Abstract
Internalization of G-protein coupled receptors is mediated by phosphorylation of the C-terminus, followed by binding with the cytosolic protein arrestin. To explore structural factors that may play a role in internalization of cannabinoid receptor 1 (CB1), we utilize a phosphorylated peptide derived from the distal C-terminus of CB1 (CB15P454-473). Complexes formed between the peptide and human arrestin-2 (wt-arr21-418) were compared to those formed with a truncated arrestin-2 mutant (tr-arr21-382) using isothermal titration calorimetry and nuclear magnetic resonance spectroscopy. The penta-phosphopeptide CB15P454-473 adopts a helix-loop conformation, whether binding to full-length arrestin-2 or its truncated mutant. This structure is similar to that of a hepta-phosphopeptide, mimicking the distal segment of the rhodopsin C-tail (Rh7P330-348), binding to visual arrestin, suggesting that this adopted structure bears functional significance. Isothermal titration calorimetry (ITC) experiments show that the CB15P454-473 peptide binds to tr-arr21-382 with higher affinity than to the full-length wt-arr21-418. As the observed structure of the bound peptides is similar in either case, we attribute the increased affinity to a more exposed binding site on the N-domain of the truncated arrestin construct. The transferred nOe data from the bound phosphopeptides are used to predict a model describing the interaction with arrestin, using the data driven HADDOCK docking program. The truncation of arrestin-2 provides scope for positively charged residues in the polar core of the protein to interact with phosphates present in the loop of the CB15P454-473 peptide.
Arrestins are cytosolic proteins that regulate the functioning of G-protein coupled receptors (GPCRs) by binding to ligand stimulated, and phosphorylated forms of the receptors (1, 2). This results in attenuation of G-protein mediated signaling and the internalization of GPCRs, with arrestin acting as a scaffold for endocytic proteins (1, 3-5). Arrestin may also facilitate signaling through pathways that are independent of G-protein activation (6, 7). Recent studies have utilized biased GPCR ligands (8-10) to show that different bound conformations of arrestin may be responsible for distinct functional outcomes (11). It has been postulated that this type of directed agonism may be due to ligand stabilization of specific GPCR conformations that promote distinct, and functionally specific, conformations in the bound arrestins (11, 12). There is evidence that the conformation of arrestin changes upon binding a receptor, however, there is limited direct structural detail available for complexes between GPCRs and arrestins(13, 14).
A key stage in the formation of a complex is thought to involve phosphorylated segments of the GPCR C-terminus and the N-domain of arrestin (15, 16). The available arrestin crystal structures show that residues near the carboxy-terminal end of the protein interact with a site on the N-domain. This association of the C-terminus of arrestin with the N-domain is thought to stabilize a basal state of the protein(17-22). In this state, arrestin exhibits a high affinity for GPCRs that are both activated by a ligand, and phosphorylated, in what is likely a multisite interaction (15, 16, 23, 24). Arrestins in this state have a far lower affinity for receptors that are activated by a ligand but unphosphorylated, or, phosphorylated and in an inactive state. Truncation of the arrestin carboxy-terminus results in a partially active mutant that binds with higher affinity to GPCRs that are phosphorylated but have not been activated by a ligand (25).
A number of studies utilizing peptides and mutagenesis have placed the phosphate sensitive elements of arrestin in the N-domain of the protein (19, 26-28). Furthermore, a peptide mimicking the distal rhodopsin C-terminus was found to adopt a helix-loop conformation upon binding to arrestin-1, however, there is little structural information with respect to other receptors and arrestins(29, 30). In this study, we have employed cannabinoid receptor 1 (CB1) as a model GPCR to probe structural factors involved in the arrestin2-GPCR interaction. Cannabinoid receptors are G-protein coupled receptors (GPCRs) that have been extensively targeted for therapeutic benefit (31-35). A number of studies have shown that CB1 exhibits a complex mechanism of activation that may extend to the nature of the interaction with arrestin (11, 36-38). It was observed in AtT20 cells that phosphorylation of the distal C-terminal tail of CB1 from Thr460-Leu473 regulates internalization (39-41). By contrast phosphorylation at Ser426 and Ser430, located upstream of the distal segment, mediated desensitization while having no effect on internalization (39, 40). In subsequent experiments using HEK293 cells, this delineation between internalization and desensitization was not as prevalent (41, 42). Nevertheless the distal C-terminal segment of CB1 was shown to play a key role in limiting internalization when unphosphorylated and allowing internalization to proceed upon phosphorylation (41).
Several studies have addressed structural aspects of the CB1 C-terminus. The formation of a helix shortly after the C-terminal end of the conserved NPxxY motif and proximal to the membrane surface is commonly referred to as helix eight, or as the fourth cytoplasmic loop. It is a common feature of rhodopsin like GPCRs (43-47) and has been implicated in ligand binding, signal transduction as well as processes such as phosphorylation and desensitization (48-50). In the case of CB1, the NPxxY motif may form a microdomain that couples helix eight to conformational changes within the trans-membrane helix bundle (44, 51, 52). A study of the C-terminus of CB1 in membrane mimetic media has identified the possible formation of a second amphipathic helix downstream of helix eight (53). There is, however, little information concerning the effects of phosphorylation and the nature of complexes formed with cytosolic partners.
The peptide we utilize in this study mimics the distal section of the C-terminus where no structure has been observed in solution(53). In a previous study we showed that the doubly phosphorylated version of a peptide corresponding to residues Thr419-ASN439 of the CB1 C-terminus, responsible for desensitization, binds to arrestin-2 and adopts helical conformations in the vicinity of the phosphorylated residues (54). This prompted us to study a phosphorylated peptide corresponding to the residues between Thr454 and Leu473 of the CB1 receptor shown to influence internalization (41). We examine the effects of phosphorylation on the solution structure of CB15P454-473 and compare the binding of this peptide to full-length arrestin-2 versus its truncated mutant.
Experimental Procedures
Peptide synthesis
The non phosphorylated peptide TVKIAKVTMSVSTDTSAEAL (CB1454-473) and a pentaphophorylated version, TVKIAKVT(p)MS(p)VS(p)TDT(p)S(p)AEAL1 (CB15P454-473), derived from the distal C-terminal sequence of the human CB1 receptor were obtained from New England Peptides at > 95% purity as shown by HPLC and MALDI-TOF mass spectral analysis.
Expression of arrestin-2 and a truncated mutant of arrestin-2
For expression of arrestin-2, we modified the pTrcHis vector (Invitrogen) by digesting with NcoI and BamH1 to remove the hexa-histidine tag and then blunted the ends with klenow polymerase to ensure that the start codon would be regenerated upon religation. We refer to this modified vector as pTrcK. All vectors and constructs were verified for integrity by sequencing (University of Connecticut Biotechnology Center, Storrs, CT).
The plasmids were transformed into Escherichia coli strain BL21 (Invitrogen) following standard procedures as outlined by the vendor. To express arrestin-2, 1 litre of LB medium was inoculated with 10 ml of overnight cell culture and induced at 30° C with 300 μM isopropyl-1-thio-β-D-galactopyranoside at a cell density of 0.6. After a twelve-hour incubation, cells were harvested by centrifugation at 7500 rpm and resuspended in lysis buffer containing 20 mM Tris, pH 8.0, 0.5 mM PMSF, 2 mM DTT, 1 mg/ml lysozyme, 10 mM EDTA and 0.2 mg/ml benzamidine. The presence of arrestin-2 was determined by SDS-PAGE and immuno-gel blot analysis using a commercially available antibody raised against an epitope found on arrestin-1 that is also present on arrestin-2 (Sigma).
Protein Purification
Purification of wt-arr21-418 and tr-arr21-382 followed methods as elaborated in several publications that involved ammonium sulfate precipitation of the cell lysate followed by affinity chromatography on a heparin column and size exclusion chromatography (18, 21, 55). Briefly, re-suspended cells were lysed using a French pressure cell and the lysate centrifuged at 12000 rpm for 30 min. Ammonium sulfate was added to the supernatant at 4° C to attain 55% saturation in small lots over 45 mins and the resultant precipitate was collected after 30 mins by centrifugation. The precipitate was re-dissolved and dialyzed against the heparin column load buffer containing 10 mM Tris pH=8.0, 0.5 mM PMSF, 2 mM DTT, 100 mM NaCl and 10% glycerol. The heparin column wash and elution buffers differed only in salt concentration (200 mM & 500 mM NaCl respectively). 12 ml of Pharmacia Heparin Fast Flow resin was washed with load buffer and equilibrated against the protein solution with gentle shaking at 4° C for two hours. The column was washed with 200 mM NaCl and eluted with 500 mM NaCl. The elutions were further subjected to size exclusion chromatography by loading them onto a pre-packed Amersham HiLoad 16/60 Superdex 75 pg column using an AKTA FPLC. The protein was eluted with a buffer containing 10 mM NaH2PO4 pH=7.2, 100 mM NaCl and 2 mM DTT. The identities of the purified proteins were confirmed by immuno-blot analysis using a commercially available antibody raised against an epitope found on arrestin-1 that is also present on arrestin-2 (Sigma). CD spectra of the purified arrestin-2 in 0.1 M sodium phosphate pH = 7.2 were recorded on a JASCO J-715 spectrometer over the range of 190-250 nm. NMR samples were prepared from these elutions by concentrating to a concentration of 0.1 mM protein using Millipore centricon tubes with 10 kDa molecular weight cutoff.
NMR analysis of CB1454-473, CB15P454-473 and arrestin-2-peptide mixtures
Peptide samples of CB1454-473 and CB15P454-473 were dissolved at 1 mM concentration in 350 μL of a buffer containing 10 mM NaH2PO4/Na2HPO4 pH=7.0, 100 mM NaCl and 10% D2O. DSS was used as the internal chemical shift reference for NMR experiments. For preparation of the arrestin-peptide mixtures, a concentrated peptide solution (10 mM PO4, 100 mM NaCl) was added to 270 μL of a wt-arr21-418 or tr-arr21-382 solution, prepared in buffer containing 10 mM NaH2PO4 pH=7.0, 100 mM NaCl and 10% D2O. The final concentration of the arrestins and peptides were 0.1 mM and 1 mM respectively (ratio of 1:10) and the final pH of the mixtures was measured at 6.9-7.1.
NMR spectra were acquired on a Varian Unity 600 MHz spectrometer equipped with a cryoprobe. TOCSY spectra (56) were collected using DIPSI spinlock (57) and watergate water suppression (58) with mixing time of 80 ms. WATERGATE-NOESY spectra were acquired with a mixing time of 200, 300, and 400 ms. Spectra were recorded with 2400 data points, 256 increments and relaxation delay of 1s. Spectra were processed using NMRPipe (59).
NMR analysis was accomplished using the Sparky NMR assignment program (http://www.cgl.ucsf.edu/home/Sparky). The chemical shift assignments of the peptides were obtained by analysis of TOCSY and NOESY spectra obtained at 10°C using standard techniques (60). TRNOESY experiments performed with mixing times of 400 ms at 10°C were analyzed using the SPARKY program as no spin diffusion was observed at this mixing time. The nOes were classified into strong, medium and weak based on peak volume calculated in SPARKY. The distance range of 1.8-2.8 Å for strong, 1.8-3.5 Å for medium and 1.8-5.0 or 6.0 Å for weak nOes was given (61, 62). The upper limits for distances involving methyl protons were increased by an additional 0.5 Å (63, 64).
Structure Calculations and Docking Studies
Structure calculations from NOESY distance restraints were carried out using nOe tables as described in the results section and a hybrid distance geometry simulated annealing protocol implemented in CNS, version 1.2 (65, 66). Starting structures were generated using NIH-XPLOR scripts (67) and the required topology and parameter files for phosphorylated residues were generated using the PRODRG server (68). Low energy structures were subjected to the water refinement protocol of CNS. Validation of structural geometry and visualization of structures were carried out using PROCHECK (69) and VMD-XPLOR programs (70).
For a better understanding of the interaction between the phosphorylated peptide, CB15P454-473, and wt-arr21-418 or tr-arr21-382, we generated models of the complexes. This was accomplished by docking families of low energy structures of CB15P454-473 onto the crystal structure of bovine arrestin-2 using the data driven HADDOCK program (71). We docked the CB15P454-473 structures to full-length arrestin-2 to explore binding sites available in the basal state of arrestin-2. Full-length arrestin-2 (wt-arr21-418) contains the C-tail from residues 383-418, a segment of which presumably dissociates from the N-domain upon binding to phosphorylated GPCRs. To account for this conformational change that exposes important residues in the N-domain of arrestin, CB15P454-473 was also docked onto a structure where C-terminal residues down to Asp383 were deleted.
Isothermal Titration Calorimetry (ITC) Experiments
ITC experiments were performed using purified wt-arr21-418 and tr-arr21-382 against CB1454-473 or CB15P454-473 in 10 mM NaH2PO4 pH 7.0 and 100 mM NaCl. All ITC measurements were performed at 283K in a VP-ITC isothermal titration calorimeter (Microcal, Northampton, MA). The sample cell (1.4 mL) was loaded with arrestin-2 (0.01 mM) and a peptide (0.5 mM) solution was loaded into the syringe. A total of 13 additions of 15 ul each were made sequentially to the sample cell containing arrestin-2. Each addition was made 210 seconds apart to ensure the titration peak returned to the baseline before the next addition. The amount of power required for maintaining the reaction cell at constant temperature after each injection was monitored as a function of time. The peptide was injected until a 2-fold excess of peptide to arrestin-2 was achieved. As a control experiment, the individual dilution heats for the peptide CB15P454-473 were determined under the same experimental conditions by carrying out identical injections of CB15P454-473 into 10 mM NaH2PO4 (pH 7.0), 100 mM NaCl buffer solution. Data were collected in high feedback mode with a filter period of 2 s and analyzed using ORIGIN 7.0 (Microcal, Northhampton, MA). The isotherms best fitted a one-site binding model assuming a 1:1 stoichiometry.
RESULTS
Expression and purification of arrestin-2
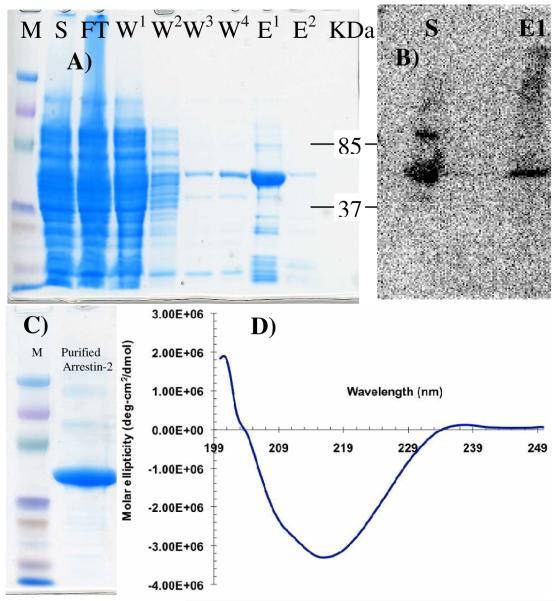
Our purification procedure was equally successful for both wild type and truncated versions of arrestin. The typical results from SDS-PAGE and western blot analysis of elutions from the heparin column are shown in figure 1. Reasonable purity is achieved from the heparin column purification step and the subsequent gel filtration step produced highly purified samples estimated at > 95% as shown by SDS-PAGE analysis (Fig. 1). The identity of the arrestin-2 bands was confirmed in western blots (Fig. 1). The CD spectrum is consistent with literature CD spectra recorded of bovine arrestin that are dominated by the beta sheet structure of the protein and a shoulder near 222 nm is evidence of the presence of some alpha-helical character as expected in the N-domain of the protein (Fig. 1D).
Figure 1.
A) SDS PAGE analysis of fractions obtained from heparin column purification of arrestin. The supernatant (S), flow through (FT), washes (W1-4) and Elutions (E1-2) are shown. B) Western blot analysis of supernatant (S) and Elution1 (E1) from panel A. C) SDS PAGE analysis of gel filtration purified E1 from panel A. The overloaded lane shows a clean product. D) CD spectrum of wt-arr21-418 showing a predominance of beta-sheet and some evidence of alpha-helical character.
Structural Analysis of Free peptides
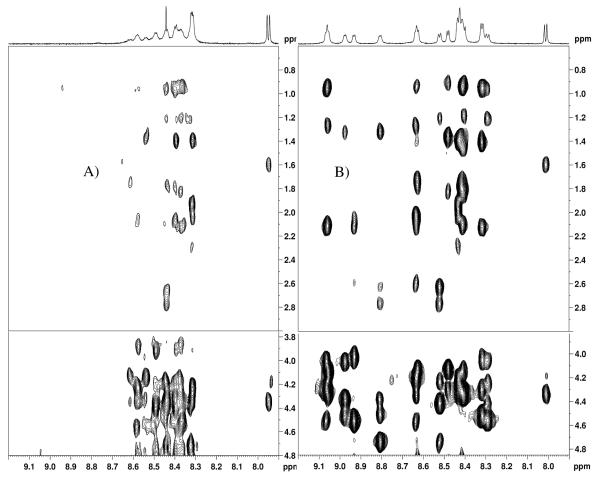
The amide 1H spectral regions of the peptides, CB1454-473 and CB15P454-473 in H2O/10%D2O are shown in figure 2. The unphosphorylated peptide shows a great degree of spectral overlap whereas the spectrum of the pentaphosphorylated peptide is more dispersed. As a result, assignment of the unphosphorylated peptide CB1454-473 was not possible. Side chain spin systems were also overlapped and in cases where the spin system of a residue such as serine was distinguishable it was not possible to assign the spin system to a specific residue. The NOESY spectra of CB1454-473 (Fig. 2), even at long mixing times, showed weak nOes. The fact that resonances do not occur over a wider frequency range combined with weak nOes suggests that the free peptide CB1454-473 is very flexible in solution and adopts a random coil structure.
Figure 2.
Fingerprint and side chain regions of the 1H NOESY spectra, with 1D projections, recorded at 10°C of A) CB1454-473, B) CB15P454-473 in 10 mM phosphate buffer, 100 mM NaCl at pH=7.0. The following long range nOes were observed: Thr461:CH3-Ser463:HN, Thr461:HN-Ser463:HN, Thr461:CH3-Val464:CHβ, Thr461:CH3-Val464:HN, Val464:CH3-Asp467:HN, Ser465:CHα-Asp467:HN, Thr466:HN-Thr468:HN, Met462:CHβ2-Thr468:HN
The spectrum of the pentaphosphorylated peptide shows great dispersion in the amide region that is critical for analysis(Fig. 2). A sequential nOe pattern was observed in the NOESY spectrum of free CB15P454-473 that allowed full assignment of the spectrum. Several (i,i+2), (i,i+3), and one (i,i+4) nOe between backbone and side chain atoms in the region from residue Thr461 to Thr468 were also observed (Fig 2). These suggest slowed dynamics of the peptide resulting in a turn in this region of the peptide as shown in figure 3.
Figure 3.
(A) Ensemble of five low energy structures of free CB15P454-473 calculated using long range nOes. These demonstrate a persistent turn in the peptide. B) Stick representation of CB15P454-473 highlighting intramolecular hydrogen bonding that may serve to restrict the flexibility of the peptide in solution.
Isothermal titration calorimetry of complexes
Using isothermal titration calorimetry (ITC), we attempted a thermodynamic characterization of the interaction between the peptides and wt-arr21-418 or tr-arr21-382. To characterize these lower affinity interactions we titrated a fifty fold concentration of peptide relative to arrestin. The unphosphorylated peptide showed no signs of interaction. For the pentaphosphorylated peptide, the binding was exothermic as shown for CB15P454-473 vs tr-arr21-382 in figure 4. The data fit a single-site model assuming 1:1 stoichiometry with Kd values of approximately 116 μm for the wt-arr21-418 complex and 2.2 μm for the tr-arr21-382 complex (Fig 4).
Figure 4.
Titration of the truncated arrestin with CB15P454-473, showing the calorimetric response as successive injections of ligand are added to the reaction cell. Panel B depicts the binding isotherm of the calorimetric titration shown in panel A. The continuous line represents the least-squares fit of the data to a single-site binding model.
Structural analysis of peptide binding to wt-arr21-418 and tr-arr21-382
When either wt-arr21-418 or tr-arr21-382 was added to a solution of CB1454-473, there are no additional peaks in the nOe spectra compared to the free peptide spectrum suggesting little or no interaction. However, when CB15P454-473, is in the presence of either wt-arr21-418 or tr-arr21-382, there are increases to both the intensity and number of nOes (Fig. 5). This observation is evidence for the formation of complexes between CB15P454-473 and the arrestin constructs, with fast-intermediate exchange rates on the NMR time scale. This fast exchange phenomenon is consistent with the results from the isothermal titration calorimetry experiments.
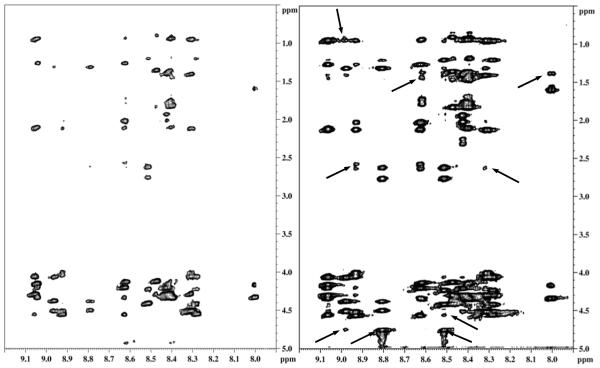
Figure 5.
2D transferred NOESY spectrum of A) Unbound 1.0 mM CB15P454-473 B) Mixture of 1.0 mM CB15P454-473 with 100 μM human arrestin-2 showing the presence of additional chemical exchange peaks. Arrows indicate short and medium range interactions and circle indicates a long range interaction in this region.
The nOes observed for CB15P454-473 in complex with each arrestin construct are summarized in figure 6. A total of 226 and 252 nOes were used for structure calculations in the case of wt-arr21-418 and tr-arr21-382 respectively. In the first step, twenty substructures were calculated by distance geometry starting from an extended random coil conformation of CB15P454-473. The distance restraints were used as input to calculate five hundred substructures with a hybrid distance geometry-simulated annealing protocol. The resulting substructures were further subjected to a simulated annealing protocol where structures were heated to 2000 K for 3 ps in 1000 steps, and then slowly cooled to 0 K for 5 ps in 1000 steps. During the cooling stage, the force constant on the van der Waals repulsion term was varied from 0.003 to 4 kcal mol−1 Å−4. In the next stage, structures were minimized. The force constants on the dihedral angle restraints were 200 kcal mol−1 rad−2 during molecular dynamics and 400 kcal mol−1 rad−2 during energy minimization. The force constant on nOe restraints was 50 kcal mol−1 Å−2 throughout all calculations. The CNS task file accept.inp, identified two hundred and eighty accepted structures satisfying experimental restraints and local geometry. Fifteen structures with lowest energies out of 180 accepted structures were selected as final structures and subjected to the water refinement protocol of CNS. Analysis and stereochemical evaluation of final structures were performed by PROCHECK (69) and VMD-XPLOR programs (70). An overlay of the six lowest energy structures for arrestin-2 bound CB15P454-473 that satisfied nOe restraints is shown in figure 7.
Figure 6.
nOe interactions observed for CB15P454-473 bound to A) wild-type arrestin-2 and B) truncated arrestin-2. Long range nOes included Met462:CHβ2-Thr468:HN and Met462:-Glu471:HN. Ambiguous nOes observed included, Thr454:CH3-Ser469:CH2, Val455:CH3-Asp467:HN Val455:CH3-Ser469:HN.
Figure 7.
A) Ensemble of the six low energy structures of full-length arrestin-2 bound CB15P454-473 with backbone superimposition. B) Lowest energy structure with phosphorylated residues shown as ball and stick C) Ramachandran plot and the structural statistics of bound CB15P454-473.
The sequential nOe patterns of weak dN-N(i,i+1) and strong dα-N(i,i+1), observed from residue Val455 to Thr461 is indicative of extended backbone structure. In each of the bound CB15P454-473 structures calculated for each arrestin construct, there is a loop formed in the peptide from Thr461-Asp466. This stable loop is supported by medium range (i, i+2) and (i, i+3) nOes, in combination with longer range nOes observed between the sidechain of Met462 to the amide protons of Thr468 and Glu471 in the wt-arr21-418 complex (Fig 6). These long range nOes did not appear in the tr-arr21-382 complex however the shorter range nOes consistently resulted in the formation of a loop in this region. The number and intensity of dα-N(i,i+2), dα-N(i,i+3), dβ-N(i,i+3) dN-N(i,i+1), and dN-N(i,i+2) nOes in the C-terminal region of CB15P454-473 from residue Asp467 to Glu471 revealed the presence of a helical conformation. The helix loop structure is similar to that observed for a nineteen residue distal fragment of the rhodopsin receptor binding to arrestin-1. In that case there were nOes between residues near the N- and C-termini of the peptide that constrained the structure of the loop. In our results, any such potential long range nOes were ambiguous due to signal overlap (Fig 6). Therefore, we cannot confirm whether the structure of the peptide is such that the extended N-terminus is constrained in a position that places the N- and C-termini near each other. Nevertheless our results do confirm the presence of a helix at the C-terminus and a stable turn in the middle of the peptide loop across the residues Thr461-Asp466.
Docking of CB15P454-473 to wt-arr21-418 and tr-arr21-382
Ambiguous interaction restraints (AIRs) were generated based on mutation data in conjugation with the coordinate files of arrestin-2 and the peptide. According to criteria of the Haddock program, the “active” residues are those that have been shown by mutations to abolish or perturb complex formation and are solvent exposed. Based on our result that truncation of the C-terminus results in an increase in affinity of the peptide, we focused our attention to the N-domain region with which the C-terminus of arrestin interacts. For arrestin-2, positively charged residues in the N-domain were defined as active residues since mutagenesis studies have shown that they interact with phosphates on the C-tail of receptor (28, 72). The surrounding residues were chosen as “passive” residues (Fig. 8), which are defined as residues that are solvent exposed neighbors of active residues. For the peptide, phosphorylated residues were chosen as active and all other residues were chosen as passive. Relative solvent accessibility of all active and passive residues was more than 60% as determined by the NACCESS program (73). The HADDOCK docking program generated the lowest energy structure via a three-stage process: an initial rigid-body docking to generate 1000 structures, followed by a semi-flexible simulated annealing in torsion angle space of 200 best structures in terms of intermolecular energy (sum of van der Waals, electrostatic, and ambiguous interaction restraints energy terms). Finally, water was included in the calculation to improve the energy of structures. The solutions were then clustered using a 3.5 Å rmsd cut-off and ranked according to their average interaction energies (sum of Eelec, Evdw, EACS) and their average buried surface area according to the HADDOCK protocol. The structures with the lowest energy and the greatest buried surface area were taken as the best fits as shown in figure 7. The lowest energy structure for CB15P454-473-wt-arr21-418 complex had total energy −1027.51kcal/mol and buried surface area of 1314.49 Å2. The lowest energy structure for the CB15P454-473-tr-arr21-382 complex had a total energy −1332.83 kcal/mol and buried surface area of 1750.49 Å2.
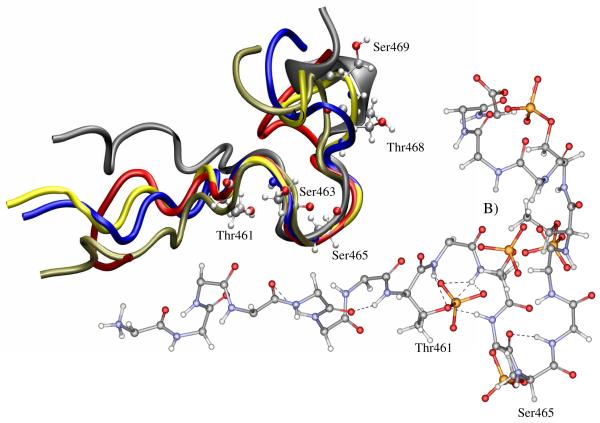
Figure 8.
A) Ribbon representation of CB15P454-473, colored in blue (at the N-terminus) to red (at the C-terminus) binding the wild type arrestin colored in green. B) Ribbon representation of CB15P454-473 shown binding to a surface representation of N-domain binding site highlighted in green. The blue line highlights the placement of the C-terminus residues that occlude the binding cavity. The positively charged residues in the binding site are labeled and colored in red. Interactions with phosphorylated residues Thr460, Ser463 and Ser465 of the peptide are labeled a, b and c respectively. C) Ribbon representation of CB15P454-473, colored in blue (at the N-terminus) to red (at the C-terminus) binding the truncated arrestin colored in green. Note the absence of the C-terminus. D) Ribbon representation of CB15P454-473 shown binding to a surface representation of the N-domain binding site highlighted in green. The positively charged residues in the binding site are labeled and colored in red. Interactions with phosphorylated residues Thr460, Ser463 and Ser465 of the peptide are labeled a, b and c respectively. The list of active and passive residues chosen to define the binding site in HADDOCK is shown in the table.
Discussion
Structure of Free Peptides
The peptide CB1454-473 mimics the distal section of the cannabinoid receptor that, upon phosphorylation, mediates internalization (39). The peptide, displayed little evidence of structure in aqueous solution as the spectrum showed poor dispersion of peaks and a low number of nOe peaks at longer mixing times, and at lower temperatures. This correlates with a recent NMR structure of a segment derived from the entire C-tail of CB1 that found little structure in aqueous solution (53).
Upon phosphorylation of the peptide we find good dispersion of resonances and more intense nOes that allowed us to assign and calculate its structure. We find evidence that the peptide is more conformationally restricted, resulting in the formation of a turn in the peptide between residues Thr460-Ser466. Intramolecular hydrogen bonding in the phosphorylated peptide may play a role in this decreased flexibility of the peptide as, shown in figure 3. In this case, the attached phosphate group of Thr460 allows hydrogen bonding with both Ser463 and Val464. There are no nOes that support structure at either the N or the C-terminus of the peptide, and this is most likely due to increased solvent exposure, and fraying at the peptide termini. However, at the C-terminus, where there is an increased level of phosphorylation, there is an increase in the number and intensity of NN(i,i+1) nOes, suggest that the C-terminus of the peptide is more conformationally restricted.
The region in which the data suggests a turn in the peptide, contains the section of the CB1 C-tail, Thr461-Ser463, shown to affect internalization (39-41). A study of the CB1 C-terminus in membrane mimetic media identified the formation of an amphipathic helical region downstream of helix eight, that overlaps with the N-terminus of the CB15P454-473 peptide. This helical structure terminates at the methionine residue within the Thr461-Ser463 region (53). We do not observe any structure at the N-terminus of the CB15P454-473 peptide however our results permit the speculation that an amphipathic helix may be formed when the C-terminus is in close proximity to the membrane surface. Phosphorylation of residues in the distal C-terminus may destabilize this helix and cause the C-terminus to dissociate from the surface of the membrane. This may allow an interaction with arrestin to proceed.
Numerous studies have shown that affinity for arrestin is driven by phosphorylation of the GPCR. Moreover, it has been shown that at least two phosphorylated residues are required in the distal C-terminus for internalization to take place (41). We have observed evidence for the formation of a turn in a doubly phosphorylated version of CB1454-473 (data not shown). The fact that nOes suggesting a turn persist in both the free and the bound structure of CB15P454-473, at a region that mediate desensitization, suggests that it may be important for the formation of a complex that leads to internalization.
Binding of CB15P454-473 to Arrestin
The Transferred NOESY method results in an increase in the number and intensity of nOe cross peaks when a peptide and a large protein interact, due to chemical exchange between the peptide in the free and protein bound state (74). The results of ITC experiments showed no indication of an interaction between the unphosphorylated peptide and wt-arr21-418 or tr-arr21-382. This correlates with the fact that transferred nOes were not observed for mixtures of the CB1454-473 peptide with either of the arrestin constructs. When the phosphorylated peptide CB15P454-473 was mixed with either of the arrestin constructs, a number of transferred nOes were observed (Fig. 5 & 6). The solution structures of the peptide binding to wt-arr21-418, and to tr-arr21-382, were calculated independently. In each case, CB15P454-473 binds in a conformation that has a helical region in the C-domain from residues Asp467 to Glu471, a turn from residues Met462 to Thr466, and an extended region in the N-terminus from residues Val455 to Thr461. The structural statistics of bound CB15P454-473 to wt-arr21-418 are shown in figure 7, with similar results obtained for binding to tr-arr21-382.
The helix loop conformation we observe resembles the conformation of a peptide mimicking a segment of the rhodopsin receptor (Rh330-348) binding with visual arrestin (or arrestin-1) (30). As in this study, the Rh330-348 peptide was a mimic for the distal C-terminus of the rhodopsin receptor, however, the amino acid sequences and the phosphorylation pattern between CB15P454-473 and Rh330-348 bear no similarity. The fact that similar structures are observed in each case suggests that there is a specific arrestin binding pocket that may hold significance to the activation of arrestins. Our ITC results show that CB15P454-473 has approximately a fifty fold greater affinity for the truncated version of arrestin. This difference in affinity, combined with the fact that the same bound peptide structure is observed for both wt-arr21-418 and tr-arr21-382 complexes suggests that the protein-binding site is in the N-domain and is occluded by the C-terminus of arrestin.
Docking studies
Our data is consistent with studies suggesting that the binding surface for the phosphorylated receptor C-tail is localized to a cavity in the N-domain of arrestin, which contains many positively charged residues (23). Moreover, our data shows that the state of the arrestin can affect binding. Mutagenesis studies suggest that Lys11 on β-strand 1 of the N-domain may be an initial point of contact, while nearby charges direct phosphates towards charged residues of the polar core (28). The most important residue implicated in binding GPCR attached phosphates, Arg169, is buried within the polar core of the protein, and forms a salt bridge with Asp290, which is a key interaction in stabilizing the basal state. In the crystal structure, arrestin residues Arg383-Arg393 shield the region of the N-domain near Lys11, and the cavity that allows access to Arg169, thereby assisting in stabilizing the inactive state (Fig. 8A&B). Studies utilizing truncated arrestins, and proteolytic cleavage of arrestin in the presence of phosphopeptides indicate that the C-terminus of arrestin may be displaced by the phosphorylated GPCR (24). The increased affinity we observed between the peptide CB15P454-473 and the truncated arrestin supports the notion that CB15P454-473 may either compete with, or displace the arrestin C-tail from a binding site in the N-domain.
To further address this issue, we docked low energy structures of CB15P454-473 to both the wild-type and truncated arrestins as shown in figure 8. We attempted to minimize assumptions regarding binding by choosing positively charged residues in the N-domain as the “active” residues, as shown in figure 8. By doing this, the Haddock program was allowed to sample a large surface area in the N-domain. The calculation utilized the bound structure of CB15P454-473 as a starting structure and initial placement and orientation of the peptide was calculated according to energies of interaction. The structure of the peptide was flexible during docking but constrained by the observed nOes. The best solutions were taken as those with the lowest energy and greatest buried surface area for the peptide.
As shown in figure 8A&B, the phosphates within the helical section of the peptide are associated with Lys160 and Arg161 residues located in a highly flexible loop region of arrestin. In the wild-type arrestin, the loop of the CB15P454-473 peptide enters into the N-domain cavity and the phosphate group of Ser465 contacts Lys11 of arrestin-2. Similarly, the Ser463 and Thr461 phosphate groups interact with Lys294 of arrestin as shown in figure 8B. The C-terminus of arrestin, highlighted in Figure 8B, is shielding Arg25 and occludes the cavity that exposes Arg169. This prevents the peptide from interacting deeper within the cavity.
Docking of the CB15P454-473 with the truncated arrestin resulted in a similar orientation of the C-terminal helix and association with Lys160 and Arg161, but a closer association between the peptide and the N-domain cavity as shown in Figure 8C. With the more exposed binding site, the phosphate of Ser465 interacts with Lys10 and the phosphate of Thr461 with Arg25 of arrestin. This allows the Ser463 residue, located in the loop region of the peptide, to interact closely with the floor of the cavity and for the phosphate group to interact with Arg169.
Thus the observed bound, conformation of the peptide can be reconciled with the binding surfaces of the N-domain cavity. Affinity is clearly governed by phosphorylation, and the associated charges, however, our results also suggest that phosphorylation serves to limit the dynamics of the peptide. This would provide a further entropic impetus for binding and possibly predispose the CB1 terminus towards conformations suitable for binding. The modeling results shown in figures 8B and 8D are consistent with models of arrestin activation in the literature (15). In docking to the wild-type arrestin, the Lys11 of arrestin is an initial point of contact for the Ser465 residue located centrally on the peptide (15, 28). One may then speculate that the association of the peptide helix with the flexible loop of arrestin provides increased affinity for this active site and allows the phosphate groups of the peptide loop-region to interact with functionally important arrestin residues. As the loop region of the phosphorylated peptide is a persistent feature, the possibility of an interaction with Lys294 in wt-Arr2 is maximized. This may be a means by which the C-tail of arrestin may be displaced to yield a more exposed cavity and allow subsequent interactions with key residues of arrestin as shown in figure 8D.
In summary, the observation of similar bound structures of the CB15P454-473 but increased affinity for the truncated arrestin suggests binding of the peptide to the N-domain. The increased affinity for the truncated arrestin can be explained by the increased exposure of positively charged residues in the N-domain cavity. The interactions modeled against the N-domain cavity are congruent with arrestin activation models (23) and also with CB1 receptor mutational studies (40, 41).
Footnotes
- GPCR
- G-Protein Coupled Receptor
- CB1
- cannabinoid receptor 1
- wt-arr21-418
- wild-type human arrestin-2
- tr-arr21-382
- a truncated human arrestin-2 mutant comprised of residues 1–382
- CB15P454-473
- a pentaphosphorylated peptide mimicking residues 454-473 of CB1
- Rh7P330-348
- a heptaphosphorylated peptide mimicking residues 330-348 of the rhodopsin receptor
- ITC
- Isothermal Titration Calorimetry
- AM411
- (−) adamantyl-Δ8-tetrahydrocannabinol
- Thr460
- threonine residue at position 460
- Leu473
- leucine residue at position 473
- AtT20 cells
- a mouse anterior pituitary-derived cell line
- Ser426
- serine residue at position 426
- Ser430
- serine residue at position 430
- HEK293 cells
- Human Embryonic Kidney 293 cells
- NPxxY
- conserved peptide sequence consisting of Asparagine and Proline separated from Tyrosine by two amino acids
- Thr419
- threonone residue at position 419
- ASN439
- asparagine residue at position 439
- Thr454
- threonine residue at position 454
- HPLC
- High Pressure Liquid Chromatography
- MALDI-TOF
- Matrix-assisted laser desorption/ionization Time of Flight mass spectrometry
- pTrcHis
- proprietary plasmid from invitrogen
- NcoI
- Restriction enzyme cleaving at NcoI site
- BamH1
- Restriction enzyme cleaving at BamH1 site
- pTrcK
- modified version of pTrcHis plasmid
- LB
- Luria Broth
- ml
- milliliters
- μM
- micromolar
- mM
- millimolar
- PMSF
- phenylmethylsulfonyl fluoride
- DTT
- dithiothreitol
- EDTA
- Ethylenediaminetetraacetic acid
- SDS-PAGE
- sodium dodecyl sulfate polyacrylamide gel electrophoresis
- rpm
- revolution per minute
- NaCl
- sodium chloride
- NMR
- nuclear magnetic resonance
- MHz
- megahertz
- DSS
- 4,4-dimethyl-4-silapentane-1-sulfonic acid
- TOCSY
- Total Correlation Spectroscopy
- DIPSI
- Decoupling In the Presence of Scalar Interactions
- ms
- millisecond
- NOESY
- nuclear Overhauser effect spectroscopy
- s
- second
- nOes
- nuclear Overhauser effect peaks
- Å
- angstrom
- K
- Kelvin
- nm
- nanometers
- ps
- picoseconds
References
- 1.Attramadal H, Arriza JL, Aoki C, Dawson TM, Codina J, Kwatra MM, Snyder SH, Caron MG, Lefkowitz RJ. Beta-arrestin2, a novel member of the arrestin/beta-arrestin gene family. J. Biol. Chem. 1992;267:17882–17890. [PubMed] [Google Scholar]
- 2.Gurevich VV, Benovic JL. Visual arrestin binding to rhodopsin. Diverse functional roles of positively charged residues within the phosphorylation-recognition region of arrestin. J. Biol. Chem. 1995;270:6010–6016. doi: 10.1074/jbc.270.11.6010. [DOI] [PubMed] [Google Scholar]
- 3.Lefkowitz RJ, Inglese J, Koch WJ, Pitcher J, Attramadal H, Caron MG. G-protein-coupled receptors: Regulatory role of receptor kinases and arrestin proteins. Cold Spring Harb. Symp. Quant. Biol. 1992;57:127–133. doi: 10.1101/sqb.1992.057.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Lohse MJ, Andexinger S, Pitcher J, Trukawinski S, Codina J, Faure JP, Caron MG, Lefkowitz RJ. Receptor-specific desensitization with purified proteins. Kinase dependence and receptor specificity of beta-arrestin and arrestin in the beta 2-adrenergic receptor and rhodopsin systems. J. Biol. Chem. 1992;267:8558–8564. [PubMed] [Google Scholar]
- 5.Mukherjee S, Palczewski K, Gurevich V, Benovic JL, Banga JP, Hunzicker-Dunn M. A direct role for arrestins in desensitization of the luteinizing hormone/choriogonadotropin receptor in porcine ovarian follicular membranes. Proc. Natl. Acad. Sci. U. S. A. 1999;96:493–498. doi: 10.1073/pnas.96.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. Beta-arrestins and cell signaling. Annu. Rev. Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 7.Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 8.Kenakin T. Agonist-receptor efficacy. II. Agonist trafficking of receptor signals. Trends Pharmacol. Sci. 1995;16:232–238. doi: 10.1016/s0165-6147(00)89032-x. [DOI] [PubMed] [Google Scholar]
- 9.Wei H, Ahn S, Shenoy SK, Karnik SS, Hunyady L, Luttrell LM, Lefkowitz RJ. Independent beta-arrestin 2 and G protein-mediated pathways for angiotensin II activation of extracellular signal-regulated kinases 1 and 2. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10782–10787. doi: 10.1073/pnas.1834556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holloway AC, Qian H, Pipolo L, Ziogas J, Miura S, Karnik S, Southwell BR, Lew MJ, Thomas WG. Side-chain substitutions within angiotensin II reveal different requirements for signaling, internalization, and phosphorylation of type 1A angiotensin receptors. Mol. Pharmacol. 2002;61:768–777. doi: 10.1124/mol.61.4.768. [DOI] [PubMed] [Google Scholar]
- 11.Shukla AK, Violin JD, Whalen EJ, Gesty-Palmer D, Shenoy SK, Lefkowitz RJ. Distinct conformational changes in beta-arrestin report biased agonism at seven-transmembrane receptors. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9988–9993. doi: 10.1073/pnas.0804246105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Violin JD, Lefkowitz RJ. Beta-arrestin-biased ligands at seven-transmembrane receptors. Trends Pharmacol. Sci. 2007;28:416–422. doi: 10.1016/j.tips.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Hanson SM, Francis DJ, Vishnivetskiy SA, Klug CS, Gurevich VV. Visual arrestin binding to microtubules involves a distinct conformational change. J. Biol. Chem. 2006;281:9765–9772. doi: 10.1074/jbc.M510738200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanson SM, Francis DJ, Vishnivetskiy SA, Kolobova EA, Hubbell WL, Klug CS, Gurevich VV. Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4900–4905. doi: 10.1073/pnas.0600733103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol. Sci. 2004;25:105–111. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Gurevich VV, Gurevich EV. The structural basis of arrestin-mediated regulation of G-protein-coupled receptors. Pharmacol. Ther. 2006;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Granzin J, Wilden U, Choe HW, Labahn J, Krafft B, Buldt G. X-ray crystal structure of arrestin from bovine rod outer segments. Nature. 1998;391:918–921. doi: 10.1038/36147. [DOI] [PubMed] [Google Scholar]
- 18.Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Crystal structure of beta-arrestin at 1.9 Å: Possible mechanism of receptor binding and membrane translocation. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch JA, Schubert C, Gurevich VV, Sigler PB. The 2.8 Å crystal structure of visual arrestin: A model for arrestin’s regulation. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 20.Milano SK, Kim YM, Stefano FP, Benovic JL, Brenner C. Nonvisual arrestin oligomerization and cellular localization are regulated by inositol hexakisphosphate binding. J. Biol. Chem. 2006;281:9812–9823. doi: 10.1074/jbc.M512703200. [DOI] [PubMed] [Google Scholar]
- 21.Milano SK, Pace HC, Kim YM, Brenner C, Benovic JL. Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis. Biochemistry. 2002;41:3321–3328. doi: 10.1021/bi015905j. [DOI] [PubMed] [Google Scholar]
- 22.Sutton RB, Vishnivetskiy SA, Robert J, Hanson SM, Raman D, Knox BE, Kono M, Navarro J, Gurevich VV. Crystal structure of cone arrestin at 2.3Å: Evolution of receptor specificity. J. Mol. Biol. 2005;354:1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 23.Gurevich VV, Benovic JL. Visual arrestin interaction with rhodopsin. Sequential multisite binding ensures strict selectivity toward light-activated phosphorylated rhodopsin. J. Biol. Chem. 1993;268:11628–11638. [PubMed] [Google Scholar]
- 24.Palczewski K, Buczylko J, Imami NR, McDowell JH, Hargrave PA. Role of the carboxyl-terminal region of arrestin in binding to phosphorylated rhodopsin. J. Biol. Chem. 1991;266:15334–15339. [PubMed] [Google Scholar]
- 25.Potter RM, Key TA, Gurevich VV, Sklar LA, Prossnitz ER. Arrestin variants display differential binding characteristics for the phosphorylated n-formyl peptide receptor carboxyl terminus. J. Biol. Chem. 2002;277:8970–8978. doi: 10.1074/jbc.M111086200. [DOI] [PubMed] [Google Scholar]
- 26.Hanson SM, Gurevich VV. The differential engagement of arrestin surface charges by the various functional forms of the receptor. J. Biol. Chem. 2006;281:3458–3462. doi: 10.1074/jbc.M512148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vishnivetskiy SA, Paz CL, Schubert C, Hirsch JA, Sigler PB, Gurevich VV. How does arrestin respond to the phosphorylated state of rhodopsin? J. Biol. Chem. 1999;274:11451–11454. doi: 10.1074/jbc.274.17.11451. [DOI] [PubMed] [Google Scholar]
- 28.Vishnivetskiy SA, Schubert C, Climaco GC, Gurevich YV, Velez MG, Gurevich VV. An additional phosphate-binding element in arrestin molecule. Implications for the mechanism of arrestin activation. J. Biol. Chem. 2000;275:41049–41057. doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- 29.Kisselev OG, Downs MA, McDowell JH, Hargrave PA. Conformational changes in the phosphorylated C-terminal domain of rhodopsin during rhodopsin arrestin interactions. J. Biol. Chem. 2004;279:51203–51207. doi: 10.1074/jbc.M407341200. [DOI] [PubMed] [Google Scholar]
- 30.Kisselev OG, McDowell JH, Hargrave PA. The arrestin-bound conformation and dynamics of the phosphorylated carboxy-terminal region of rhodopsin. FEBS Lett. 2004;564:307–311. doi: 10.1016/S0014-5793(04)00226-1. [DOI] [PubMed] [Google Scholar]
- 31.Hwang J, Adamson C, Butler D, Janero DR, Makriyannis A, Bahr BA. Enhancement of endocannabinoid signaling by fatty acid amide hydrolase inhibition: A neuroprotective therapeutic modality. Life Sci. 2010;86:615–623. doi: 10.1016/j.lfs.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makriyannis A, Goutopoulos A. Cannabinergics: Old and new therapeutic possibilities. In: Makriyannis A, Biegel D, editors. Drug discovery strategies and methods. Marcel Dekker, Inc.; New York: 2004. pp. 89–128. [Google Scholar]
- 33.Pavlopoulos S, Thakur GA, Nikas SP, Makriyannis A. Cannabinoid receptors as therapeutic targets. Curr. Pharm. Des. 2006;12:1751–1769. doi: 10.2174/138161206776873743. [DOI] [PubMed] [Google Scholar]
- 34.Price MR, Baillie GL, Thomas A, Stevenson LA, Easson M, Goodwin R, McLean A, McIntosh L, Goodwin G, Walker G, Westwood P, Marrs J, Thomson F, Cowley P, Christopoulos A, Pertwee RG, Ross RA. Allosteric modulation of the cannabinoid CB1 receptor. Mol. Pharmacol. 2005;68:1484–1495. doi: 10.1124/mol.105.016162. [DOI] [PubMed] [Google Scholar]
- 35.Tam J, Vemuri VK, Liu J, Batkai S, Mukhopadhyay B, Godlewski G, Osei-Hyiaman D, Ohnuma S, Ambudkar SV, Pickel J, Makriyannis A, Kunos G. Peripheral CB1 cannabinoid receptor blockade improves cardiometabolic risk in mouse models of obesity. J. Clin. Invest. 2010;120:2953–2966. doi: 10.1172/JCI42551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonhaus DW, Chang LK, Kwan J, Martin GR. Dual activation and inhibition of adenylyl cyclase by cannabinoid receptor agonists: Evidence for agonist-specific trafficking of intracellular responses. J. Pharmacol. Exp. Ther. 1998;287:884–888. [PubMed] [Google Scholar]
- 37.Chatterjee C, Mukhopadhyay C. Interaction and structural study of kinin peptide bradykinin and ganglioside monosialylated 1 micelle. Biopolymers. 2005;78:197–205. doi: 10.1002/bip.20278. [DOI] [PubMed] [Google Scholar]
- 38.Luk T, Jin W, Zvonok A, Lu D, Lin XZ, Chavkin C, Makriyannis A, Mackie K. Identification of a potent and highly efficacious, yet slowly desensitizing CB1 cannabinoid receptor agonist. Br. J. Pharmacol. 2004;142:495–500. doi: 10.1038/sj.bjp.0705792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin W, Brown S, Roche JP, Hsieh C, Celver JP, Kovoor A, Chavkin C, Mackie K. Distinct domains of the CB1 cannabinoid receptor mediate desensitization and internalization. J. Neurosci. 1999;19:3773–3780. doi: 10.1523/JNEUROSCI.19-10-03773.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsieh C, Brown S, Derleth C, Mackie K. Internalization and recycling of the CB1 cannabinoid receptor. J. Neurochem. 1999;73:493–501. doi: 10.1046/j.1471-4159.1999.0730493.x. [DOI] [PubMed] [Google Scholar]
- 41.Daigle TL, Kwok ML, Mackie K. Regulation of CB1 cannabinoid receptor internalization by a promiscuous phosphorylation-dependent mechanism. J. Neurochem. 2008;106:70–82. doi: 10.1111/j.1471-4159.2008.05336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daigle TL, Kearn CS, Mackie K. Rapid CB1 cannabinoid receptor desensitization defines the time course of ERK1/2 map kinase signaling. Neuropharmacology. 2008;54:36–44. doi: 10.1016/j.neuropharm.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi G, Guo J, Makriyannis A. The conformation of the cytoplasmic helix 8 of the CB1 cannabinoid receptor using NMR and circular dichroism. Biochim. Biophys. Acta. 2005;1668:1–9. doi: 10.1016/j.bbamem.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 45.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A g protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 46.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 47.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AG, Tate CG, Schertler GF. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anavi-Goffer S, Fleischer D, Hurst DP, Lynch DL, Barnett-Norris J, Shi S, Lewis DL, Mukhopadhyay S, Howlett AC, Reggio PH, Abood ME. Helix 8 Leu in the CB1 cannabinoid receptor contributes to selective signal transduction mechanisms. J. Biol. Chem. 2007;282:25100–25113. doi: 10.1074/jbc.M703388200. [DOI] [PubMed] [Google Scholar]
- 49.Gehret AU, Jones BW, Tran PN, Cook LB, Greuber EK, Hinkle PM. Role of helix 8 of the thyrotropin-releasing hormone receptor in phosphorylation by G protein-coupled receptor kinase. Mol. Pharmacol. 2010;77:288–297. doi: 10.1124/mol.109.059733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnston CA, Willard FS, Jezyk MR, Fredericks Z, Bodor ET, Jones MB, Blaesius R, Watts VJ, Harden TK, Sondek J, Ramer JK, Siderovski DP. Structure of Galpha(i1) bound to a GDP-selective peptide provides insight into guanine nucleotide exchange. Structure. 2005;13:1069–1080. doi: 10.1016/j.str.2005.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tiburu EK, Bowman AL, Struppe JO, Janero DR, Avraham HK, Makriyannis A. Solid-state NMR and molecular dynamics characterization of cannabinoid receptor-1 (CB1) helix 7 conformational plasticity in model membranes. Biochim. Biophys. Acta. 2009;1788:1159–1167. doi: 10.1016/j.bbamem.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tyukhtenko S, Tiburu EK, Deshmukh L, Vinogradova O, Janero DR, Makriyannis A. NMR solution structure of human cannabinoid receptor-1 helix 7/8 peptide: Candidate electrostatic interactions and microdomain formation. Biochem. Biophys. Res. Commun. 2009 doi: 10.1016/j.bbrc.2009.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahn KH, Pellegrini M, Tsomaia N, Yatawara AK, Kendall DA, Mierke DF. Structural analysis of the human cannabinoid receptor one carboxyl-terminus identifies two amphipathic helices. Biopolymers. 2009;91:565–573. doi: 10.1002/bip.21179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bakshi K, Mercier RW, Pavlopoulos S. Interaction of a fragment of the cannabinoid CB1 receptor c-terminus with arrestin-2. FEBS Lett. 2007;581:5009–5016. doi: 10.1016/j.febslet.2007.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schubert C, Hirsch JA, Gurevich VV, Engelman DM, Sigler PB, Fleming KG. Visual arrestin activity may be regulated by self-association. J. Biol. Chem. 1999;274:21186–21190. doi: 10.1074/jbc.274.30.21186. [DOI] [PubMed] [Google Scholar]
- 56.Braunschweiler L, Ernst RR. Coherence transfer by isotropic mixing: Application to proton correlation spectroscopy. J. Magn. Reson. 1983;53:521–528. [Google Scholar]
- 57.Shaka AJ, Lee CJ, Pines A. Iterative schemes for bilinear operators; application to spin decoupling. J. Magn. Res. B. 1988;77:274–293. [Google Scholar]
- 58.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 59.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRpipe: A multidimensional spectral processing system based on unix pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 60.Wuthrich K. NMR of proteins and nucleic acids. New York: 1986. [Google Scholar]
- 61.Clore GM, Brunger AT, Karplus M, Gronenborn AM. Application of molecular dynamics with interproton distance restraints to three-dimensional protein structure determination. A model study of crambin. J. Mol. Biol. 1986;191:523–551. doi: 10.1016/0022-2836(86)90146-4. [DOI] [PubMed] [Google Scholar]
- 62.Clore GM, Nilges M, Sukumaran DK, Brunger AT, Karplus M, Gronenborn AM. The three-dimensional structure of alpha1-purothionin in solution: Combined use of nuclear magnetic resonance, distance geometry and restrained molecular dynamics. Embo J. 1986;5:2729–2735. doi: 10.1002/j.1460-2075.1986.tb04557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clore GM, Gronenborn AM, Nilges M, Ryan CA. Three-dimensional structure of potato carboxypeptidase inhibitor in solution. A study using nuclear magnetic resonance, distance geometry, and restrained molecular dynamics. Biochemistry. 1987;26:8012–8023. doi: 10.1021/bi00398a069. [DOI] [PubMed] [Google Scholar]
- 64.Wagner G, Braun W, Havel TF, Schaumann T, Go N, Wuthrich K. Protein structures in solution by nuclear magnetic resonance and distance geometry. The polypeptide fold of the basic pancreatic trypsin inhibitor determined using two different algorithms, DISGEO and DISMAN. J. Mol. Biol. 1987;196:611–639. doi: 10.1016/0022-2836(87)90037-4. [DOI] [PubMed] [Google Scholar]
- 65.Brunger AT. Version 1.2 of the crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 66.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 67.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The XPLOR-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 68.Schuttelkopf AW, van Aalten DM. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 69.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. PROCHECK - a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 70.Schwieters CD, Clore GM. The vmd-xplor visualization package for NMR structure refinement. J. Magn. Reson. 2001;149:239–244. doi: 10.1006/jmre.2001.2300. [DOI] [PubMed] [Google Scholar]
- 71.Dominguez C, Boelens R, Bonvin AM. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 72.Vishnivetskiy SA, Hosey MM, Benovic JL, Gurevich VV. Mapping the arrestin-receptor interface - structural elements responsible for receptor specificity of arrestin proteins. J. Biol. Chem. 2004;279:1262–1268. doi: 10.1074/jbc.M308834200. [DOI] [PubMed] [Google Scholar]
- 73.Hubbard S, Thornton J. NACCESS. 2.1.1 ed. Dept of Biochemistry and Molecular Biology, University College; London: 1993. [Google Scholar]
- 74.Clore GM, Gronenborn AM. The two-dimensional transferred nuclear overhauser effect. J. Magn. Reson. 1982;48:402–417. [Google Scholar]