Abstract
Background & Aims
Pharmacological approaches can potentially improve fatty liver condition in alcoholic and non-alcoholic fatty liver diseases. The salutary effects of reducing lipid synthesis or promoting lipid oxidation have been well reported, but the benefits of increasing lipid degradation have yet to be well explored. Macroautophagy is a cellular degradation process that can remove subcellular organelles including lipid droplets. We thus investigated whether pharmacological modulation of macroautophagy could be an effective approach to alleviate fatty liver condition and liver injury.
Methods
C57BL/6 mice were given ethanol via intraperitoneal injection (acute) or by a 4-week oral feeding regime (chronic), or given high fat diet for 12 weeks. An autophagy enhancer, carbamazepine or rapamycin, or an autophagy inhibitor, chloroquine, was given before sacrifice. Activation of autophagy, level of hepatic steatosis, and blood levels of triglycerides, liver enzyme, glucose and insulin were measured.
Results
In both acute and chronic ethanol condition macroautophagy were activated. Carbamazepine, as well as rapamycin, enhanced ethanol-induced macroautophagy in hepatocytes in vitro and in vivo. Hepatic steatosis and liver injury were exacerbated by chloroquine, but alleviated by carbamazepine. The protective effects of carbamazepine and rapamycin in reducing steatosis and in improving insulin sensitivity were also demonstrated in high fat diet-induced non-alcoholic fatty liver condition.
Conclusions
These findings indicate that pharmacological modulation of macroautophagy in the liver can be an effective strategy for reducing fatty liver condition and liver injury.
Keywords: macroautophagy, hepatic steatosis, alcohol, high fat diet, carbamazepine
Introduction
Fatty liver is the most common early response of the liver to heavy alcohol consumption, which can render the liver more susceptible to inflammatory mediators or other toxic agents, leading to alcoholic fatty liver disease (AFLD) with the presentation of steatohepatitis, fibrosis, and cirrhosis [1–4]. Nonalcoholic fatty liver disease (NAFLD) is a condition ranging from benign lipid accumulation to steatohepatitis and cirrhosis. NAFLD may be considered the hepatic event in an overall disturbed metabolic status and is therefore linked with common metabolic syndrome risk factors such as obesity, insulin resistance, dyslipidemia, and hypertension [5–8]. Fatty liver diseases, whether caused by alcoholic or nonalcoholic factors, seem to share similar mechanisms and pathologic features [4, 9]. Alcoholic and nonalcoholic steatosis can be a reversible presentation of the liver pathology and reduction of steatosis could halt or slow the progression to inflammation and fibrosis.
Although life-style changes could be most beneficial in handling AFLD or NAFLD, this may not be always practical or sufficient Hence, it would be important to develop specific pharmacological approaches to control hepatic steatosis. Consequently, pharmacological interventions have been attempted for NAFLD [10–12]. While the mechanisms could be multiple, the main results seem to be related to an increased utilization of the fatty acids and/or their efflux out of the liver. Alternative approaches, such as that aiming at the promoting of lipid degradation had not been well explored.
Macroautophagy (referred to here as autophagy) is an essential cellular degradation process with important pathophysiological significance [13]. Autophagy involves the formation of double-membraned autophagosomes, which envelop the substrates and fuse with the lysosomes for degradation. Autophagy is now known to be widely involved in the pathogenesis of many human diseases and is activated under a variety of stress conditions. Autophagy actively participates in liver physiology and pathogenesis [14, 15]. Autophagy can constitute an effective defense mechanism against multiple pathological insults, including alcohol and non-alcoholic fatty liver conditions [13, 16–19].
Autophagy can regulate the intracellular level of lipids through its function in removing lipid droplets (lipophagy) [16]. Inhibition of autophagy in cells cultured with lipids or ethanol increases intracellular lipid content [16, 19] Loss of the autophagy capacity in vivo also alters fatty liver condition and liver injury induced by high fat diet or acute alcohol exposure [16, 18]. Conversely, genetically enhancing autophagy by over-expressing an autophagy gene, Atg7, could improve hepatic steatosis and insulin resistance in ob/ob mice and in high fat diet (HFD)-fed mice [17]. These observations led us to postulate that autophagy modulation via pharmacological agents may offer a new strategy for treating fatty liver conditions.
While many agents could simulate autophagy in vitro, few have been tested in vivo, particularly in mammals. Rapamycin is a well-established autophagy inducer by inhibiting mTORC1, which control the activation of the core autophagy machinery [20]. Rapamycin could reduce the level of Malory-Denk bodies, one of the hall markers of chronic ethanol intoxication, in genetically susceptible mice through autophagy degradation [21], and the level of steatosis in mice given binge alcohol drinking [18]. Another FDA-approved drug, carbamazepine (CBZ), has been used as an anti-epileptic drug, but can induce autophagy by disturbing inositol metabolism [20]. Although direct evidence lacks that CBZ can induce autophagy in the liver, its application in mice was found to promote degradation of α1-antitrypsin mutant proteins in a transgenic model [22].
We thus investigated whether pharmacological modulation of autophagy in the liver with CBZ and rapamycin could be effective for alleviating fatty liver conditions caused by an alcoholic or non-alcoholic etiology. Indeed, we found that the autophagy-enhancing drugs alleviated liver steatosis, liver injury, and dyslipidemia in both alcohol-fed and HFD-fed mice. These findings indicate that the autophagy-promoting drugs have important implications as a new genre of therapeutic agents for fatty livers.
Materials and Methods
Reagents
Ethanol was purchased from Pharmaco, Inc. (Shelbyville, KY). Other chemicals were from Sigma, Invitrogen, or Calbiochem. Antibodies used were anti-LC3 [23], anti-β-actin (Sigma, St. Louis, MO), and horseradish peroxidase-labeled secondary antibodies (Jackson ImmunoResearch Lab., West Grove, PA).
Animal models
Wild type C57BL/6 mice and GFP-LC3 transgenic mice [24] were used in this study. All procedures were approved by the Institutional Animal Care and Use Committee. For the acute ethanol treatment, mice were intraperitoneally given ethanol (33%, v/v in normal saline) at a dose of 1.2 g/kg body weight [25]. Control mice received the same volume of normal saline. For chronic ethanol treatment, mice were given the Lieber-DeCarli low fat liquid diet for 4 weeks as previously described [26].
Ethanol-containing diet was prepared with ethanol added to the liquid diet base (DYETS, #710261, Bethlehem, Philadelphia, PA) accounting for 29% or 36% of the total calorie intake (1 kcal/ml). Control diet substitutes ethanol with maltose dextrin to account for the same level of calorie requirement. In the HFD model, mice were fed with a regular chow diet (Fat 14%, Lab Diet, #5P76) or a HFD (60% kcal in fat; Research Diets, #D12492) for 12 weeks. Pharmacological modulation of autophagy is achieved by intraperitoneal administration of chloroquine (60 mg/kg), rapamycin (2 mg/kg) or CBZ (25 mg/kg) unless otherwise indicated in the figure legends.
Cell culture
Murine hepatocytes were isolated and cultured in William’s medium with 10% FCS as previously described [18]. For modulation of autophagy hepatocytes were cultured in Earle’s Balanced Salt Solution (EBSS) or treated with ethanol (80 mM), CBZ (50 μM), rapamycin (5 μM) or 3-methyladenine (10 mM). Autophagy was assessed as previously described [18, 27] by long lived protein degradation assay and GFP-LC3 quantification, in which cells were pre-infected with adenoviral GFP-LC3 the night before the indicated treatment.
Biochemical analyses
Hepatic and plasma triglycerides (TG) were determined using a commercial kit (Pointe Scientific, MI) [18]. Levels of blood ALT, glucose and insulin were measured using a kit from Biotron Diagnostics (Hemet, CA), an Ascensia Contour glucose meter (Bayer HealthCare, IN), and a commercial kit from Millipore (Billerica, MA), respectively. Levels of insulin resistance were determined by Homeostatic Model Assessment (HOMA = [fasting glucose (mmol/l) × fasting insulin (μU/ml)]/22.5) with a conversion factor of 1 μU/ml = 6 pmol/l for insulin measurement [28].
Immunoblotting and fluorescence microscopic analyses
Immunoblot assays with liver lysates were performed as previously described [23]. Cryosections of livers were stained with Bodipy 493/503 (0.1 μM) for 15 minutes for quantification of lipid droplets [18]. GFP-LC3 puncta were quantified for autophagy status. All fluorescence images were digitally acquired with a Nikon epifluorescence microscope (Nikon TE200). At least 150 cells from different fields were examined for each mouse and the number of mice used per treatment group was indicated in the figure legend.
Quantitative RT-PCR
Total RNA was extracted from livers and quantitative RT-PCR was performed as previously described [29] using cyclophilin A as a housekeeping gene control, which remains stable in the liver under different conditions.
Statistical analysis
Quantitative data (mean±SEM) are subjected to Student’s t test or one way ANOVA with Holm-Sidak post-hoc analysis, using SigmaStat 3.5 (Systat Software, Inc., San Jose, CA).
Results
Carbamazepine promoted autophagy in hepatocytes
CBZ has been shown to possess a pro-autophagy activity in several cell lines [20]. It also promotes an effective clearance of aggregated α1-antitrypsin mutant proteins in mouse livers [22]. In order to determine whether CBZ could be used to alleviate fatty liver condition through autophagy induction we first examined whether CBZ could directly induce autophagy in hepatocytes.
Mice expressing GFP-LC3 in the liver have been successfully validated to determine the response to autophagy stimuli [18, 24]. We found that an injection of CBZ into the GFP-LC3 transgenic mice caused a significant accumulation of GFP-LC3 puncta in the liver (Fig. 1A–B). Treatment of isolated primary hepatocytes with CBZ (Fig. 1C) confirmed the induction of autophagy to the same level caused by rapamycin, a well-studied autophagy inducer that works by inhibiting mTOR [20] (Fig. S1). Importantly, co-injection with chloroquine (CQ) in vivo (Fig. 1A–B), or co-treatment with baflomycin A1 in vitro (data not shown), both of which can block autophagy degradation in the lysosome, further elevated the punctuation, indicating that CBZ-induced autophagosomes were degraded in the lysosome [18, 30]. The autophagic flux level in the presence of CBZ was about 75% more than that in the absence of CBZ (Fig. 1B). Consistent with the involvement of the core autophagy machinery, CBZ and rapamycin-induced autophagy in hepatocytes were suppressed by 3-methyladenine (3-MA), which inhibits the class III PI-3 kinase required for the induction (Fig. 1C). Finally, CBZ, like rapamycin and starvation (culture in EBSS), elevated 3-MA-suppressible long-lived protein degradation in hepatocytes (Fig. 1D), which is a key functional index of autophagy induction [27]. Taken together, these observations indicated that CBZ, like rapamycin, could clearly activate autophagy in hepatocytes.
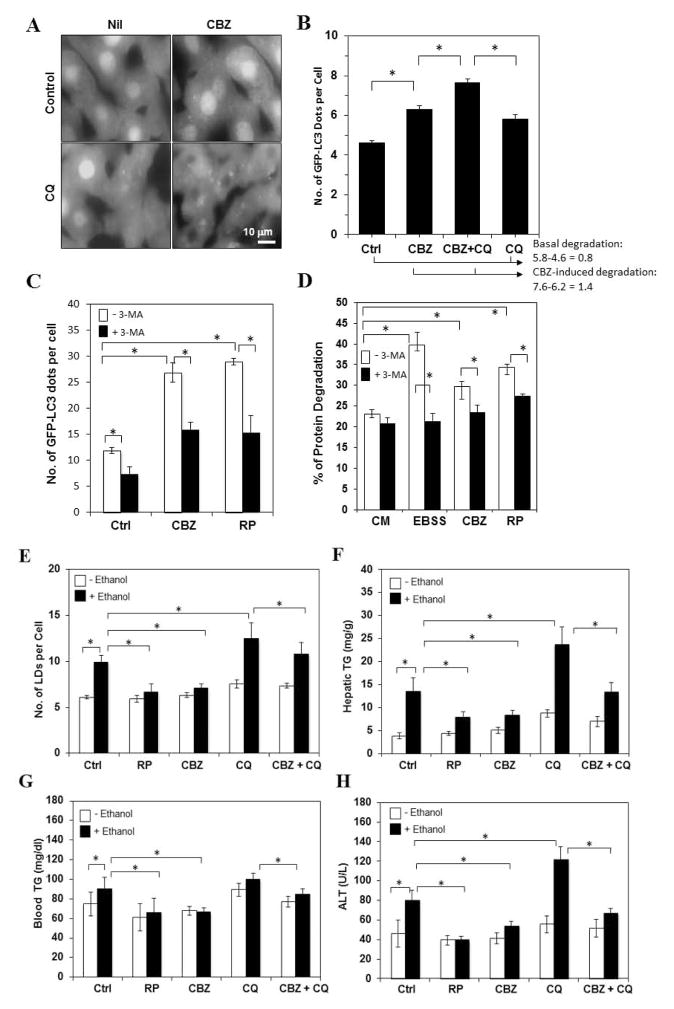
Figure 1. Carbamazepine induces autophagy in hepatocytes and alleviates acute ethanol-induced liver steatosis and liver injury whereas chloroquine inhibits both.
(A–B). GFP-LC3 transgenic mice were treated with CBZ with or without pretreatment of CQ for 16 hours. GFP-LC3 puncta were quantified. (C–D) Primary hepatocytes were pre-incubated with adenoviral GFP-LC3 overnight and then cultured in complete medium (CM), or in EBSS, or in CM with CBZ, rapamycin or 3-MA for 6 hours followed by fluorescence microscopy for LC3 dot formation (C) or for 16 hours for long-lived protein degradation assay (D). (E). Wild type mice were given ethanol (E) with or without rapamycin, or CBZ. Mice were sacrificed 16 hours later and the numbers of Bodipy-positive LDs were quantified. (F–H). The levels of hepatic (F) and plasma (G) TG, and plasma ALT (H) were measured. *: p<0.01 (n=3–7).
Carbamazepine alleviated acute ethanol-induced liver injury
Acute ethanol administration in mice through oral gavage [18] or intraperitoneally (data not shown) induced autophagy in the liver, which depends on its metabolism and reactive oxygen species, and involves mTOR inhibition [18](Fig. S1). Consistent with the enhanced induction of autophagy co-treatment with CBZ or rapamycin significantly reduced ethanol-induced hepatic steatosis as measured by the number of LDs and the level of TG in the liver and plasma (Fig. 1E–G). These changes were accompanied with a reduced liver injury as shown by the decreased ALT level in the blood (Fig. 1H), indicating an overall protective effect of autophagy. In contrast, CQ inhibits autophagic degradation and promoted ethanol-induced steatosis and injury (Fig. 1E–H). Notably, CBZ could antagonize the detrimental effects of CQ as measured by these parameters. This observation suggested that CBZ was potent in causing a larger extent of autophagy than what could be suppressed by CQ at the dose used, thus leading to a net gain of autophagy capacity.
We further showed that at this dose CBZ did not have major effects on the expression of several genes important for lipogenesis or fatty acid oxidation (Table 1). This observation supported the notion that the effect of CBZ could be mainly mediated by its autophagy-enhancing effects. Interestingly, rapamycin could noticeably, although not significantly, alter the expression of Acox1 and Cpt1a, suggesting that its effects on hepatic steatosis could be also mediated by regulating lipid oxidation.
Table 1.
Fold of change in the expression level of genes involved in lipogenesis and fatty acid oxidation
| CBZ# | Rapamycin# | 29% ED$ | HFD* | |
|---|---|---|---|---|
| Srebp-1c | 1.18±0.13 | 1.45±0.46 | 1.33±0.33 | 2.77±0.29a |
| Fasn | 0.97±0.17 | 1.53±0.61 | 3.41±1.72 | 1.88±0.11a |
| Acc1 | 0.84±0.11 | 0.94±0.21 | 0.48±0.06a | 0.82±0.08 |
| Acox1 | 0.80±0.34 | 1.80±0.80 | 1.27±0.13 | 1.31±0.08b |
| Cpt1a | 1.18±0.25 | 3.67±1.58 | 0.58±0.07a | 1.21±0.06b |
Hepatic RNA were isolated and subjected to quantitative RT-PCR for the genes indicated. The expression levels were normalized with cyclophilin A and compared to untreated controls, which is set at 1.0. Srebp-1c, sterol regulatory element binding protein-1c, Fasn: fatty acid synthase, Acc1: acetyl-CoA carboxylase 1, Acox1: acyl-CoA oxidase 1, Cpt1a: carnitine-palmitoyl transferase, 1a. ND: not determined. Values shown are mean±SEM.
Letter a indicates p<0.01, letter b indicates p<0.05 (t test).
Mice were given CBZ (25 mg/kg) or rapamycin (2 mg/kg) by i.p. and sacrificed 16 hours later. Expression levels are compared to control mice without treatment (n=3–6).
Mice were given 29% ethanol liquid diet for 4 weeks. Expression levels were compared to mice receiving control liquid diet (n=3).
Mice were given HFD for 20 weeks. Expression levels were compared to mice receiving regular chow diet (n=4–6).
Autophagy remained active and protective against liver pathology caused by long-term ethanol administration
Pharmacological augment of autophagy may be particularly beneficial in long-term ethanol treatment where autophagy function may be affected more significantly than in the acute condition [31]. We thus examined the autophagy status in the liver of mice given the Lieber-DeCarli ethanol diet for 4 weeks. Using GFP-LC3 mice we found that there were more autophagosomes in the livers of mice given ethanol diet than in the livers of mice given control diet (Fig. 2A–B). This difference was particularly notable when the degradation was inhibited by CQ (Fig. 2A–B), which would indicate an increased autophagosome formation [18, 30]. In addition, a higher value in the presence of CQ within the same diet group (either the control or the ethanol diet group) indicated that there was an autophagy flux resulting in enhanced degradation. That this difference was larger in the ethanol diet group than the control diet group indicated an increased autophagosome degradation stimulated by ethanol. In addition, immunoblot analysis of the lipidated LC3 (LC3-II) indicated a significant increased formation and degradation in the ethanol diet group (Fig. 2C). Moreover, the level of p62, an autophagy target, was reduced following ethanol treatment, which could be reversed by CQ (Fig. S2). Finally, the activities of some of the key lysosomal enzymes were noticeably increased in the ethanol groups, although variations were observed (Fig. S3).
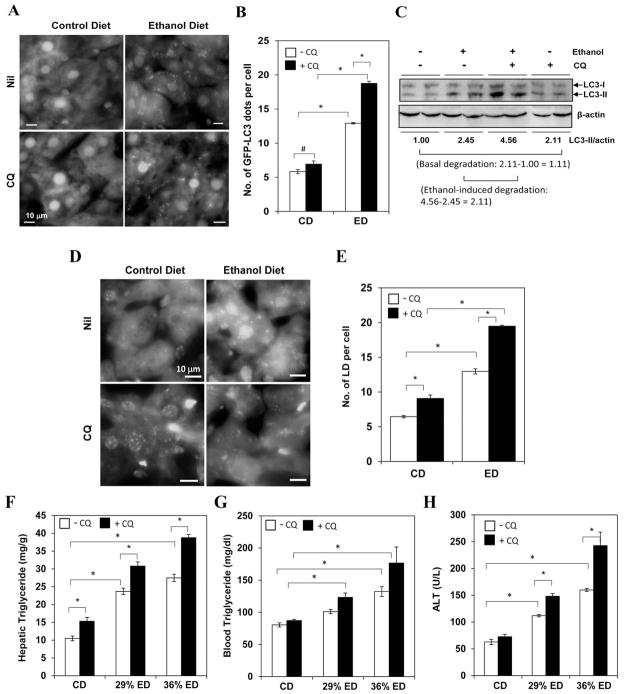
Figure 2. Autophagy helps to control hepatic steatosis and liver injury under chronic ethanol condition.
(A–B) GFP-LC3 mice were given control diet (CD) or 29% ethanol diet (ED) for 4 weeks, followed by CQ administrated 16 hr before sacrifice. (C) Wild type mice were fed with CD or 36% ED for 4 weeks. A subgroup of mice was given CQ before sacrifice. The levels of LC3-II quantified by densitometry (normalized to the loading level and to the CD without CQ) were shown below the blot. (D-–E). Wild type mice were treated with CD or 29% ED for 4 weeks and given CQ 16 hr before sacrifice. Liver cryosections were stained with Bodipy (D) and the number of LDs were quantified (E). (F–H). Mice were treated as indicated and analyzed for hepatic TG (F), blood TG (G) and blood ALT (H) levels. #: p<0.05, *: p<0.01 (n=3–5 for A–E, 2–10 for F–H).
Functionally, suppression of autophagic degradation with CQ further increased hepatic steatosis induced by ethanol diet as measured by Bodipy staining (Fig. 2D–E) and by the level of TG in the liver and the blood (Fig. 2F–G), indicating that lipophagy remained active in these chronically treated mice. Consistent with the acute ethanol model, administration of CQ increased liver injury significantly (Fig. 2H) and slightly increased liver inflammation (Fig. S4), suggesting that the autophagy function was required for cellular protection against ethanol toxicity in the chronic condition.
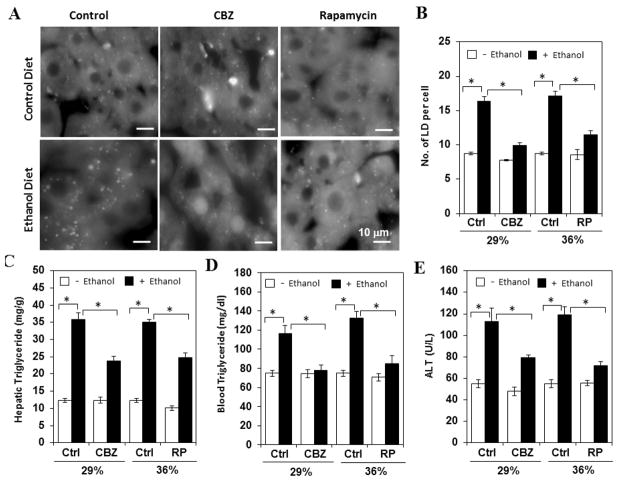
We then reasoned that if autophagy function could be further enhanced the liver pathology would be improved accordingly. Indeed when these mice were given CBZ or rapamycin we observed a reduced level of steatosis as measured by the level of LDs (Fig. 3A–B) and hepatic TG (Fig. 3C). The blood TG level was also reduced (Fig. 3D). Accompanied with these improvements, the blood ALT level was significantly lower in autophagy-enhanced groups than in the control group (Fig. 3E). As in the case of autophagy inhibition (Fig. 2) the impact of autophagy enhancement was observed in mice receiving ethanol diet. Taken together, these observations indicate that pharmacological agents aiming to enhance autophagy could reduce chronic ethanol-induced liver injury.
Figure 3. Enhanced autophagy alleviated chronic ethanol-induced liver steatosis and liver injury.
Mice were treated with control diet, 29% (A–E) or 36% (B–E) ethanol diet and then given rapamycin every other day for three times in the week before sacrifice, or CBZ (12.5 mg/kg) once on the day before sacrifice. Cryosections of the livers were stained with Bodipy 493/503 (A). The numbers of LDs were quantified (B). Hepatic TG (C), blood TG (D) and ALT (E) levels were determined. *: p<0.01 (n=3–13).
Enhanced autophagy could improve fatty liver condition in HFD-fed mice
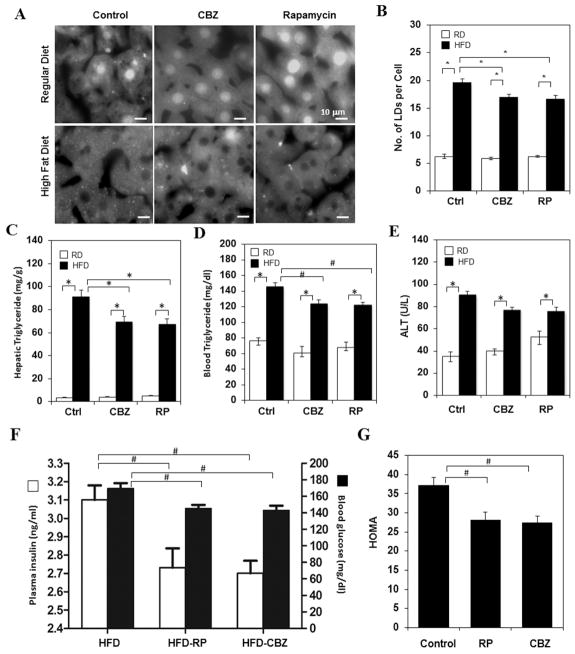
Fatty livers are also commonly observed in non-alcoholic conditions and one well established case is that caused by HFD. To determine whether pharmacological modulation of autophagy could also affect non-alcoholic fatty liver condition, we treated HFD-fed mice with CBZ or rapamycin. The agents were given two or three times in the last week of a 12-week feeding scheme with the intention of a transient application to avoid potential side effects. We found that this short-term treatment nevertheless could significantly reduce hepatic steatosis (Fig. 4A–B) and hepatic and blood TG levels (Fig. 4C–D). The plasma ALT level was also noticeably, although not statistically significantly, reduced (Fig. 4E). Interestingly, insulin resistance was improved as well, as measured by the level of blood glucose and insulin (Fig. 4F–G). These observations were consistent with a previous finding that over-expressing an autophagy gene, Atg7, in the liver of HFD-fed mice or the ob/ob mice improved fatty liver condition and insulin resistance [17]. Thus it seems that pharmacological modulation of autophagy could be also beneficial in NAFLD.
Figure 4. Enhanced autophagy alleviated HFD-induced steatosis.
Wild type mice were fed on a regular chew diet or a HFD for 12 weeks. CBZ or Rapamycin was injected every other day for two or three times, respectively, in the week before sacrifice. (A–B) Cryosections of the livers were stained with Bodipy 493/503 and the numbers of LDs were quantified. Hepatic (C) and blood (D) TG and blood ALT levels (E) were determined. (F–G) Blood glucose and insulin levels were measured in mice fed with HFD with or without rapamycin or CBZ treatment (F), from which the HOMA values were calculated (G). #: p<0.05, *: p<0.01(n=3–7).
Discussion
Acute ethanol exposure through binge drinking [18] or intraperitoneal injection (this study) could clearly activate cytoprotective autophagy. Our present studies also showed that autophagy remained active under chronic ethanol treatment administrated through a widely adopted 4-week regimen of the Lieber-DeCarli liquid diet with the final ethanol content accounting for 29% or 36% of the total calorie of the diet. Flux analysis based on either imaging or biochemical analyses showed that there was a net increase of autophagic degradation. Suppression of the degradation with lysosome inhibitors led to increased liver injury and steatosis, suggesting that the autophagy function remains to be pathologically relevant.
However, since prolonged ethanol treatment can affect vesicular trafficking and lysosome function, which could adversely affect autophagy [31], it has to be cautioned that our present results do not rule out the possibility that the capacity of autophagy degradation could be at its suboptimal condition under the 4-week Lieber-DeCarli regime, despite that it is at a level higher than that at the basal condition. It is also possible that in a more severe regimen and/or over a longer period autophagy function could eventually deteriorate although different components of the autophagy pathway may have different susceptibility. It is thus interesting to note that the cathepsin B activity, but not that of several other lysosomal enzymes, was reduced in the 36% ethanol group, but not in the 29% ethanol group (Fig. S3), reflecting the above considerations. While these issues have yet to be fully explored, it does seem clear that it would be beneficial to maintain or enhance the autophagy capacity in the liver to improve the pathological condition. The need of such modulations is further precipitated by the current lack of effective remedies treating AFLD.
The present study demonstrated that pharmacological enhancement of autophagy with either CBZ or rapamycin reduced liver injury. We have found that CBZ and rapamycin can induce autophagy in hepatocytes. Unlike in non-hepatocytes [32], rapamycin seems to be able to fully suppress mTOR activity in hepatocytes as indicated by the dephosphorylation of both p70S6 kinase and 4E-BP1 [18](Fig. S1). While enhanced autophagy could result in increased degradation of multiple targets, one of the key targets examined in this study is the lipid droplets. Earlier studies have demonstrated that autophagy is important in controlling hepatic lipid level in both alcohol and non-alcohol conditions [16–19]. We found that both CBZ and rapamycin stimulated the reduction of hepatic steatosis and blood TG levels, which could in turn lead to an overall reduction of total cellular free fatty acids content and thus contribute to the overall reduction of liver injury [4, 5, 9]. Although not examined in this study, other effects of autophagy, such as removing damaged mitochondria, would be also expected to play a role in enhancing the protection against liver injury [13, 18].
Notably CBZ and rapamycin were also effective in reducing hepatic steatosis and dyslipidemia in mice fed with high fat diet for 12 weeks. Conversely, chloroquine treatment would further enhance hepatic steatosis and liver injury (data not shown). As for AFLD, there are also very limited options in mitigating NAFLD although several clinical trials are now showing promising results [10–12]. NAFLD and AFLD seem to share similar pathogenesis that involves progression from simple steatosis to steatohepatitis, fibrosis and cirrhosis [4, 5, 9]. In addition, NAFLD can be part of the metabolic syndrome that also presents with dyslipidemia, insulin resistance and Type II diabetes [6–8].
Although it is limited, emerging evidence now indicates that the progression of NAFLD is associated with a reduced function of autophagy in both ob/ob and HFD-fed mice [16, 17]. Genetically over-expressing an autophagy gene, Atg7, reduced hepatic steatosis and the associated endoplasmic reticulum stress presentation [17]. Furthermore, this genetic manipulation also resulted in reduced insulin resistance, consistent with the notion that treatment of liver presentation could lead to improvement of metabolic syndrome overall [6, 7, 12, 17]. Our present study provides the first example of achieving the results using available pharmacological agents that simulate autophagy, indicating that this strategy could be considered as a novel approach for alleviating the liver condition in NAFLD as well as in AFLD.
Pharmacological reduction of hepatic steatosis in both AFLD and NAFLD by CBZ and rapamycin would be most likely related to the enhanced autophagy degradation of lipids, or lipophagy, as supported by our present studies. This notion is consistent with previous reports using rapamycin to reduce Malory-Denk bodies [21] and using CBZ to reduce mutant α1-antitrypsin proteins [22] in the livers of genetically altered mice. However, it cannot be ruled out that in different regimens these drugs could have additional effects on other mechanisms affecting lipid turnover. This may be relevant in the case of rapamycin, which seem to be able to up-regulate genes like Acox1 and Cpt1a (Table 1). It is possible that rapamycin can promote clearance of steatosis through its stimulating effects on both autophagy and fatty acid oxidation although the relative contribution of the latter is not clear.
CBZ has been used in clinic to treat seizures and trigeminal neuralgia and is well tolerated. The mechanism is not entirely clear but it can inhibit myo-inositol-1-phosphate synthase and reduce inositol and IP3 levels, which could be also related to how it induces autophagy [20]. This is different from rapamycin, which induces autophagy by inhibiting mTOR complex 1. There is an advantage of CBZ over rapamycin in clinic in that the latter could have adverse effects in other systems, such as the immune system. There are still several more well-tolerated drugs, such as sodium valproate and verapamil [20], that are currently used in clinic for other purposes, but are known to stimulate autophagy in vitro. All these agents could be potentially explored for their autophagy-enhancing effects in fatty liver conditions. Taken together, pharmacological modulation of autophagy for AFLD and/or NAFLD could be a novel strategy that can take the advantage of existing medicines that have a high safety profile.
Supplementary Material
Acknowledgments
Financial support: This work was in part supported by NIH (CA83817, CA111456 to X-M.Y)
List of abbreviations
- AFLD
alcoholic fatty liver disease
- ALT
alanine aminotransferase
- CBZ
carbamazepine
- CQ
chloroquine
- EBSS
Earle’s Balanced Salt Solution
- HFD
high fat diet
- LD
lipid droplets
- NAFLD
nonalcoholic fatty liver disease
- RP
rapamycin
- TG
triglycerides
Footnotes
Conflict of interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zakhari S, Li TK. Determinants of alcohol use and abuse: Impact of quantity and frequency patterns on liver disease. Hepatology. 2007;46:2032–2039. doi: 10.1002/hep.22010. [DOI] [PubMed] [Google Scholar]
- 2.Lieber CS. Alcoholic fatty liver: its pathogenesis and mechanism of progression to inflammation and fibrosis. Alcohol. 2004;34:9–19. doi: 10.1016/j.alcohol.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Lumeng L, Crabb DW. Alcoholic liver disease. Curr Opin Gastroenterol. 2000;16:208–218. doi: 10.1097/00001574-200005000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Hall PD. Pathological spectrum of alcoholic liver disease. Alcohol Alcohol Suppl. 1994;2:303–313. [PubMed] [Google Scholar]
- 5.Angulo P. Nonalcoholic fatty liver disease. N Engl J Med. 2002;346:1221–1231. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- 6.Fabbrini E, Sullivan S, Klein S. Obesity and nonalcoholic fatty liver disease: biochemical, metabolic, and clinical implications. Hepatology. 2009;51:679–689. doi: 10.1002/hep.23280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choudhury J, Sanyal AJ. Insulin resistance and the pathogenesis of nonalcoholic fatty liver disease. Clin Liver Dis. 2004;8:575–594. ix. doi: 10.1016/j.cld.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Jou J, Choi SS, Diehl AM. Mechanisms of disease progression in nonalcoholic fatty liver disease. Semin Liver Dis. 2008;28:370–379. doi: 10.1055/s-0028-1091981. [DOI] [PubMed] [Google Scholar]
- 9.Diehl AM, Goodman Z, Ishak KG. Alcohollike liver disease in nonalcoholics. A clinical and histologic comparison with alcohol-induced liver injury. Gastroenterology. 1988;95:1056–1062. [PubMed] [Google Scholar]
- 10.Sanyal AJ, Chalasani N, Kowdley KV, McCullough A, Diehl AM, Bass NM, et al. Pioglitazone, vitamin E, or placebo for nonalcoholic steatohepatitis. N Engl J Med. 2010;362:1675–1685. doi: 10.1056/NEJMoa0907929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Socha P, Horvath A, Vajro P, Dziechciarz P, Dhawan A, Szajewska H. Pharmacological interventions for nonalcoholic fatty liver disease in adults and in children: a systematic review. J Pediatr Gastroenterol Nutr. 2009;48:587–596. doi: 10.1097/MPG.0b013e31818e04d1. [DOI] [PubMed] [Google Scholar]
- 12.Ratziu V, Charlotte F, Bernhardt C, Giral P, Halbron M, Lenaour G, et al. Long-term efficacy of rosiglitazone in nonalcoholic steatohepatitis: results of the fatty liver improvement by rosiglitazone therapy (FLIRT 2) extension trial. Hepatology. 2010;51:445–453. doi: 10.1002/hep.23270. [DOI] [PubMed] [Google Scholar]
- 13.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin XM, Ding WX, Gao W. Autophagy in the liver. Hepatology. 2008;47:1773–1785. doi: 10.1002/hep.22146. [DOI] [PubMed] [Google Scholar]
- 15.Rautou PE, Mansouri A, Lebrec D, Durand F, Valla D, Moreau R. Autophagy in liver diseases. J Hepatol. 2010;53:1123–1134. doi: 10.1016/j.jhep.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding WX, Li M, Chen X, Ni HM, Lin CW, Gao W, et al. Autophagy reduces acute ethanol-Induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu D, Wang X, Zhou R, Cederbaum A. CYP2E1 enhances ethanol-induced lipid accumulation but impairs autophagy in HepG2 E47 cells. Biochem Biophys Res Commun. 2010;402:116–122. doi: 10.1016/j.bbrc.2010.09.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleming A, Noda T, Yoshimori T, Rubinsztein DC. Chemical modulators of autophagy as biological probes and potential therapeutics. Nat Chem Biol. 2011;7:9–17. doi: 10.1038/nchembio.500. [DOI] [PubMed] [Google Scholar]
- 21.Harada M, Hanada S, Toivola DM, Ghori N, Omary MB. Autophagy activation by rapamycin eliminates mouse Mallory-Denk bodies and blocks their proteasome inhibitor-mediated formation. Hepatology. 2008;47:2026–2035. doi: 10.1002/hep.22294. [DOI] [PubMed] [Google Scholar]
- 22.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 23.Ding WX, Ni HM, Gao W, Hou YF, Melan MA, Chen X, et al. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem. 2007;282:4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- 24.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He J, de la Monte S, Wands JR. Acute ethanol exposure inhibits insulin signaling in the liver. Hepatology. 2007;46:1791–1800. doi: 10.1002/hep.21904. [DOI] [PubMed] [Google Scholar]
- 26.You M, Fischer M, Deeg MA, Crabb DW. Ethanol induces fatty acid synthesis pathways by activation of sterol regulatory element-binding protein (SREBP) J Biol Chem. 2002;277:29342–29347. doi: 10.1074/jbc.M202411200. [DOI] [PubMed] [Google Scholar]
- 27.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinemann L. Insulin assay standardization: leading to measures of insulin sensitivity and secretion for practical clinical care: response to Staten et al. Diabetes Care. 2010;33:e83. doi: 10.2337/dc09-1206. author reply e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao R, Wei D, Gao H, Liu Y, DePinho RA, Dong XC. Hepatic FoxOs regulate lipid metabolism via modulation of expression of the nicotinamide phosphoribosyltransferase gene. The Journal of biological chemistry. 2011;286:14681–14690. doi: 10.1074/jbc.M110.201061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk V, Kaushik S, et al. In search of an “autophagomometer”. Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 31.Donohue TM., Jr Autophagy and ethanol-induced liver injury. World J Gastroenterol. 2009;15:1178–1185. doi: 10.3748/wjg.15.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.