Abstract
PAX2 is one of nine PAX genes that regulate tissue development and cellular differentiation in embryos. However, the functional role of PAX2 in ovarian cancer is not known. Twenty-six ovarian cancer cell lines with different histology origins were screened for PAX2 expression. Two ovarian cancer cell lines: RMUGL (mucinous) and TOV21G (clear cell), with high PAX2 expression were chosen for further study. Knockdown PAX2 expression in these cell lines was achieved by lentiviral shRNAs targeting the PAX2 gene. PAX2 stable knockdown cells were characterized for cell proliferation, migration, apoptosis, protein profiles, and gene expression profiles. The result indicated that these stable PAX2 knockdown cells had reduced cell proliferation and migration. Microarray analysis indicated that several genes involved in growth inhibition and motility, such as G0S2, GREM1, and WFDC1, were up-regulated in PAX2 knockdown cells. On the other hand, over-expressing PAX2 in PAX2-negative ovarian cell lines suppressed their cell proliferation. In summary, PAX2 could have both oncogenic and tumor suppression functions, which might depend on the genetic content of the ovarian cancer cells. Further investigation of PAX2 in tumor suppression and mortality is warranted.
Keywords: PAX2, ovarian cancer, G0S2, WFDC1, GREM1, shRNA
1. Introduction
PAX2 is one of nine PAX genes; all of these genes have a conserved DNA sequence motif called the paired box that comprises a 128-amino acid domain in the amino-terminal portion of the protein [1]. During embryogenesis, PAX2 is abundantly expressed in the kidney [2,3], ureter [4], eye [5], cochlea [6], pancreas [7], and central nervous system [8,9] and is crucial to embryogenic development, morphogenesis, and organogenesis [10,11]. Embryonic PAX2 gene expression is largely attenuated in adult tissue although continued expression can be detected in female genital tract, breast and other tissues [12]. PAX2 deficiency has been associated with various growth defects, such as kidney hypoplasia, optic coloboma, and vesicoureteral reflux [13]. Conversely, PAX2 overexpression is associated with cystic or tumorous epithelial overgrowth [14], such as renal cystic dysplasia, renal cell carcinoma, Wilms’ tumor, nephrogenic adenoma, prostate cancer, breast cancer and ovarian cancer [15–19]. Expression of PAX2 in these cells appears to be important for tumor cell survival [17,20,21]. However, recent studies indicated that loss of PAX2 expression correlates with the development of serous carcinoma in the fallopian tube [22–24]. Similarly, the loss of PAX2 expression also correlates with the development of endometrial precancer and cancer [25]. Thus, it is possible that PAX2 could be an oncogene or tumor suppressor [12]. The function of PAX2 in the development of ovarian cancer is still unknown. In this study, using both PAX2 positive and negative ovarian cancer cell lines, we investigated the potential functional roles of PAX2 in ovarian cancer.
2. Results
2.1. Ovarian Cancer Cell Lines Expressed Different Level of PAX2
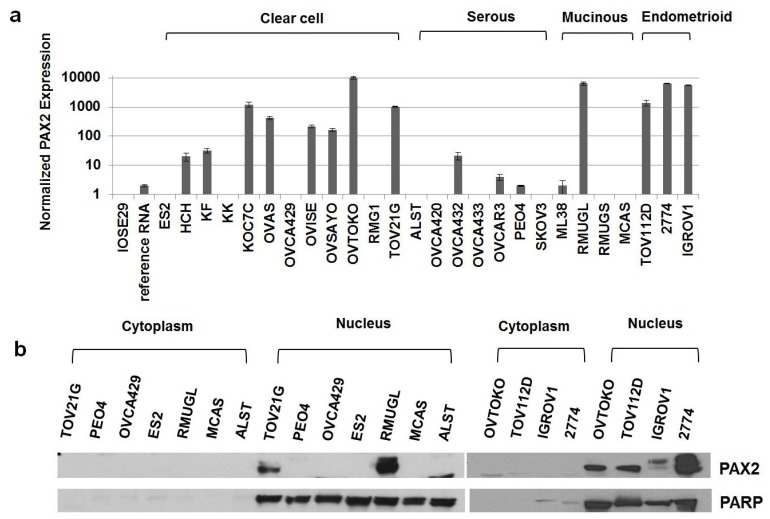
Twenty-six ovarian cancer cell lines (8 serous ovarian cancer cell lines, 12 clear ovarian cancer cell lines, 3 mucinous ovarian cancer cell lines, 3 endometrioid ovarian cancer cell lines) and one immortalized normal ovarian surface epithelium cell line (IOSE29) were screened for PAX2 expression by real-time RT-PCR. Sixteen of the cell lines (HCH, KF, KOC7C, OVAS, OVISE, OVSAYO, OVTOKO, TOV21G, OVCA 432, OVCAR3, PEO4, ML38, RMUGL, TOV112D, 2774, and IGROV1) exhibited 2 to 10270 times higher PAX2 mRNA expression than IOSE29 cells did. PAX2 was found to be highly expressed in mostly non-serous ovarian cancer cell lines (Figure 1A). RMUGL and OVTOKO had the highest expression level of PAX2 mRNA followed by 2774, IGROV1, KOC7C, TOV112D and TOV21G. Figure 1B showed the nuclear protein expression of PAX2 expression by Western blot in a few selected ovarian cancer cell lines with different level of mRNA expression. There is a strong correlation of PAX2 protein expression with the mRNA expression.
Figure 1.
Expression of PAX2 in various ovarian cancer cell lines. (a) Real-time RT-PCR analysis of PAX2 mRNA expression in twenty-six ovarian cancer cell lines with different histology origins; (b) Western blot analysis of PAX2 protein expression level in seven selected ovarian cancer cell lines.
2.2. PAX2 Knockdown Is Associated with Reduced Cell Proliferation
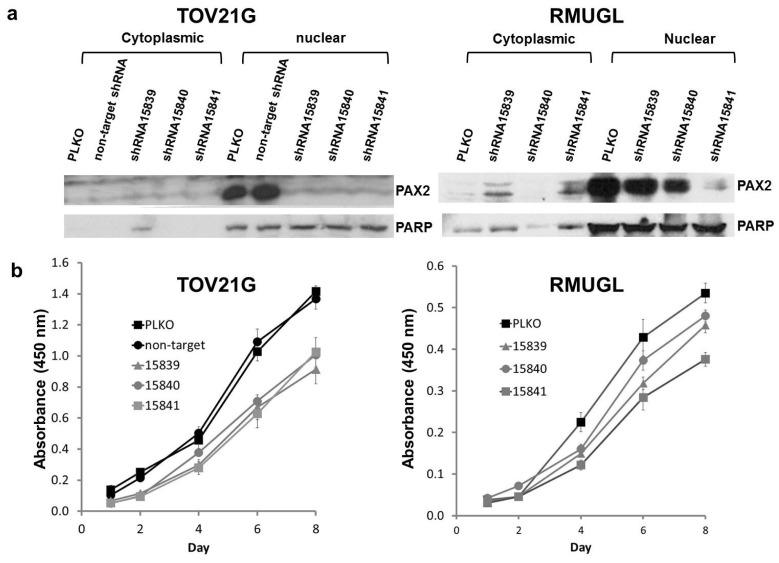
Two PAX2 positive cell lines of different histology origins (RMUGL and TOV21G) and of different levels of PAX2 expression were chosen for PAX2 knockdown experiments. MISSION TRC shRNA lentiviral particles (three independent shRNAs—15839, 15840 and 15841) were used to transduce the ovarian cancer cell lines RMUGL and TOV21G. After selecting stably transfected cells by puromycin, Western blotting was used to evaluate the PAX2 knockdown efficiency (Figure 2). For RMUGL cell line, PAX2 expression was partially knockdown in shRNA 15839- and shRNA 15840-stably transfected cells, but almost completely knockdown in shRNA 15841-stably transfected cell (Figure 2a). For TOV21G cell line, PAX2 expression was completely knockdown in all PAX2 shRNA stably transfected cells (Figure 2a). The knockdown efficiency was especially robust using shRNA 15841. The difference in PAX2 knockdown efficiency is likely due to a 10-fold higher expression of PAX2 in parental RMUGL cell line than TOV21G cell line.
Figure 2.
(a) Western blot analysis of TOV21G clear cell line and RMUGL mucinous cell line stably transfected with PAX2-targeted shRNAs 15839, 15840, and 15841. PLKO and non-target are vector controls; (b) Cell proliferation of PAX2 shRNAs stably transfected cells measured by WST-1 assay.
Cell proliferation assays were performed on both the original cell lines and the PAX2 stably knockdown cell lines. As shown in Figure 2b, the cell proliferation rate of the most PAX2 knockdown RMUGL cell line (shRNA 15841) had a significant decrease in cell proliferation rate in comparison to the PLKO vector-transfected control cells while the decrease in cell proliferation in other two shRNAs (15839 and 15840) stably transfected RMUGL was not significant. This suggested a near complete knockdown of PAX2 might be necessary to affect cell proliferation. The effect of PAX2 knockdown on cell proliferation was significant (p < 0.05) in the clear cell ovarian cancer cell line TOV21G (Figure 2b).
2.3. PAX2 Knockdown Is Associated with Reduced Cell Motility
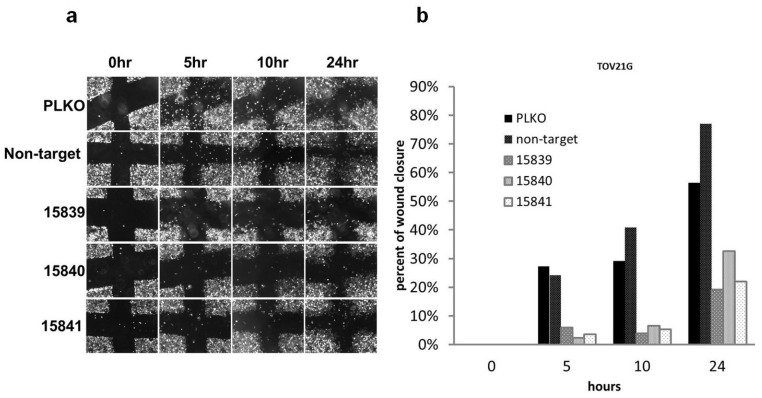
To test whether PAX2 plays a role in cell migration, we conducted a wound healing assay using the TOV21G cell lines. Compared with control cell lines with positive PAX2 expression, PAX2 knockdown cell lines had reduced cell motility (Figure 3). Similar results were obtained with PAX2 knockdown RMUGL cells by shRNA 15841 (data not shown).
Figure 3.
(a) Images of wound closure assay of TOV21G cell line stably transfected with PAX2 specific shRNAs during a 24 h period. PLKO and non-target shRNA were the negative controls, and shRNAs 15839, 15840 and 15841 were PAX2-targeted shRNAs; (b) the images were analyzed by the TScratch program [26] to estimate the percentage of wound closure. PAX2 stable knockdown TOV21G cells had slower rate of wound closure.
2.4. Cell Signaling Protein Expression Analysis by Reverse Phase Protein Array (RPPA) Suggested PAX2 Knockdown Might Enhance Apoptotic Signaling
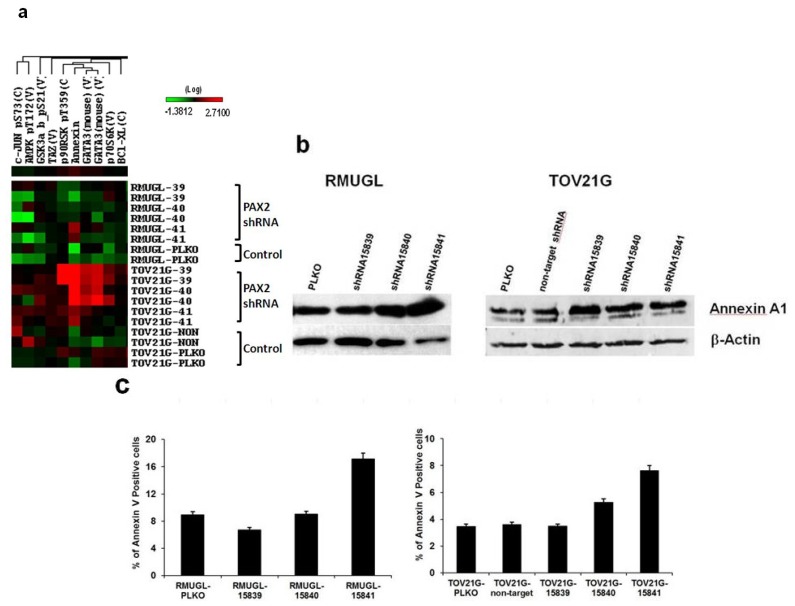
RPPA was used to analyze the effects of PAX2 knockdown on the expression of 207 signaling proteins in ovarian cancer cell lines RMUGL and TOV21G. Two independent protein lysates replicate were analyzed. RPPA sample preparation, slide printing, staining and data analysis were processed as described [27]. From the RPPA analysis, PAX2 knockdown cell lines had increased expression of Annexin A1, a marker of early stage apoptosis (Figure 4a). Annexin A1 expression was further confirmed by Western blot (Figure 4b). To evaluate the potential role of PAX2 in apoptosis, allophycocyanin-Annexin V staining, which detects an increase of phosphatidylserine residues in the outer plasma membrane leaflet during the early stages of apoptosis was used to measure apoptotic cells in RMUGL and TOV21G ovarian cancer cell lines with PAX2 knockdown (Figure 4c). In RMUGL, the percentage of apoptotic cells was 6.73%, 9.01%, and 17.15% for cells transfected with shRNAs 15839, 15841, and 15841, respectively, compared with 8.95% in the PLKO control (Figure 4c). In TOV21G, the percentage of apoptotic cells was 3.48%, 5.27% and 7.63% for cells transfected with shRNAs 15839, 15841, and 15841, respectively, compared with 3.47% in the PLKO control (Figure 4c). Thus, there was an association between the extent of PAX2 knockdown and the percentage of apoptotic cells.
Figure 4.
Up-regulation of Annexin A1 and increase in apoptotic cells in PAX2 knockdown cell lines. (a) Reverse phase protein array (RPPA) analysis showed PAX2 knockdown ovarian cancer cell lines had a higher expression of Annexin A1 than control cells; (b) Western blot analysis was used to measure Annexin A1 expression in RMUGL and TOV21G ovarian cancer cell lines with or without PAX2 knockdown; (c) Flow cytometric analysis of percentage of apoptotic cells using APC-Annexin V staining.
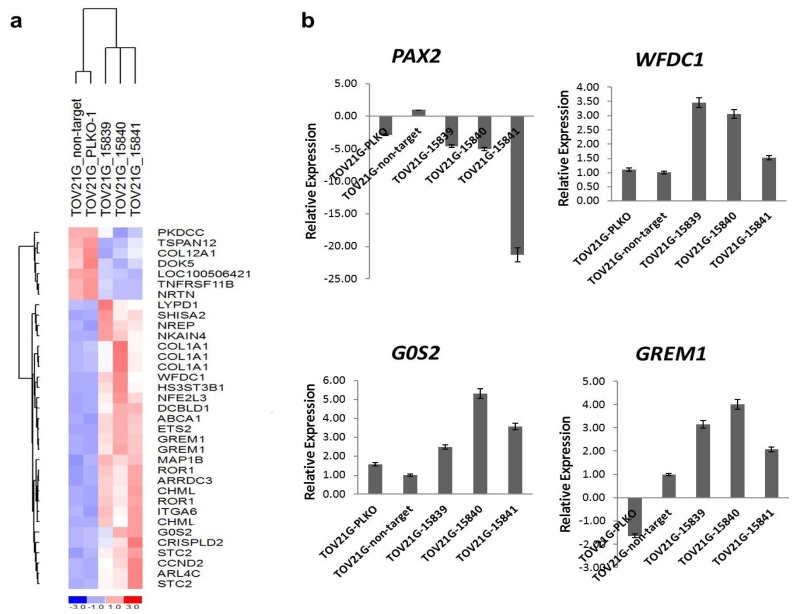
2.5. G0S2, WFDC1 and GREM1 Were Up-Regulated in PAX2 Knockdown TOV21G Cell Lines
Potential genes affected by PAX2 knockdown were identified by gene expression profiling of TOV21G cells with PAX2 knockdown by shRNA 15839, 15840, and 15841; these expression profiles were compared to that of TOV21G-PLKO amd TOV21G-non-target control cells. Gene expression data has been deposited in the public database (Gene Expression Omnibus, GSE30501). Genes that are highly differentially expressed between PAX2 knockdown TOV21G cells and control cells were shown (Figure 5a). The up-regulation of three genes involved in suppressing cell proliferation and/or cellular movement (G0S2, WFDC1 and GREM1) was further validated by real-time RT-PCR (Figure 5b). The top 100 differentially expressed genes were listed in the Table S1.
Figure 5.
(a) The most differentially expressed genes in the PAX2 stable knockdown TOV21G clear cell ovarian cancer cell lines identified by microarray analysis; (b) Validation of down-regulation of PAX2 and the up-regulation of G0S2, WFDC1, and GREM1 in PAX2 knockdown TOV21G cell lines by RT-PCR.
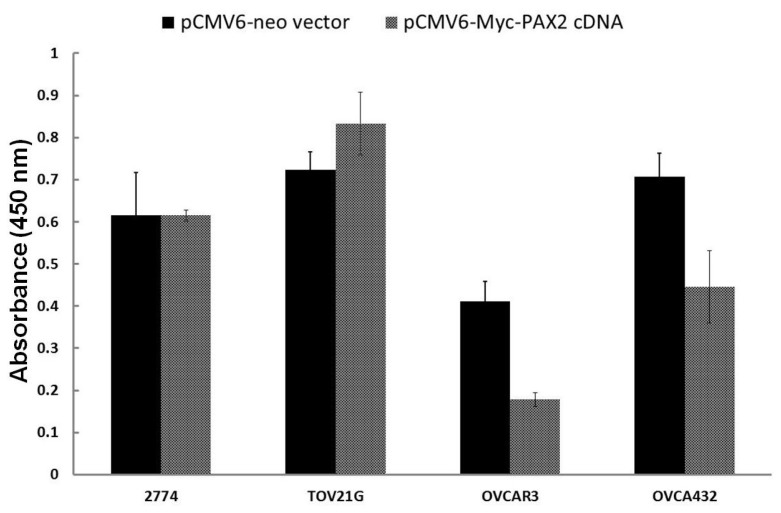
2.6. Over-Expressing PAX2 Protein in PAX2-Negative Ovarian Cancer Cell Lines Is Associated with Reduced Cell Proliferation
Two PAX2 negative serous ovarian cancer cell lines (OVCAR3 and OVCA432) after transfected with pCMV6-Myc-PAX2, cell proliferation was significantly suppressed in comparison to the vector transfection control (Figure 6). The cell numbers of pCMV6-Myc-PAX2 transfected OVCA432 cells were only 43% of pCMV6-Neo Vector transfected OVCA432. Similarly, pCMV6-Myc-PAX2 transfected OVCAR3 cells were only 63% of pCMV6-Neo vector transfected OVCAR3. On the other hand, there was no growth inhibition for two PAX2-positive cell lines (2774 and TOV21G) transfected by pCMV6-Myc-PAX2 plasmid DNA (Figure 6). Transfection experiments were repeated at least three times. Over-expression of PAX2 protein in the nuclear extract from transfected 2774 and TOV21G cells by pCMV6-Myc-PAX2 were confirmed by Western blot (data not shown).
Figure 6.
Cell viability of pCMV6-Myc-PAX2 transfected ovarian cancer cells. 2774 and TOV21G are PAX2-positive cells, while OVCA432 and OVCAR3 are PAX2-negative cells. The reduction in cell viability of OVCAR3 and OVCA432 cells after transfected with pCMV6-Myc-PAX2 plasmid in comparison to pCMV6-Neo vector control was significant (p < 0.05).
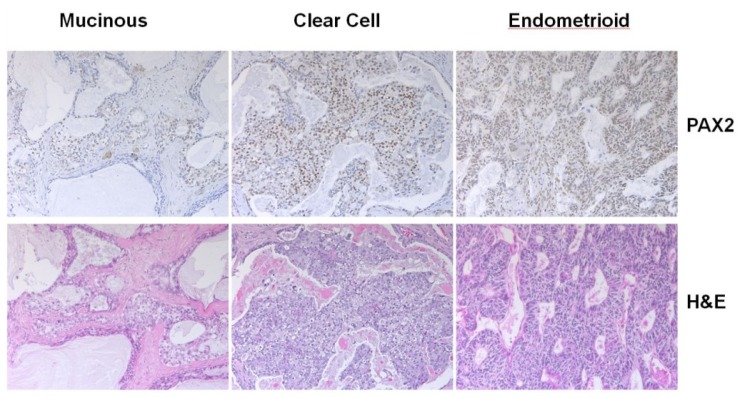
2.7. Analysis of PAX2 Protein Expression in Non-Serous Ovarian Cancer Patient Samples by Immunohistochemistry
Immunostaining of PAX2 was performed on paraffin sections from 55 non-serous ovarian carcinomas. Nine out of 26 clear cell carcinomas were stained positive (9/26 = 35%), four out of 12 endometrioid carinomas were stained positive (4/12 = 33%), and two out of 17 mucinous carcinomas were stained positive (2/17 = 12%). Examples of positive nuclear staining were shown (Figure 7). For the clear carcinomas with PAX2 nuclear positive staining, the percentage of tumor cells with positive staining varied from 10% to over 75%. On the other hand, endometrioid carcinomas with PAX2 nuclear positive staining had percentage of tumor cells with positive staining from 50% to more than 75%. For the two mucinous carcinomas with positive staining, one had 5% tumor cells with positive staining, and the other one had more than 50% tumor cells with positive staining. Thus, the expression of PAX2 in clear cell and endometrioid carcinomas were significantly higher than that of serous carcinomas (9%) that we had reported previously [19]. The gene expression level of PAX2 was estimated from microarray data (Figure S1), which indicated that non-serous ovarian carcinomas also had a higher gene expression of PAX2.
Figure 7.
Examples of nuclear immunostains for PAX2 in a clear cell, mucinous and endometrioid ovarian carcinomas.
3. Discussion
In this study, we found that PAX2 is frequently expressed in ovarian cancer cell lines, especially for those derived from non-serous type ovarian cancers (clear cell, mucinous, and endometrioid). This is consistent with the PAX2 expression in non-serous ovarian cancer from the gene expression data of ovarian cancer patient samples with different histological subtypes (GSE6008) and our own microarray data (Figure S1). This may reflect the different molecular pathogenesis or cellular origin of serous and non-serous ovarian carcinomas [28]. We have previously shown that PAX2 did not express in normal ovarian surface epithelia but expressed in ciliated epithelial inclusion in the ovaries and epithelia cells of the fallopian tube [19]. It is possible that some non-serous carcinomas are derived from ciliated epithelial inclusions. Ovarian clear cell and endometrioid carcinomas frequently carry ARID1A mutations but none of the high grade ovarian serous carcinomas have mutation in ARID1A [29]. On the other hand, almost all high grade ovarian serous carcinomas have mutated p53 but most ovarian clear cell and endometrioid carcinomas have wild-type p53 [28]. Recent study has shown that PAX2 promoter activity is stimulated by wild-type p53 but inhibited by a dominant negative mutant p53 [30]. The binding of p53 to the chromatin regions of the PAX2 promoter was identified by ChIP-Seq using developing kidneys in mice [30]. Thus, it is possible that the prevalent expression of PAX2 in non-serous carcinomas could be partly due to its activation by wild-type p53. However, further investigation will be necessary.
To determine the functional role of PAX2, knockdown PAX2 expression in PAX2-expressing ovarian cancer cells is associated with reduced cell proliferation and cell motility. The potential oncogenic role of PAX2 in these ovarian cancer cells is consistent with previous findings in prostate cancer, colon cancer, breast cancer and renal cancer [17,18,31,32]. Wound healing assays showed that PAX2 knockdown ovarian cancer cell lines had decreased cell motility in comparison to the parental PAX2-expressing cell lines. The reduced cell motility is further supported by the identification of more than 40 differentially expressed genes involved in cell proliferation and cell movement between the control cell lines and PAX2 knockdown cell lines. Three highly up-regulated genes (G0S2, WFDC1, and GREM1) in PAX2 knockdown cells were further validated by RT-PCR. G0S2 is a small basic nuclear phosphoprotein, one of the G0/G1 switch (G0S) genes that are differentially expressed in lymphocytes during lectin-induced switch from the G0 to the G1 phase of the cell cycle [33]. The expression of G0S2 is required to commit cells to enter the G1 phase of the cell cycle [34]. G0S2 specifically interacts with Bcl-2 and promotes apoptosis through preventing the formation of protective Bcl-2/Bax heterodimers [35]. WFDC1 (whey acidic protein four-disulfide core domain 1), a secreted protease inhibitor, and has been found it to be down-regulated in various cancers including fibrosarcomas, lung, bladder, and brain tumors [36]. Overexpression of WFDC1 has been shown to inhibit the growth rate of the fibrosarcoma HT1080 cell line [37]. GREM1 (Gremlin 1) is a member of the BMP (bone morphogenic protein) antagonist family [38]. Expression of GREM1 can regulate cancer cell growth positively [39] or negatively [40]. Further investigation of the roles of these up-regulated genes in PAX2 knockdown cells will be necessary to clarify their potential tumor suppression function in PAX2-expressing ovarian cancer cells.
The effects of PAX2 knockdown on cell signaling protein expression were studied using RPPA analysis. Annexin A1 was found to be the most significant up-regulated molecule detected by RPPA in PAX2 knockdown cell lines. Annexin A1 is proapoptotic by binding to the p65 subunit of NF-κB and thus inhibiting the NF-κB signal transduction pathway [41]. The increase in the percentage of apoptic cells in PAX2 knockdown cell lines were further demonstrated by flow cytometry with Allophycocyanin-Annexin V staining. Thus, the increased expression of Annexin A1 supports the notion that PAX2 may promote tumor growth in TOV21G ovarian cancer cell line by inhibiting the early stages of apoptosis through down-regulation of Annexin A1.
On the other hand, we have also shown for the first time that over-expressing PAX2 in PAX2-negative ovarian cancer cell lines suppresses their cell proliferation. Thus, PAX2 could have both oncogenic and tumor suppression functions which will depend on the genetic content of the cancer cells. However, the molecular mechanism is still unknown and will need further investigation.
4. Experimental Section
4.1. Cell Culture
Twenty-six human ovarian carcinoma cell lines (ES-2, HCH, KF, KK, KOC7C, OVAS, OVCA429, OVISE, OVSAYO, OVTOKO, RMG1, TOV21G, ALST, OVCA420, OVCA432, OVCA433, OVCAR3, PEO4, SKOV-3, ML38, RMUGL, RMUGS, MCAS, TOV112D, 2774, and IGROV1) and one immortalized ovarian surface epithelium cell line (IOSE29) were obtained from The University of Texas MD Anderson Cancer Center (Houston, TX, USA). All cells were grown in Roswell Park Memorial Institute-1640 (RPMI-1640) with 10% fetal bovine serum and 1% penicillin/streptomycin in 5% CO2 at 37 °C.
4.2. Stable PAX2 Knockdown Cell Lines
Twenty-six human ovarian carcinoma cell lines were screened for PAX2 expression by Western blots. Two cell lines (TOV21G, RMUGL) with robust PAX2 expression were used for generating stably PAX2 knockdown cell lines with PAX2 targeted shRNAs. MISSION TRC shRNA Lentiviral Particles (Sigma-Aldrich, St. Louis, MO, USA) targeting various regions of PAX2 (shRNA 15839, CCGGCGTCTCTTCCATCAACAGAATCTCGAGATTCTGTTGATGGAAGAGACGTTTTT; hRNA 15840, CCGGCCCAAAGTGGTGGACAAGATTCTCGAGAATCTTGTCCACCACTTTGGGTT TTT; shRNA 15841, CCGGGATGAAGTCAAGTCGAGTCTACTCGAGTAGACTCGACTTGACT TCATCTTTTT) were used to infect the ovarian cancer cell lines that expressed PAX2. The control lentivirus PLKO-puro (no insert sequence) (Sigma-Aldrich, St. Louis, MO, USA) and shRNA non-target control (insert sequence: CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGG TGCTCTTCATCTTGTTGTTTTT) (Sigma-Aldrich, St. Louis, MO, USA) were used as negative controls. Puromycin was used to select stable PAX2 knockdown cells. After stable knockdown, Western blot and real-time reverse transcription polymerase chain reaction (RT-PCR) were used to confirm the efficacy of PAX2 knockdown.
4.3. Gene Expression Profiling
TOV21G PLKO vector control, TOV21G non-target control, and TOV21G PAX2 knockdowns were profiled using Affymetrix Human Genome U133 plus 2.0 GeneChips (Affymetrix, Santa Clara, CA, USA) as previously described [42]. Raw expression images (CEL files) were processed using dChip software [43], and analysis was performed as described previously [44].
4.4. WST-1 Assay
The cell proliferation reagent WST-1 (Roche Applied Science, IN, USA) was used to analyze cell viability. Cells were seeded at 8000 cells per well in 96-well plates. Cell viability of the parental ovarian cancer cell lines with PAX2 expression and PAX2 knockdown ovarian cancer cell lines at 1, 2, 4, 6, 7, 8 days were measured by adding 10 μL of WST-1 reagent to each wells. The plates were incubated from 0.5 to 4 h in a humidified atmosphere (37 °C, 5% CO2). Plates were thoroughly shaken for 1 min, and then their light absorbance at 450 nm was measured against background controls using a microtiter plate reader.
4.5. Wound Healing Assay
Parental ovarian cancer cells (RMUGL, TOV21G) and PAX2 stable knockdown ovarian cancer cells were cultured to confluence or near confluence (>90%) in a 6-well dish. Cells were subsequently rinsed with phosphate-buffered saline and starved overnight in low serum medium (1.5 mL; 0.5%–0.1% serum in Dulbecco’s modified Eagle’s medium). On the day of the assay, a sterile 200 μL pipette tip was used to scratch a cross-shaped wound through the cell lawn. Cells were rinsed with phosphate-buffered saline, and the low serum medium was replaced with 1.5 mL of medium containing 10% fetal bovine serum. After the wounds were created, the cultures were photographed using phase contrast at 10× magnification at 0, 5, 10 and 24 h. The TScratch program (Computational Science and Engineering Laboratory, Zurich, Switzerland) [26] was used to measure the open areas and analyze the data.
4.6. Taqman Real-Time RT-PCR
One microgram of total RNA from each sample was used for first-strand cDNA synthesis using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Carlsbad, CA, USA). Real-time PCR was performed on the synthesized cDNA using G0S2, WFDC1, GREM1, and PAX2 Taqman gene expression assay mixes on the Bio-Rad CFX96 system (Bio-Rad Laboratories, Hercules, CA, USA). All results were normalized using cyclophilin A.
4.7. Western Blot Analysis
Cytoplasmic and nuclear proteins were extracted as described previously [45]. Western blots were performed with rabbit polyclonal anti-PAX2 antibody (Zymed Laboratories, San Francisco, CA, USA) at a 1:500 dilution; rabbit polyclonal anti-Annexin AI antibody 71-3400 (invitrogen, Camarillo, CA, USA) at a 1:1000 dilution; rabbit polyclonal PARP-1/2 (H-250) antibody sc-7150 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at a 1:10,000 dilution; mouse monoclonal anti-beta-actin antibody (Sigma-aldrich Inc., St. Louis, MO, USA) at a 1:10,000 dilution; goat anti-mouse IgG-horseradish peroxidase sc-2005 (Santa Cruz Biotechnology) at a 1:10,00 dilution; and goat anti-rabbit IgG-horseradish peroxidase sc-2004 (Santa Cruz Biotechnology) at a 1:10,000 dilution. The bound antibodies were detected using an Amersham ECL Western blot detection reagent kit (GE Healthcare, Fairfield, CT, USA). Nuclear expression of PAX2 was normalized with nuclear expression PARP-1/2. Total protein expression of Annexin AI was normalized with beta-actin expression.
4.8. Reverse Phase Protein Array (RPPA)
Cell lysates were extracted by using lysis buffer and were serially diluted four times from undiluted to 1:16 dilution before they were arrayed on nitrocellulose-coated slides in an 11 × 11 format. Samples were probed with antibodies by a catalyzed signal amplification system and visualized by a diaminobenzadine colorimetric reaction. Slides were scanned on a flatbed scanner to produce a 16-bit tiff image. Spots from the tiff images were identified, and their density was quantified using MicroVigene (VigeneTech Inc., Carlisle, MA, USA). Relative protein levels for each sample were determined by interpolation of each dilution curve from the “standard curve” (supercurve) of the slide (antibody). All the data points were normalized for protein loading and transformed to linear values designated as “linear after normalization”. The “linear after normalization” values were then transformed to natural log values and median-centered for hierarchical cluster analysis. Samples were probed with 217 antibodies. Based on our QC samples which were defined by the software, only 207 antibodies were included in the data analysis. A heat map was used to express overall patterns.
4.9. Allophycocyanin-Annexin V Staining
One million cells were aliquoted into centrifuge tubes. Cells were centrifuged, and the supernatant was decanted. One hundred microliters of diluted (1:20 dilution) Annexin V (BD Pharmingen, Bedford, MA, USA) were added to each sample, followed by incubation at room temperature in the dark for 15 min. Precipitates were washed with the Annexin V binding buffer and resuspended in 400 μL binding buffer. Annexin V expression was determined using a FACSCalibur flow cytometer (Becton Dickinson, Mountain View, CA, USA), and single color samples were used to set compensation on the flow cytometer. Data were analyzed using the Becton Dickinson CellQuest Pro software package.
4.10. Transfection of Ovarian Cancer Cell Lines with PAX2 Full-Length cDNA Clone
Full-length PAX2 cDNA (pCMW-Myc-PAX2) clone and vector (pCMV-Neo) were purchased from Origene (Rockville, MD, USA). Ten thousands cells in each well of a 96 well plate were transfected with 0.1 μg DNA using Lipofectamin 2000 reagent (Life Technologies, Grand Island, NY, USA). After 3 days, cell survival was measured with WST-1 assay.
4.11. Immunostaining of PAX2 in Non-Serous Ovarian Carcinomas Paraffin Sections
Immunohistochemistry of paraffin embedded tissue was conducted to determine PAX2 protein expression in patient samples. Paraffin-embedded specimens were sliced into 5-μm sections and the histologic subtypes were confirmed by a pathologist (MD) with specialty in gynecologic malignancies. Following deparaffinization and rehydration, antigen retrieval was performed using citrate buffer in a decloaking chamber at the following settings: 121 °C for 3 min and 95 °C for 1 min. (Biocare Medical, Concord, CA, USA). Staining of the slides was conducted using the Lab Vision Autostainer 360 (Thermo Scientific, Waltham, CA, USA). A PAX2 rabbit polyclonal antibody (Invitrogen, Camarillo, CA, USA) was used along with the Envision + System-HRP Labelled Polymer Anti-Rabbit (Dako, Carpinteria, CA, USA). Slides were also counterstained with hematoxylin.
5. Conclusions
In this study, we demonstrated that knockdown PAX2 expression in ovarian cancer cells with high level of PAX2 expression is associated with reduced cell proliferation and motility. However, over-expressing PAX2 in PAX2-negative ovarian cancer cells suppressed their growth. In summary, PAX2 could have both oncogenic or tumor suppression functions, which will depend on the genetic content of ovarian cancer cells. Further investigation is warranted.
Acknowledgments
We thank Wendy Schober at The University of Texas MD Anderson Cancer Center Flow Cytometry and Cellular Imaging Core Laboratory for help with flow cytometry. Author ERK and CST are supported by the National Cancer Institute-Department of Health and Human Services-National Institutes of Health Training of Academic Oncologists Grant (T32 CA101642). This research was also supported in part by the Blanton-Davis Ovarian Cancer Research Program; the Sara Brown Musselman Fund for Serous Ovarian Cancer Research; the Molbeck Fund for Clear cell ovarian carcinomas, and the National Institutes of Health through The University of Texas MD Anderson Cancer Center Specialized Program of Research Excellence in Ovarian Cancer (P50 CA08369), grant R01-CA133057, and MD Anderson’s Cancer Center Support Grant (CA016672).
Supplementary Files
Supplementary Information (DOC, 229 KB)
Table S1. Differentially expressed genes between PAX2 knockdown (XLSX, 18 KB)
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Eccles M.R., He S., Legge M., Kumar R., Fox J., Zhou C., French M., Tsai R.W. PAX genes in development and disease: The role of PAX2 in urogenital tract development. Int. J. Dev. Biol. 2002;46:535–544. [PubMed] [Google Scholar]
- 2.Chan-Ling T., Chu Y., Baxter L., Weible Ii M., Hughes S. In vivo characterization of astrocyte precursor cells (APCs) and astrocytes in developing rat retinae: Differentiation, proliferation, and apoptosis. Glia. 2009;57:39–53. doi: 10.1002/glia.20733. [DOI] [PubMed] [Google Scholar]
- 3.Wasco M.J., Pu R.T. Comparison of PAX-2, RCC antigen, and antiphosphorylated H2AX antibody (gamma-H2AX) in diagnosing metastatic renal cell carcinoma by fine-needle aspiration. Diagn. Cytopathol. 2008;36:568–573. doi: 10.1002/dc.20839. [DOI] [PubMed] [Google Scholar]
- 4.Sims-Lucas S., Cusack B., Baust J., Eswarakumar V.P., Masatoshi H., Takeuchi A., Bates C.M. Fgfr1 and the IIIc isoform of Fgfr2 play critical roles in the metanephric mesenchyme mediating early inductive events in kidney development. Dev. Dyn. 2011;240:240–249. doi: 10.1002/dvdy.22501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdel-Hakeem A.K., Henry T.Q., Magee T.R., Desai M., Ross M.G., Mansano R.Z., Torday J.S., Nast C.C. Mechanisms of impaired nephrogenesis with fetal growth restriction: Altered renal transcription and growth factor expression. Am. J. Obstet. Gynecol. 2008;199:252.e251–252.e257. doi: 10.1016/j.ajog.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton Q., Cole L.K., Mulheisen M., Chang W., Wu D.K. The role of Pax2 in mouse inner ear development. Dev. Biol. 2004;272:161–175. doi: 10.1016/j.ydbio.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 7.Ritz-Laser B., Estreicher A., Gauthier B., Philippe J. The paired homeodomain transcription factor Pax-2 is expressed in the endocrine pancreas and transactivates the glucagon gene promoter. J. Biol. Chem. 2000;275:32708–32715. doi: 10.1074/jbc.M005704200. [DOI] [PubMed] [Google Scholar]
- 8.Fletcher J., Hu M., Berman Y., Collins F., Grigg J., McIver M., Juppner H., Alexander S.I. Multicystic dysplastic kidney and variable phenotype in a family with a novel deletion mutation of PAX2. J. Am. Soc. Nephrol. 2005;16:2754–2761. doi: 10.1681/ASN.2005030239. [DOI] [PubMed] [Google Scholar]
- 9.Sanyanusin P., Schimmenti L.A., McNoe L.A., Ward T.A., Pierpont M.E., Sullivan M.J., Dobyns W.B., Eccles M.R. Mutation of the PAX2 gene in a family with optic nerve colobomas, renal anomalies and vesicoureteral reflux. Nat. Genet. 1995;9:358–364. doi: 10.1038/ng0495-358. [DOI] [PubMed] [Google Scholar]
- 10.Soofi A., Levitan I., Dressler G.R. Two novel EGFP insertion alleles reveal unique aspects of Pax2 function in embryonic and adult kidneys. Dev. Biol. 2012;365:241–250. doi: 10.1016/j.ydbio.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soukkarieh C., Agius E., Soula C., Cochard P. Pax2 regulates neuronal-glial cell fate choice in the embryonic optic nerve. Dev. Biol. 2007;303:800–813. doi: 10.1016/j.ydbio.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 12.Li C.G., Eccles M.R. PAX genes in cancer; Friends or foes? Front. Genet. 2012;3:6. doi: 10.3389/fgene.2012.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber S., Taylor J.C., Winyard P., Baker K.F., Sullivan-Brown J., Schild R., Knuppel T., Zurowska A.M., Caldas-Alfonso A., Litwin M., et al. SIX2 and BMP4 mutations associate with anomalous kidney development. J. Am. Soc. Nephrol. 2008;19:891–903. doi: 10.1681/ASN.2006111282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dressler G.R., Woolf A.S. Pax2 in development and renal disease. Int. J. Dev. Biol. 1999;43:463–468. [PubMed] [Google Scholar]
- 15.Tong G.X., Chiriboga L., Hamele-Bena D., Borczuk A.C. Expression of PAX2 in papillary serous carcinoma of the ovary: Immunohistochemical evidence of fallopian tube or secondary Mullerian system origin? Mod. Pathol. 2007;20:856–863. doi: 10.1038/modpathol.3800827. [DOI] [PubMed] [Google Scholar]
- 16.Gnarra J.R., Dressler G.R. Expression of Pax-2 in human renal cell carcinoma and growth inhibition by antisense oligonucleotides. Cancer Res. 1995;55:4092–4098. [PubMed] [Google Scholar]
- 17.Khoubehi B., Kessling A.M., Adshead J.M., Smith G.L., Smith R.D., Ogden C.W. Expression of the developmental and oncogenic PAX2 gene in human prostate cancer. J. Urol. 2001;165:2115–2120. doi: 10.1097/00005392-200106000-00080. [DOI] [PubMed] [Google Scholar]
- 18.Silberstein G.B., Dressler G.R., van Horn K. Expression of the PAX2 oncogene in human breast cancer and its role in progesterone-dependent mammary growth. Oncogene. 2002;21:1009–1016. doi: 10.1038/sj.onc.1205172. [DOI] [PubMed] [Google Scholar]
- 19.Tung C.S., Mok S.C., Tsang Y.T., Zu Z., Song H., Liu J., Deavers M.T., Malpica A., Wolf J.K., Lu K.H., Gershenson D.M., Wong K.K. PAX2 expression in low malignant potential ovarian tumors and low-grade ovarian serous carcinomas. Mod. Pathol. 2009;22:1243–1250. doi: 10.1038/modpathol.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muratovska A., Zhou C., He S., Goodyer P., Eccles M.R. Paired-box genes are frequently expressed in cancer and often required for cancer cell survival. Oncogene. 2003;22:7989–7997. doi: 10.1038/sj.onc.1206766. [DOI] [PubMed] [Google Scholar]
- 21.Doberstein K., Pfeilschifter J., Gutwein P. The transcription factor PAX2 regulates ADAM10 expression in renal cell carcinoma. Carcinogenesis. 2011;32:1713–1723. doi: 10.1093/carcin/bgr195. [DOI] [PubMed] [Google Scholar]
- 22.Quick C.M., Ning G., Bijron J., Laury A., Wei T.S., Chen E.Y., Vargas S.O., Betensky R.A., McKeon F.D., Xian W., Crum C.P. PAX2-null secretory cell outgrowths in the oviduct and their relationship to pelvic serous cancer. Mod. Pathol. 2012;25:449–455. doi: 10.1038/modpathol.2011.175. [DOI] [PubMed] [Google Scholar]
- 23.Roh M.H., Yassin Y., Miron A., Mehra K.K., Mehrad M., Monte N.M., Mutter G.L., Nucci M.R., Ning G., McKeon F.D., et al. High-grade fimbrial-ovarian carcinomas are unified by altered p53, PTEN and PAX2 expression. Mod. Pathol. 2010;23:1316–1324. doi: 10.1038/modpathol.2010.119. [DOI] [PubMed] [Google Scholar]
- 24.Chen E.Y., Mehra K., Mehrad M., Ning G., Miron A., Mutter G.L., Monte N., Quade B.J., McKeon F.D., Yassin Y., Xian W., Crum C.P. Secretory cell outgrowth, PAX2 and serous carcinogenesis in the Fallopian tube. J. Pathol. 2010;222:110–116. doi: 10.1002/path.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monte N.M., Webster K.A., Neuberg D., Dressler G.R., Mutter G.L. Joint loss of PAX2 and PTEN expression in endometrial precancers and cancer. Cancer Res. 2010;70:6225–6232. doi: 10.1158/0008-5472.CAN-10-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geback T., Schulz M.M., Koumoutsakos P., Detmar M. TScratch: A novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques. 2009;46:265–274. doi: 10.2144/000113083. [DOI] [PubMed] [Google Scholar]
- 27.Hong B., Lui V.W., Hui E.P., Lu Y., Leung H.S., Wong E.Y., Cheng S.H., Ng M.H., Mills G.B., Chan A.T. Reverse phase protein array identifies novel anti-invasion mechanisms of YC-1. Biochem. Pharmacol. 2010;79:842–852. doi: 10.1016/j.bcp.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 28.Kurman R.J., Shih Ie M. The origin and pathogenesis of epithelial ovarian cancer: A proposed unifying theory. Am. J. Surg. Pathol. 2010;34:433–443. doi: 10.1097/PAS.0b013e3181cf3d79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiegand K.C., Shah S.P., Al-Agha O.M., Zhao Y., Tse K., Zeng T., Senz J., McConechy M.K., Anglesio M.S., Kalloger S.E., et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saifudeen Z., Liu J., Dipp S., Yao X., Li Y., McLaughlin N., Aboudehen K., El-Dahr S.S. A p53-Pax2 pathway in kidney development: Implications for nephrogenesis. PLoS One. 2012;7:e44869. doi: 10.1371/journal.pone.0044869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H.S., Yan B., Li X.B., Fan L., Zhang Y.F., Wu G.H., Li M., Fang J. Pax2 induces expression of Cyclin D1 through activating AP-1 and promotes proliferation of colon cancer cells. J. Biol. Chem. 2012;287:44164–44172. doi: 10.1074/jbc.M112.401521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hueber P.A., Iglesias D., Chu L.L., Eccles M., Goodyer P. In vivo validation of PAX2 as a target for renal cancer therapy. Cancer Lett. 2008;265:148–155. doi: 10.1016/j.canlet.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 33.Russell L., Forsdyke D.R. A human putative lymphocyte G0/G1 switch gene containing a CpG-rich island encodes a small basic protein with the potential to be phosphorylated. DNA Cell Biol. 1991;10:581–591. doi: 10.1089/dna.1991.10.581. [DOI] [PubMed] [Google Scholar]
- 34.Cristillo A.D., Heximer S.P., Russell L., Forsdyke D.R. Cyclosporin A inhibits early mRNA expression of G0/G1 switch gene 2 (G0S2) in cultured human blood mononuclear cells. DNA Cell Biol. 1997;16:1449–1458. doi: 10.1089/dna.1997.16.1449. [DOI] [PubMed] [Google Scholar]
- 35.Welch C., Santra M.K., El-Assaad W., Zhu X., Huber W.E., Keys R.A., Teodoro J.G., Green M.R. Identification of a protein, G0S2, that lacks Bcl-2 homology domains and interacts with and antagonizes Bcl-2. Cancer Res. 2009;69:6782–6789. doi: 10.1158/0008-5472.CAN-09-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouchard D., Morisset D., Bourbonnais Y., Tremblay G.M. Proteins with whey-acidic-protein motifs and cancer. Lancet Oncol. 2006;7:167–174. doi: 10.1016/S1470-2045(06)70579-4. [DOI] [PubMed] [Google Scholar]
- 37.Madar S., Brosh R., Buganim Y., Ezra O., Goldstein I., Solomon H., Kogan I., Goldfinger N., Klocker H., Rotter V. Modulated expression of WFDC1 during carcinogenesis and cellular senescence. Carcinogenesis. 2009;30:20–27. doi: 10.1093/carcin/bgn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu D.R., Economides A.N., Wang X., Eimon P.M., Harland R.M. The Xenopus dorsalizing factor Gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol. Cell. 1998;1:673–683. doi: 10.1016/s1097-2765(00)80067-2. [DOI] [PubMed] [Google Scholar]
- 39.Mulvihill M.S., Kwon Y.W., Lee S., Fang L.T., Choi H., Ray R., Kang H.C., Mao J.H., Jablons D., Kim I.J. Gremlin is overexpressed in lung adenocarcinoma and increases cell growth and proliferation in normal lung cells. PLoS One. 2012;7:e42264. doi: 10.1371/journal.pone.0042264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen B., Athanasiou M., Gu Q., Blair D.G. Drm/Gremlin transcriptionally activates p21(Cip1) via a novel mechanism and inhibits neoplastic transformation. Biochem. Biophys. Res. Commun. 2002;295:1135–1141. doi: 10.1016/s0006-291x(02)00828-8. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Z., Huang L., Zhao W., Rigas B. Annexin 1 induced by anti-inflammatory drugs binds to NF-kappaB and inhibits its activation: Anticancer effects in vitro and in vivo. Cancer Res. 2010;70:2379–2388. doi: 10.1158/0008-5472.CAN-09-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gokden N., Kemp S.A., Gokden M. The utility of Pax-2 as an immunohistochemical marker for renal cell carcinoma in cytopathology. Diagn. Cytopathol. 2008;36:473–477. doi: 10.1002/dc.20842. [DOI] [PubMed] [Google Scholar]
- 43.Li C. Automating dChip: Toward reproducible sharing of microarray data analysis. BMC Bioinform. 2008;9:231. doi: 10.1186/1471-2105-9-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park D.C., Yeo S.G., Wilson M.R., Yerbury J.J., Kwong J., Welch W.R., Choi Y.K., Birrer M.J., Mok S.C., Wong K.K. Clusterin interacts with Paclitaxel and confer Paclitaxel resistance in ovarian cancer. Neoplasia. 2008;10:964–972. doi: 10.1593/neo.08604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baby J., Pickering B.F., Vashisht Gopal Y.N., van Dyke M.W. Constitutive and inducible nuclear factor-kappaB in immortalized normal human bronchial epithelial and non-small cell lung cancer cell lines. Cancer Lett. 2007;255:85–94. doi: 10.1016/j.canlet.2007.03.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information (DOC, 229 KB)
Table S1. Differentially expressed genes between PAX2 knockdown (XLSX, 18 KB)