Abstract
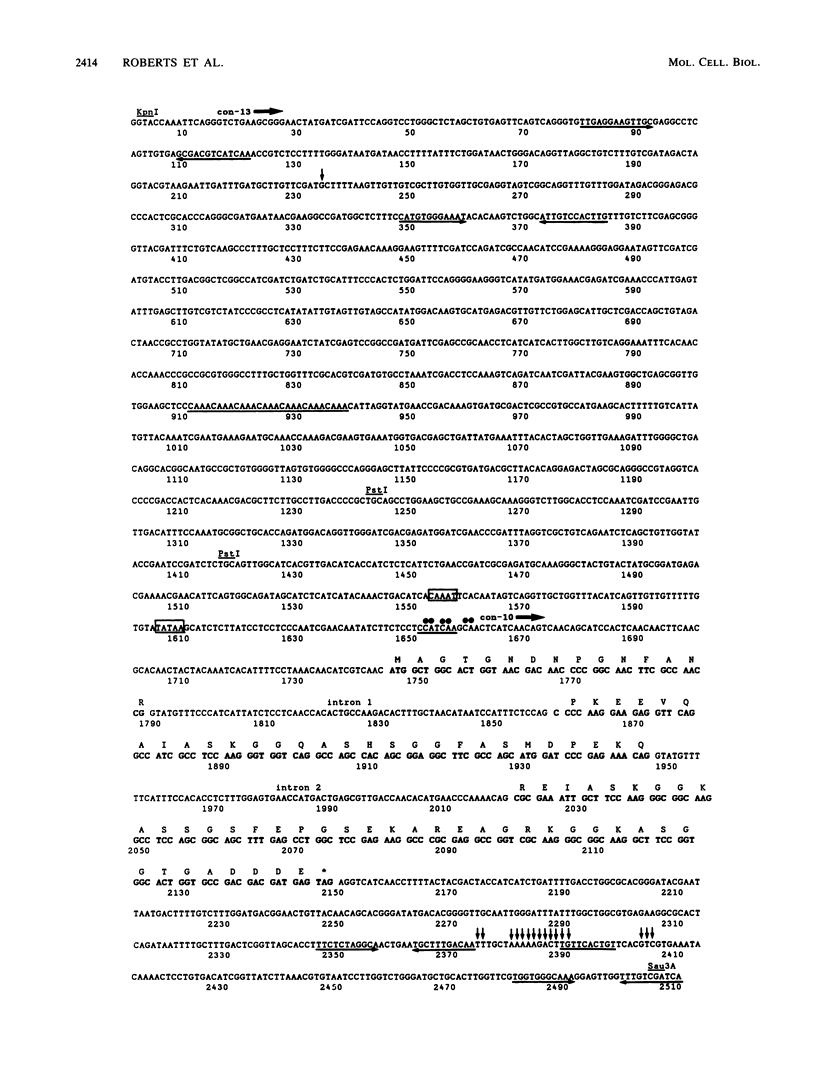
The asexual developmental pathway in the life cycle of the filamentous fungus Neurospora crassa culminates in the formation of spores called conidia. Several clones of genomic Neurospora DNA have been isolated that correspond to mRNA species expressed during conidiation and not during mycelial growth (V. Berlin and C. Yanofsky, Mol. Cell. Biol. 5:849-855, 1985). In this paper we describe the characterization of one of these clones, named pCon-10a. This clone contains two genes, con-10 and con-13, which are induced coordinately during the later stages of conidiation. The two genes are separated by 1.4 kilobases of DNA; they are located on linkage group IV and are transcribed from the same strand of DNA. The molecular organization and sequence of one of these genes, con-10, and its flanking regions are presented. Full-length cDNA clones for con-10 also were isolated and sequenced, and transcription-initiation and polyadenylation sites were defined. The con-10 gene contains an open reading frame interrupted by two small introns and encodes an 86-amino-acid residue polypeptide that is both hydrophilic and weakly acidic. Expression of the con-10 gene in various mutants defective at different stages of conidiation indicates that it plays a role after aerial hyphal development. Possible functions, organization, and regulation of conidiation-specific genes are discussed.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arends H., Sebald W. Nucleotide sequence of the cloned mRNA and gene of the ADP/ATP carrier from Neurospora crassa. EMBO J. 1984 Feb;3(2):377–382. doi: 10.1002/j.1460-2075.1984.tb01815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty J. T., Cohen S. N. Hybridization of cloned Rhodopseudomonas capsulata photosynthesis genes with DNA from other photosynthetic bacteria. J Bacteriol. 1983 Jun;154(3):1440–1445. doi: 10.1128/jb.154.3.1440-1445.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beever R. E., Dempsey G. P. Function of rodlets on the surface of fungal spores. Nature. 1978 Apr 13;272(5654):608–610. doi: 10.1038/272608a0. [DOI] [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Berlin V., Yanofsky C. Isolation and characterization of genes differentially expressed during conidiation of Neurospora crassa. Mol Cell Biol. 1985 Apr;5(4):849–855. doi: 10.1128/mcb.5.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin V., Yanofsky C. Protein changes during the asexual cycle of Neurospora crassa. Mol Cell Biol. 1985 Apr;5(4):839–848. doi: 10.1128/mcb.5.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan M. T., Mirabito P. M., Willett C. E., Zimmerman C. R., Timberlake W. E. Isolation and physical characterization of three essential conidiation genes from Aspergillus nidulans. Mol Cell Biol. 1987 Sep;7(9):3113–3118. doi: 10.1128/mcb.7.9.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn A., Bollekens J., Staub A., Benoist C., Mathis D. A multiplicity of CCAAT box-binding proteins. Cell. 1987 Sep 11;50(6):863–872. doi: 10.1016/0092-8674(87)90513-7. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- George D. G., Barker W. C., Hunt L. T. The protein identification resource (PIR). Nucleic Acids Res. 1986 Jan 10;14(1):11–15. doi: 10.1093/nar/14.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Gwynne D. I., Miller B. L., Miller K. Y., Timberlake W. E. Structure and regulated expression of the SpoC1 gene cluster from Aspergillus nidulans. J Mol Biol. 1984 Nov 25;180(1):91–109. doi: 10.1016/0022-2836(84)90432-7. [DOI] [PubMed] [Google Scholar]
- Hager K. M., Mandala S. M., Davenport J. W., Speicher D. W., Benz E. J., Jr, Slayman C. W. Amino acid sequence of the plasma membrane ATPase of Neurospora crassa: deduction from genomic and cDNA sequences. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7693–7697. doi: 10.1073/pnas.83.20.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnisch U., Weiss H., Sebald W. The primary structure of the iron-sulfur subunit of ubiquinol-cytochrome c reductase from Neurospora, determined by cDNA and gene sequencing. Eur J Biochem. 1985 May 15;149(1):95–99. doi: 10.1111/j.1432-1033.1985.tb08898.x. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Huiet L., Giles N. H. The qa repressor gene of Neurospora crassa: wild-type and mutant nucleotide sequences. Proc Natl Acad Sci U S A. 1986 May;83(10):3381–3385. doi: 10.1073/pnas.83.10.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone I. L., Hughes S. G., Clutterbuck A. J. Cloning an Aspergillus nidulans developmental gene by transformation. EMBO J. 1985 May;4(5):1307–1311. doi: 10.1002/j.1460-2075.1985.tb03777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnaird J. H., Fincham J. R. The complete nucleotide sequence of the Neurospora crassa am (NADP-specific glutamate dehydrogenase) gene. Gene. 1983 Dec;26(2-3):253–260. doi: 10.1016/0378-1119(83)90195-6. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987 Oct 26;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kreader C. A., Heckman J. E. Isolation and characterization of a Neurospora crassa ribosomal protein gene homologous to CYH2 of yeast. Nucleic Acids Res. 1987 Nov 11;15(21):9027–9042. doi: 10.1093/nar/15.21.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Legerton T. L., Yanofsky C. Cloning and characterization of the multifunctional his-3 gene of Neurospora crassa. Gene. 1985;39(2-3):129–140. doi: 10.1016/0378-1119(85)90306-3. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Matsuyama S. S., Nelson R. E., Siegel R. W. Mutations specifically blocking differentiation of macroconidia Neurospora crassa. Dev Biol. 1974 Dec;41(2):278–287. doi: 10.1016/0012-1606(74)90306-6. [DOI] [PubMed] [Google Scholar]
- Miller B. L., Miller K. Y., Roberti K. A., Timberlake W. E. Position-dependent and -independent mechanisms regulate cell-specific expression of the SpoC1 gene cluster of Aspergillus nidulans. Mol Cell Biol. 1987 Jan;7(1):427–434. doi: 10.1128/mcb.7.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münger K., Germann U. A., Lerch K. Isolation and structural organization of the Neurospora crassa copper metallothionein gene. EMBO J. 1985 Oct;4(10):2665–2668. doi: 10.1002/j.1460-2075.1985.tb03985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Orbach M. J., Porro E. B., Yanofsky C. Cloning and characterization of the gene for beta-tubulin from a benomyl-resistant mutant of Neurospora crassa and its use as a dominant selectable marker. Mol Cell Biol. 1986 Jul;6(7):2452–2461. doi: 10.1128/mcb.6.7.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr W. C., Timberlake W. E. Clustering of spore-specific genes in Aspergillus nidulans. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5976–5980. doi: 10.1073/pnas.79.19.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. D., Radford A., Newmeyer D., Björkman M. Chromosomal loci of Neurospora crassa. Microbiol Rev. 1982 Dec;46(4):426–570. doi: 10.1128/mr.46.4.426-570.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Rodriguez J. R., Pikielny C. W., Rosbash M. In vivo characterization of yeast mRNA processing intermediates. Cell. 1984 Dec;39(3 Pt 2):603–610. doi: 10.1016/0092-8674(84)90467-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Schechtman M. G., Yanofsky C. Structure of the trifunctional trp-1 gene from Neurospora crassa and its aberrant expression in Escherichia coli. J Mol Appl Genet. 1983;2(1):83–99. [PubMed] [Google Scholar]
- Schmit J. C., Brody S. Biochemical genetics of Neurospora crassa conidial germination. Bacteriol Rev. 1976 Mar;40(1):1–41. doi: 10.1128/br.40.1.1-41.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selitrennikoff C. P., Nelson R. E., Siegel R. W. Phase-specific genes for macroconidiation in Neurospora crassa. Genetics. 1974 Oct;78(2):679–690. doi: 10.1093/genetics/78.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Staden R. A computer program to enter DNA gel reading data into a computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):499–503. doi: 10.1093/nar/12.1part2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sures I., Lowry J., Kedes L. H. The DNA sequence of sea urchin (S. purpuratus) H2A, H2B and H3 histone coding and spacer regions. Cell. 1978 Nov;15(3):1033–1044. doi: 10.1016/0092-8674(78)90287-8. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Villa V. D., Storck R. Nucleotide composition of nuclear and mitochondrial deoxyribonucleic acid of fungi. J Bacteriol. 1968 Jul;96(1):184–190. doi: 10.1128/jb.96.1.184-190.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudt L. P., Pastink A., Kempers-Veenstra A. E., Jansen A. E., Mager W. H., Planta R. J. The genes coding for histone H3 and H4 in Neurospora crassa are unique and contain intervening sequences. Nucleic Acids Res. 1983 Aug 25;11(16):5347–5360. doi: 10.1093/nar/11.16.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]