. These tag SNPs have previously been shown to be in linkage disequilibrium with 2 functional variants in the ITPA gene, which cause ITPase deficiency. (B) The two functional ITPA variants rs1127354 and rs7270101 entirely explained the GWAS association signals detected in the region. The adjusted p value (*) was obtained for each SNP in a linear regression model in which the two ITPA functional variants are incorporated.
| A | ||||
|---|---|---|---|---|
|
| ||||
| Wk 4 Pl reduction | European Americans | African Americans | Hispanics | Combined p value |
| Top discovery SNPs (Illumina 610 chip) | ||||
| rs965469 | 9.02×10−10 | 0.1818 | 0.0792 | 1.29×10−9 |
| rs3310 | 1.30×10−9 | 0.4035 | 0.0816 | 3.91×10−9 |
| rs6051702 | 1.30×10−9 | 0.4621 | 0.0812 | 4.41×10−9 |
| rs6051762 | 2.76×10−9 | 0.5050 | 0.1118 | 1.28×10−8 |
| rs6051841 | 2.16×10−8 | 0.0858 | 0.1424 | 2.09×10−8 |
| rs6051693 | 2.21×10−8 | 0.3207 | 0.0953 | 4.96×10−8 |
|
| ||||
| ITPA variants | ||||
| rs1127354 (P32T) | 1.70×10−10 | 0.0005 | 0.0600 | 1.38×10−12 |
| rs7270101 (IVS2) | 9.95×10−6 | 0.0038 | 0.0231 | 3.39×10−7 |
| ITPase deficiency variable | 2.05×10−16 | 0.00002 | 0.0021 | 8.42×10−20 |
| B | ||||
|---|---|---|---|---|
|
| ||||
| GWAS hit | Population | GWAS p value | Adjusted p value* | |
| rs965469 | European Americans | 9.02×10−10 | 0.9204 | |
| rs3310 | European Americans | 1.30×10−9 | 0.7914 | |
| rs6051702 | European Americans | 1.30×10−9 | 0.7914 | |
| rs6051762 | European Americans | 2.76×10−9 | 0.8065 | |
| rs6051841 | European Americans | 2.16×10−8 | 0.9204 | |
| rs6051693 | European Americans | 2.21×10−8 | 0.8876 | |
Whereas the ITPA variants associated with ITPase deficiency have previously been shown to protect against RBV-induced hemolytic anemia [12], in this study they were associated with more pronounced reduction of Pl counts. The decline in platelet counts that occurs during antiviral therapy is known to be less pronounced when IFN is combined with RBV than in the setting of IFN monotherapy [26,27]. This has been attributed to a relative thrombocytosis occurring in response to RBV-induced hemolysis. In the current cohort, a negative correlation was noted between week 4 hemoglobin (Hb) reduction and Pl reduction (European Americans, r = −0.28, p value = 10−17, Fig. 2). Inclusion of week 4 Hb reduction in the same model with the ITPase deficiency variable largely attenuated the strength of the association with Pl reduction (European Americans, from p = 10−16 to p = 10−6, Supplementary Table 5).
Fig. 2.
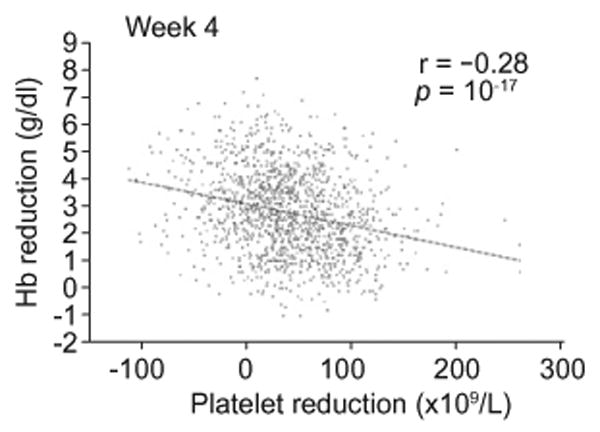
Correlation of Pl reduction at week 4 (× 109/L) with hemoglobin reduction at week 4 (g/dl), limited to the patients of European American ancestry.
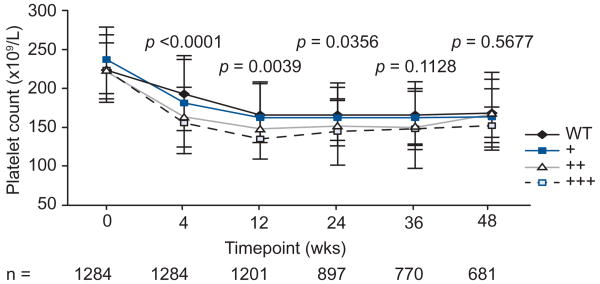
In order to evaluate the clinical relevance of this observation we considered the relationship between the ITPase deficiency variable and reductions of Pl count over the course of therapy. The ITPase deficiency variable was significantly associated with more profound reductions in Pl count at week 4, 12, and 24 (Fig. 3). Beyond week 24, there were non-significant trends in the same direction. Despite this, the number of patients in whom Pl levels fell to below 50 × 109/L, the level at which dose reduction is indicated, was low (<1.5% at any time point) and there were no significant differences in the frequency of Pl <50 × 109/ L according to predicted ITPase deficiency (data not shown). This was true both for the overall cohort, as well as an analysis limited just to those patients treated with pegIFN-alfa-2a 180 μg/week or pegIFN-alfa-2b 1.5 μg/kg/week.
Fig. 3. Median platelet count over time (× 109/L) according to predicted ITPase deficiency in the overall population.
All patients included in the analysis were >80% adherent to week 4 (n = 1284); for time points beyond week 4, patients were included if they remained on treatment, and a platelet count was available. WT = wildtype (normal ITPase activity); + = mild ITPase deficiency; ++ = moderate ITPase deficiency; +++ = severe ITPase deficiency.
Finally, genetic variation in the region of the IL28B gene on chromosome 19 is strongly associated with the pegIFN and RBV response rate [8,10,11,28]. No relationship between IL28B genotype and week 4 thrombocytopenia was noted in the 3 ethnic populations.
Interferon-alfa-mediated neutropenia
We performed a second genome-wide analysis focused on the genetic determinants of week 4 reductions in ANC as a continuous variable. The final analysis included 1292 patients (European Americans = 991, African Americans = 203, Hispanics = 98). At baseline, median ANC were lower in the African American population (European Americans = 3.65 (2.96–4.68), African Americans = 3.04 (2.14–4.04), Hispanics = 3.36 (2.77–4.24), p = 10−12). Median ANC reduction at week 4 was then less prominent in the AA population (European Americans = 2.0 (1.34–2.68), African Americans = 1.22 (0.61–1.97), Hispanics = 1.72 (1.0–2.38), p = 10−18). We tested for genetic determinants of week 4 ANC reduction using linear regression models including the covariates age, gender, BMI, hepatic fibrosis stage (F0–2 vs. F3–4), pegIFN dose (binary: alfa-2b 1.0 μg/kg/week vs. 1.5 μg/kg/week or alfa-2a 180 μg/week) and baseline neutrophil level. No common genetic variants were associated with treatment-related reduction in ANC at week 4 at the level of genome-wide significance. In particular, IL28B polymorphism was not associated with IFN-related neutropenia.
A genome-wide analysis of baseline ANC was also negative. In the AA population, we noted associations between baseline ANC and DARC gene polymorphism but these did not meet genome-wide significance criteria (top SNp rs3027041, p = 10−6, Supplementary Material VI).
Genome-wide analysis for variants associated with other leucopenia
We were also interested in identifying common genetic variants associated with baseline and pegIFN-related week 4 reductions in other white cell counts. Lymphocyte, monocyte, basophil, and eosinophil count were all considered separately. No significant associations were observed in any of these analyses (data not shown).
Discussion
To our knowledge this is the first study to consider genetic determinants of treatment-related cytopenia using a genome-wide approach in chronic hepatitis C patients. We have identified an association between ITPA variants causing ITPase deficiency and treatment related thrombocytopenia. We did not detect any common genetic variants that influenced IFN-related neutropenia or leukopenia, an important negative finding. Of note, IL28B polymorphisms, recently identified to be strongly associated with pegIFN plus RBV treatment outcome, were not associated with IFN-related cytopenia.
Two functional variants in the ITPA gene that cause ITPase deficiency, red cell ITp accumulation and protection against RBV-induced HA [12,14] were associated with more profound pegIFN-induced thrombocytopenia. This association was largely explained by a relative, reactive thrombocytosis in response to RBV-induced HA in those patients with wildtype ITPase activity. Thus the RBV-induced anemia attenuated the pegIFN effect to reduce Pl counts. Thrombocytosis is well-described as a consequence of hemolytic anemia [29], which is in keeping with the original observation in the late 1990s that on-treatment reductions of Pl counts were less marked following the addition of RBV to standard-of-care HCV therapy [26,27]. This therefore represents an indirect genetic association, where wildtype ITPase activity is associated with more profound RBV-related anemia, which in turn stimulates Pl production, manifesting as less pronounced pegIFN-induced thrombocytopenia. The ITPA variants, which protect against RBV-hemolysis, are therefore associated with greater IFN-induced thrombocytopenia. The biological mechanism underlying this relationship between Hb levels and Pl counts is not clearly understood, but may involve stimulation of the bipotent erythroid/megakaryocyte progenitor cell by erythropoietin [30,31]. Although adjustment for Hb reduction in the linear regression model largely attenuated the association between the ITPA variants and Pl counts, a residual association with the combined ‘low activity’ allele persisted (European Americans, p = 10−16 reduced to p = 10−6). Although this association was not genome-wide significant, we cannot exclude the possibility of two separate phenomena, with a weaker secondary effect due to a biological relationship between ITPA variants, exogenous IFN and Pl levels. This will require further mechanistic studies.
Despite the strong statistical association between ITPA variants, Hb reduction and Pl counts, the clinical relevance of this finding remains uncertain. Relatively few patients decreased their Pl counts to levels requiring dose reduction. It is likely that ITPA genotyping may find a role in predicting RBV-induced anemia in high risk individuals [12,13], but on the basis of the current data, there does not appear to be great clinical utility for predicting severe thrombocytopenia. We note that the current dataset did not include significant numbers of patients with advanced stage fibrosis, and it will be important to assess whether ITPA variants may predict treatment-limiting Pl reductions in this population.
No common genetic variants were associated with pegIFN-induced neutropenia or leucopenia. It was interesting that the hematological complications of IFN therapy were not associated with IL28B variants. Although a negative result, this has important implications for our understanding of the biology of the IL28B–pegIFN interaction. The data suggest that the biology of the IL28B–pegIFN treatment response association in HCV is not directly relevant to pegIFN-induced bone marrow suppression. IL28B polymorphism is strongly associated with on-treatment viral kinetics and pegIFN plus RBV treatment outcome [9]. Although the mechanism by which IL28B variation effects pegIFN sensitivity remains unclear, there is evidence that levels of intrahepatic ISG expression are important [32,33] and the effect is believed to primarily reflect sensitivity to exogenous IFN. The current data suggest that this is a liver-specific phenomenon. IFN-λ is induced by similar stimuli to type 1 IFN, and shares a common downstream signaling pathway, however the expression of the IFN-λ-receptor (IFNLR) is more restricted than that of the ubiquitous IFN-α-receptor (IFNABR). Although the IFNLR has been shown to be expressed by hepatocytes, IFNLR gene expression is not expressed in hematopoietic cells, with the exception of B lymphocytes [34,35]. Consistent with this, minimal bone marrow suppression was observed in a recent early phase clinical trial using IFN-λ-1 for the treatment of HCV, despite good antiviral potency [36]. The IL28B polymorphism may therefore act to regulate IFN-α signaling, which is dependent on co-expression of the IFNLR and the IFNABR within the same tissue.
In conclusion, two functional variants in the ITPA gene that are strongly associated with protection from RBV-induced HA are also associated with greater thrombocytopenia in chronic hepatitis C patients. This association is largely explained by a relative reactive thrombocytosis in response to RBV-induced HA, which attenuates IFN-related thrombocytopenia.
Supplementary Material
Acknowledgments
We are indebted to the IDEAL principal investigators, the study coordinators, nurses and patients involved in the study.
Financial support: This study was funded by Schering-Plough Research Institute, Kenilworth, New Jersey. Dr. Thompson received funding support from the Duke Clinical Research Institute, a generous research gift from the Richard B. Boebel Family Fund, the National Health and Medical Research Council of Australia, the Gastroenterology Society of Australia and the Royal Australasian College of Physicians.
Abbreviations
- HCV
hepatitis C virus
- pegIFN
pegylated-interferon-alfa
- RBV
ribavirin
- DARC
Duffy Antigen Receptor for Chemokines
- IL28B
Iinterleukin 28B
- ITPA
inosine triphosphatase gene
- ITPase
inosine triphosphatase
- ANC
absolute neutrophil count
- Pl
platelet
- Hb
hemoglobin
- SNP
single nucleotide polymorphism
- GWAS
genome-wide association study
- BMI
body mass index
- HA
he-molytic anemia
- IFNLR
IFN-λ-receptor
- IFNABR
IFN-α-receptor
Footnotes
Conflict of interest: Drs. McHutchison, Goldstein, Muir, Afdhal, Jacobson, Esteban, Poordad, Lawitz, McCone, Shiffman, King, Kwo, Patel and Sulkowski report having received research and grant support from Schering-Plough. Drs. McHutchison, Goldstein, Muir, Afdhal, Jacobson, Esteban, Poordad, Lawitz, Shiffman, Kwo, and Sulkowski have reported receiving consulting fees or acted in an advisory capacity for Schering-Plough. Drs. Noviello, Pedicone, Brass, Pedicone, and Albrecht are employees of Schering-Plough (now Merck & Co., Inc.) and are stock holders in this entity. Drs. Thompson, Goldstein, McHutchison, Ge, Fellay, Shianna, and Urban are co-inventors of a patent application based on the ITPA finding.
Author contributions: A.J.T. performed the primary data analysis and wrote the first draft of the manuscript with assistance and revision from P.J.C., D.G., A.S., J.F., M.F., Q.Z., A.J.M., and J.G.M. D.G., J.F., K.V.S., T.U., and D.B.G. were responsible for genotyping the ITPA variants. J.G.M. and M.S.S. were the principal investigators for the IDEAL study, and together with D.B.G., S.N., L.D.P., C.A.B., and J.K.A., developed the pharmacogenomic study protocol. A.J.M., N.H.A., I.M.J., F.P., E.J.L., J.M., M.L.S., G.W.G., J.W.K., P.Y.K., and K.P. were site investigators for the IDEAL study. All authors had full access to the data in the study and contributed to the interpretation of the results. All authors reviewed the manuscript and provided further contributions and suggestions. All authors read and approved the final manuscript.
Supplementary data: Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jhep.2011.04.021.
References
- 1.Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29:74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- 2.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Goncales FL, Jr, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 3.Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 4.McHutchison JG, Manns M, Patel K, Poynard T, Lindsay KL, Trepo C, et al. Adherence to combination therapy enhances sustained response in genotype-1-infected patients with chronic hepatitis C. Gastroenterology. 2002;123:1061–1069. doi: 10.1053/gast.2002.35950. [DOI] [PubMed] [Google Scholar]
- 5.Tillmann HL, Patel K, McHutchison JG. Role of growth factors and throm-bopoietic agents in the treatment of chronic hepatitis C. Curr Gastroenterol Rep. 2009;11:5–14. doi: 10.1007/s11894-009-0002-x. [DOI] [PubMed] [Google Scholar]
- 6.Haddy TB, Rana SR, Castro O. Benign ethnic neutropenia: what is a normal absolute neutrophil count? J Lab Clin Med. 1999;133:15–22. doi: 10.1053/lc.1999.v133.a94931. [DOI] [PubMed] [Google Scholar]
- 7.Reich D, Nalls MA, Kao WH, Akylbekova EL, Tandon A, Patterson N, et al. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS Genet. 2009;5:e1000360. doi: 10.1371/journal.pgen.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 9.Thompson AJ, Muir A, Sulkowski M, et al. IL28B polymorphism improves viral kinetics and is the strongest pre-treatment predictor of SVR in HCV-1 patients gastroenterology. 2010. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 10.Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 12.Fellay J, Thompson AJ, Ge D, Gumbs CE, Urban TJ, Shianna KV, et al. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature. 2010;464:405–408. doi: 10.1038/nature08825. [DOI] [PubMed] [Google Scholar]
- 13.Thompson AJ, Fellay J, Patel K, Tillmann HL, Naggie S, Ge D, et al. Variants in the ITPA gene protect against ribavirin-induced hemolytic anemia and decrease the need for ribavirin dose reduction. Gastroenterology. 2010;139:1181–1189. doi: 10.1053/j.gastro.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hitomi Y, Cirulli ET, Fellay J, McHutchison JG, Thompson AJ, Gumbs CE, et al. Inosine triphosphate protects against ribavirin-induced adenosine triphos-phate loss by restoring adenylosuccinate synthase function. Gastroenterology. 2011;140:1314–1321. doi: 10.1053/j.gastro.2010.12.038. [DOI] [PubMed] [Google Scholar]
- 15.McHutchison JG, Lawitz EJ, Shiffman ML, Muir AJ, Galler GW, McCone J, et al. Peginterferon Alfa-2b or Alfa-2a with ribavirin for treatment of hepatitis C infection. N Engl J Med. 2009;361:580–593. doi: 10.1056/NEJMoa0808010. [DOI] [PubMed] [Google Scholar]
- 16.Muir AJ, Bornstein JD, Killenberg PG. Peginterferon alfa-2b and ribavirin for the treatment of chronic hepatitis C in blacks and non-Hispanic whites. N Engl J Med. 2004;350:2265–2271. doi: 10.1056/NEJMoa032502. [DOI] [PubMed] [Google Scholar]
- 17.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitlock MC. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J Evol Biol. 2005;18:1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 20.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 21.Fellay J, Thompson AJ, Ge D, Gumbs CE, Urban TJ, Shianna KV, et al. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature. 2010;464:405–408. doi: 10.1038/nature08825. [DOI] [PubMed] [Google Scholar]
- 22.Sumi S, Marinaki AM, Arenas M, Fairbanks L, Shobowale-Bakre M, Rees DC, et al. Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency. Hum Genet. 2002;111:360–367. doi: 10.1007/s00439-002-0798-z. [DOI] [PubMed] [Google Scholar]
- 23.Shipkova M, Lorenz K, Oellerich M, Wieland E, von Ahsen N. Measurement of erythrocyte inosine triphosphate pyrophosphohydrolase (ITPA) activity by HPLC and correlation of ITPA genotype-phenotype in a Caucasian population. Clin Chem. 2006;52:240–247. doi: 10.1373/clinchem.2005.059501. [DOI] [PubMed] [Google Scholar]
- 24.Maeda T, Sumi S, Ueta A, Ohkubo Y, Ito T, Marinaki AM, et al. Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency in the Japanese population. Mol Genet Metab. 2005;85:271–279. doi: 10.1016/j.ymgme.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Atanasova S, Shipkova M, Svinarov D, Mladenova A, Genova M, Wieland E, et al. Analysis of ITPA phenotype–genotype correlation in the Bulgarian population revealed a novel gene variant in exon 6. Ther Drug Monit. 2007;29:6–10. doi: 10.1097/FTD.0b013e3180308554. [DOI] [PubMed] [Google Scholar]
- 26.Poynard T, Marcellin P, Lee SS, Niederau C, Minuk GS, Ideo G, et al. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. International Hepatitis Interventional Therapy Group (IHIT) Lancet. 1998;352:1426–1432. doi: 10.1016/s0140-6736(98)07124-4. [DOI] [PubMed] [Google Scholar]
- 27.Davis GL, Esteban-Mur R, Rustgi V, Hoefs J, Gordon SC, Trepo C, et al. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. International Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1493–1499. doi: 10.1056/NEJM199811193392102. [DOI] [PubMed] [Google Scholar]
- 28.Thompson AJ, Muir AJ, Sulkowski MS, Ge D, Fellay J, Shianna KV, et al. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;139:120–129. e118. doi: 10.1053/j.gastro.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 29.Schafer AI. Thrombocytosis. N Engl J Med. 2004;350:1211–1219. doi: 10.1056/NEJMra035363. [DOI] [PubMed] [Google Scholar]
- 30.Cardier JE, Erickson-Miller CL, Murphy MJ., Jr Differential effect of erythropoietin and GM-CSF on megakaryocytopoiesis from primitive bone marrow cells in serum-free conditions. Stem Cells. 1997;15:286–290. doi: 10.1002/stem.150286. [DOI] [PubMed] [Google Scholar]
- 31.Broudy VC, Lin NL, Kaushansky K. Thrombopoietin (c-mpl ligand) acts synergistically with erythropoietin, stem cell factor, and interleukin-11 to enhance murine megakaryocyte colony growth and increases megakaryocyte ploidy in vitro. Blood. 1995;85:1719–1726. [PubMed] [Google Scholar]
- 32.Honda M, Sakai A, Yamashita T, Nakamoto Y, Mizukoshi E, Sakai Y, et al. Hepatic ISG expression is associated with genetic variation in IL28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology. 2010;139:499–509. doi: 10.1053/j.gastro.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 33.Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, Cronin KD, et al. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010;52:1888–1896. doi: 10.1002/hep.23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doyle SE, Schreckhise H, Khuu-Duong K, Henderson K, Rosler R, Storey H, et al. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology. 2006;44:896–906. doi: 10.1002/hep.21312. [DOI] [PubMed] [Google Scholar]
- 35.Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muir AJ, Shiffman ML, Zaman A, Yoffe B, de la Torre A, Flamm S, et al. Phase 1b study of pegylated interferon lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology. 2010;52(3):822–832. doi: 10.1002/hep.23743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.